Proteomic Changes during the Dermal Toxicity Induced by Nemopilema nomurai Jellyfish Venom in HaCaT Human Keratinocyte
Abstract
:1. Introduction
2. Results
2.1. NnV Induces Cytotoxicity in HaCaT Cell Lines
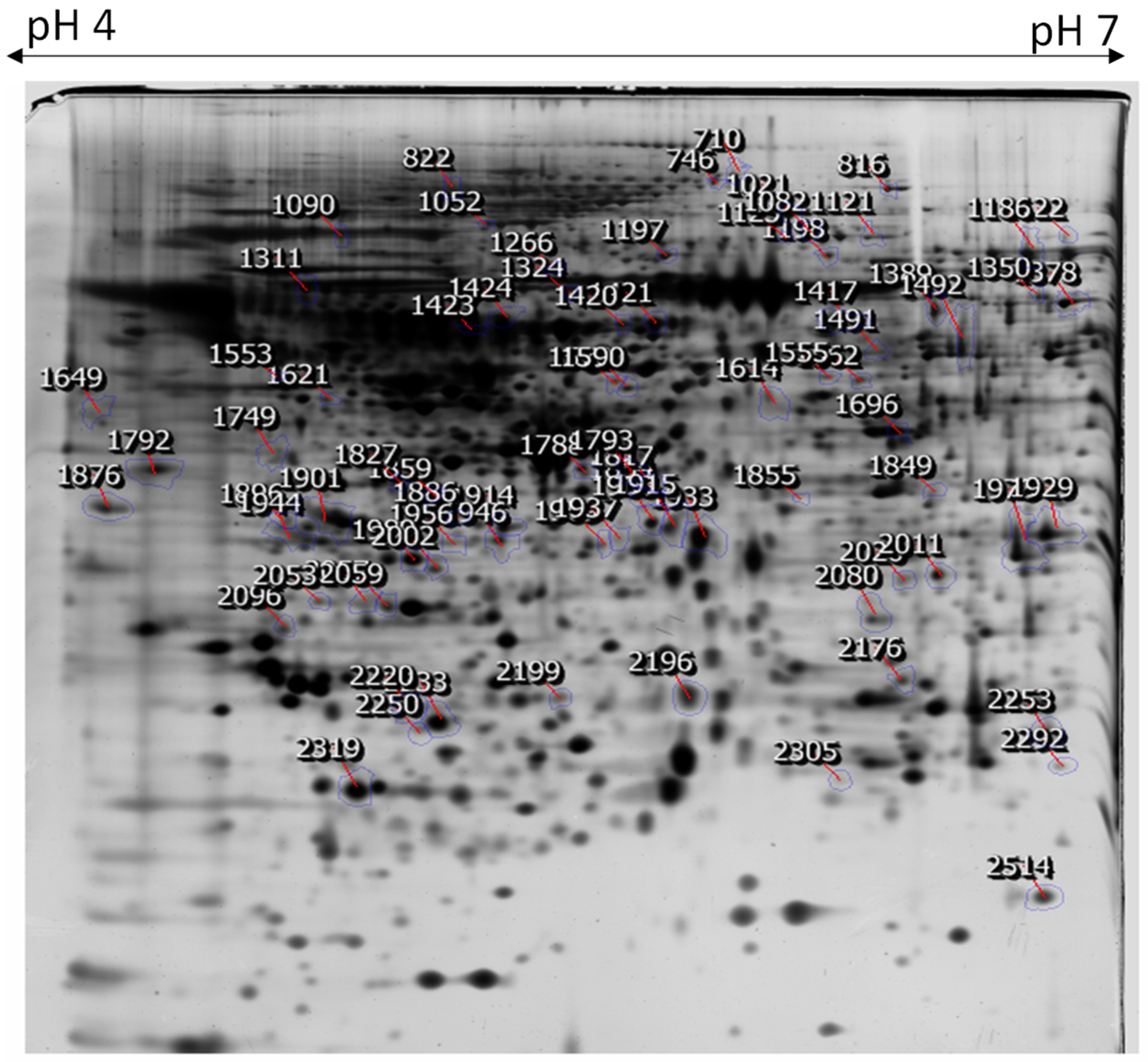
2.2. Two-Dimensional Gel Electrophoresis of Cellular Proteins from NnV-Treated HaCaT Cells
2.3. Ontological Classification of Differentially Abundant Proteins
2.4. Protein-Protein Interactions
3. Discussion
3.1. Peroxisome Proliferator-Activated Receptor γ Coactivator 1α (PGC-1α)
3.2. Elastin Microfibril Interface Located Protein 1 (EMILIN-1)
3.3. Basonuclin 2
3.4. Glucocorticoid (GC)
3.5. Plasminogen
3.6. Vinculin
3.7. Focal Adhesion Kinase 1
3.8. Myocardin Related Transcription Factor-A (MRTF-A)
3.9. Toll-Like Receptors (TLRs)
3.10. Minichromosome Maintenance (MCM)
3.11. APN/CD13 (Aminopeptidase N)
4. Conclusions
5. Remarks
6. Materials and Methods
6.1. Chemicals and Reagents
6.2. Sample Collection and Preparation
6.3. Venom Extraction and Preparation
6.4. Cell Culture
6.5. Conditions for MTT Assay for Cell Viability
6.6. DAPI Staining for Nuclear Morphological Analysis
6.7. Protein Extraction and Sample Preparation
6.8. Two-Dimensional Gel Electrophoresis and Image Analysis
6.9. MALDI-TOF/MS Analysis and Database Searching
6.10. Statistical and Bioinformatics Analysis of Protein Identified by MALDI-TOF/MS
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, Z.; Liu, D.; Keesing, J.K. Jellyfish blooms in China: Dominant species, causes and consequences. Mar. Pollut. Bull. 2010, 60, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Omori, M.; Kitamura, M. Taxonomic review of three Japanese species of edible jellyfish(Scyphozoa: Rhizostomeae). Plankton Biol. Ecol. 2004, 51, 36–51. [Google Scholar]
- Kang, C.; Munawir, A.; Cha, M.; Sohn, E.-T.; Lee, H.; Kim, J.-S.; Yoon, W.D.; Lim, D.; Kim, E. Cytotoxicity and hemolytic activity of jellyfish Nemopilema nomurai (Scyphozoa: Rhizostomeae) venom. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 150, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Lee, S.; Kim, J.-S.; Yoon, W.D.; Lim, D.; Hart, A.J.; Hodgson, W.C. Cardiovascular effects of Nemopilema nomurai (Scyphozoa: Rhizostomeae) jellyfish venom in rats. Toxicol. Lett. 2006, 167, 205–211. [Google Scholar] [CrossRef]
- Choudhary, I.; Lee, H.; Pyo, M.-J.; Heo, Y.; Bae, S.K.; Kwon, Y.C.; Yoon, W.D.; Kang, C.; Kim, E. Proteomics approach to examine the cardiotoxic effects of Nemopilema nomurai jellyfish venom. J. Proteom. 2015, 128, 123–131. [Google Scholar] [CrossRef]
- Kawahara, M.; Uye, S.-I.; Ohtsu, K.; Iizumi, H. Unusual population explosion of the giant jellyfish Nemopilema nomurai (Scyphozoa: Rhizostomeae) in East Asian waters. Mar. Ecol. Prog. Ser. 2006, 307, 161–173. [Google Scholar] [CrossRef]
- Teragawa, C.K.; Bode, H.R. Migrating interstitial cells differentiate into neurons in hydra. Dev. Biol. 1995, 171, 286–293. [Google Scholar] [CrossRef] [Green Version]
- Hausmann, K.; Holstein, T. Bilateral symmetry in the cnidocil-nematocyst complex of the freshwater medusaCraspedacusta sowerbii Lankester (Hydrozoa, Limnomedusae). J. Ultrastruct. Res. 1985, 90, 89–104. [Google Scholar] [CrossRef]
- Pyo, M.-J.; Lee, H.; Bae, S.K.; Heo, Y.; Choudhary, I.; Yoon, W.D.; Kang, C.; Kim, E. Modulation of jellyfish nematocyst discharges and management of human skin stings in Nemopilema nomurai and Carybdea mora. Toxicon 2016, 109, 26–32. [Google Scholar] [CrossRef]
- Burnett, J.W.; Calton, G.J. Venomous pelagic coelenterates: Chemistry, toxicology, immunology and treatment of their stings. Toxicon 1987, 25, 581–602. [Google Scholar] [CrossRef]
- Drury, J.; Noonan, J.; Pollock, J.; Reid, W.H. Jelly fish sting with serious hand complications. Injury 1980, 12, 66–68. [Google Scholar] [CrossRef]
- Nimorakiotakis, B.; Winkel, K. Marine envenomations: Part 1-jellyfish. Aust. Fam. Phys. 2003, 32, 969. [Google Scholar]
- Tibballs, J. Australian venomous jellyfish, envenomation syndromes, toxins and therapy. Toxicon 2006, 48, 830–859. [Google Scholar] [CrossRef]
- Brinkman, D.L.; Aziz, A.; Loukas, A.; Potriquet, J.; Seymour, J.; Mulvenna, J. Venom proteome of the box jellyfish Chironex fleckeri. PLoS ONE 2012, 7, e47866. [Google Scholar] [CrossRef] [Green Version]
- Brinkman, D.L.; Jia, X.; Potriquet, J.; Kumar, D.; Dash, D.; Kvaskoff, D.; Mulvenna, J. Transcriptome and venom proteome of the box jellyfish Chironex fleckeri. BMC Genom. 2015, 16, 407. [Google Scholar] [CrossRef] [Green Version]
- Thaikruea, L.; Siriariyaporn, P. Severe Dermatonecrotic Toxin and Wound Complications Associated With Box Jellyfish Stings 2008-2013. J. Wound Ostomy Cont. Nurs. Off. Publ. Wound Ostomy Cont. Nurses Soc. 2015, 42, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Silva-de-França, F.; Villas-Boas, I.M.; Serrano, S.M.T.; Cogliati, B.; Chudzinski, S.A.A.; Lopes, P.H.; Kitano, E.S.; Okamoto, C.K.; Tambourgi, D.V. Naja annulifera Snake: New insights into the venom components and pathogenesis of envenomation. PLoS Negl. Trop. Dis. 2019, 13, e0007017. [Google Scholar] [CrossRef] [Green Version]
- Blaylock, R.S. Antibacterial properties of KwaZulu natal snake venoms. Toxicon 2000, 38, 1529–1534. [Google Scholar] [CrossRef]
- Hornbeak, K.B.; Auerbach, P.S. Marine Envenomation. Emerg. Med. Clin. N. Am. 2017, 35, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, A.L.; de Macedo Mora, J.; Haddad Junior, V. Sea urchin granuloma. Rev. Inst. Med. Trop. Sao Paulo 2006, 48, 303–306. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Azofeifa, D.; Sasa, M.; Lomonte, B.; Diego-García, E.; Ortiz, N.; Bonilla, F.; Murillo, R.; Tytgat, J.; Díaz, C. Biochemical characterization of the venom of Central American scorpion Didymocentrus krausi Francke, 1978 (Diplocentridae) and its toxic effects in vivo and in vitro. Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP 2019, 217, 54–67. [Google Scholar] [CrossRef]
- Pelin, M.; Zanette, C.; De Bortoli, M.; Sosa, S.; Loggia, R.D.; Tubaro, A.; Florio, C. Effects of the marine toxin palytoxin on human skin keratinocytes: Role of ionic imbalance. Toxicology 2011, 282, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Jin, Y.B.; Kwak, J.; Jung, H.; Yoon, W.D.; Yoon, T.-J.; Kim, J.-S.; Kim, E. Protective effect of tetracycline against dermal toxicity induced by Jellyfish venom. PLoS ONE 2013, 8, e57658. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.H.; Lee, H.; Choudhary, I.; Kang, C.; Chae, J.; Kim, E. Protective effect of epigallocatechin-3-gallate (EGCG) on toxic metalloproteinases-mediated skin damage induced by Scyphozoan jellyfish envenomation. Sci. Rep. 2020, 10, 18644. [Google Scholar] [CrossRef]
- Choudhary, I.; Hwang, D.H.; Lee, H.; Yoon, W.D.; Chae, J.; Han, C.H.; Yum, S.; Kang, C.; Kim, E. Proteomic Analysis of Novel Components of Nemopilema nomurai Jellyfish Venom: Deciphering the Mode of Action. Toxins 2019, 11, 153. [Google Scholar] [CrossRef] [Green Version]
- Senff-Ribeiro, A.; Henrique da Silva, P.; Chaim, O.M.; Gremski, L.H.; Paludo, K.S.; Bertoni da Silveira, R.; Gremski, W.; Mangili, O.C.; Veiga, S.S. Biotechnological applications of brown spider (Loxosceles genus) venom toxins. Biotechnol. Adv. 2008, 26, 210–218. [Google Scholar] [CrossRef]
- da Silveira, R.B.; Chaim, O.M.; Mangili, O.C.; Gremski, W.; Dietrich, C.P.; Nader, H.B.; Veiga, S.S. Hyaluronidases in Loxosceles intermedia (Brown spider) venom are endo-beta-N-acetyl-d-hexosaminidases hydrolases. Toxicon Off. J. Int. Soc. Toxinol. 2007, 49, 758–768. [Google Scholar] [CrossRef]
- Ombati, R.; Wang, Y.; Du, C.; Lu, X.; Li, B.; Nyachieo, A.; Li, Y.; Yang, S.; Lai, R. A membrane disrupting toxin from wasp venom underlies the molecular mechanism of tissue damage. Toxicon Off. J. Int. Soc. Toxinol. 2018, 148, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Goswami, P.C. Down-regulation of peroxisome proliferator activated receptor γ coactivator 1α induces oxidative stress and toxicity of 1-(4-Chlorophenyl)-benzo-2,5-quinone in HaCaT human keratinocytes. Toxicol. In Vitro. Int. J. Publ. Assoc. Bibra 2015, 29, 1332–1338. [Google Scholar] [CrossRef] [Green Version]
- Zanetti, M.; Braghetta, P.; Sabatelli, P.; Mura, I.; Doliana, R.; Colombatti, A.; Volpin, D.; Bonaldo, P.; Bressan, G.M. EMILIN-1 deficiency induces elastogenesis and vascular cell defects. Mol. Cell. Biol. 2004, 24, 638–650. [Google Scholar] [CrossRef] [Green Version]
- Nakatomi, Y.; Tsuruga, E.; Nakashima, K.; Sawa, Y.; Ishikawa, H. EMILIN-1 regulates the amount of oxytalan fiber formation in periodontal ligaments in vitro. Connect. Tissue Res. 2011, 52, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Doliana, R.; Mongiat, M.; Bucciotti, F.; Giacomello, E.; Deutzmann, R.; Volpin, D.; Bressan, G.M.; Colombatti, A. EMILIN, a component of the elastic fiber and a new member of the C1q/tumor necrosis factor superfamily of proteins. J. Biol. Chem. 1999, 274, 16773–16781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spessotto, P.; Cervi, M.; Mucignat, M.T.; Mungiguerra, G.; Sartoretto, I.; Doliana, R.; Colombatti, A. beta 1 Integrin-dependent cell adhesion to EMILIN-1 is mediated by the gC1q domain. J. Biol. Chem. 2003, 278, 6160–6167. [Google Scholar] [CrossRef] [Green Version]
- Bressan, G.M.; Daga-Gordini, D.; Colombatti, A.; Castellani, I.; Marigo, V.; Volpin, D. Emilin, a component of elastic fibers preferentially located at the elastin-microfibrils interface. J. Cell Biol. 1993, 121, 201–212. [Google Scholar] [CrossRef] [Green Version]
- Vanhoutteghem, A.; Delhomme, B.; Hervé, F.; Nondier, I.; Petit, J.M.; Araki, M.; Araki, K.; Djian, P. The importance of basonuclin 2 in adult mice and its relation to basonuclin 1. Mech. Dev. 2016, 140, 53–73. [Google Scholar] [CrossRef] [PubMed]
- Romano, R.A.; Li, H.; Tummala, R.; Maul, R.; Sinha, S. Identification of Basonuclin2, a DNA-binding zinc-finger protein expressed in germ tissues and skin keratinocytes. Genomics 2004, 83, 821–833. [Google Scholar] [CrossRef]
- Tseng, H.; Green, H. Association of basonuclin with ability of keratinocytes to multiply and with absence of terminal differentiation. J. Cell Biol. 1994, 126, 495–506. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, K.; Inoue, H.; Kumagai, N. Re-epithelialisation and the possible involvement of the transcription factor, basonuclin. Int. Wound J. 2004, 1, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Bigas, J.; Sevilla, L.M.; Carceller, E.; Boix, J.; Pérez, P. Epidermal glucocorticoid and mineralocorticoid receptors act cooperatively to regulate epidermal development and counteract skin inflammation. Cell Death Dis. 2018, 9, 588. [Google Scholar] [CrossRef]
- Lesovaya, E.; Agarwal, S.; Readhead, B.; Vinokour, E.; Baida, G.; Bhalla, P.; Kirsanov, K.; Yakubovskaya, M.; Platanias, L.C.; Dudley, J.T.; et al. Rapamycin Modulates Glucocorticoid Receptor Function, Blocks Atrophogene REDD1, and Protects Skin from Steroid Atrophy. J. Investig. Dermatol. 2018, 138, 1935–1944. [Google Scholar] [CrossRef] [Green Version]
- Reichrath, J. Ancient friends, revisited: New aspects on the important role of nuclear receptor signalling for skin physiology and for the treatment of skin diseases. Derm. Endocrinol. 2011, 3, 121–124. [Google Scholar] [CrossRef]
- Pérez, P. Glucocorticoid receptors, epidermal homeostasis and hair follicle differentiation. Derm. Endocrinol. 2011, 3, 166–174. [Google Scholar] [CrossRef] [Green Version]
- Sevilla, L.M.; Latorre, V.; Sanchis, A.; Pérez, P. Epidermal inactivation of the glucocorticoid receptor triggers skin barrier defects and cutaneous inflammation. J. Investig. Dermatol. 2013, 133, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Rossignol, P.; Ho-Tin-Noé, B.; Vranckx, R.; Bouton, M.C.; Meilhac, O.; Lijnen, H.R.; Guillin, M.C.; Michel, J.B.; Anglés-Cano, E. Protease nexin-1 inhibits plasminogen activation-induced apoptosis of adherent cells. J. Biol. Chem. 2004, 279, 10346–10356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulniute, R.; Shen, Y.; Guo, Y.Z.; Fallah, M.; Ahlskog, N.; Ny, L.; Rakhimova, O.; Broden, J.; Boija, H.; Moghaddam, A.; et al. Plasminogen is a critical regulator of cutaneous wound healing. Thromb. Haemost. 2016, 115, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Tam, S.W.; Demissie-Sanders, S.; Filler, S.A.; Oh, C.K. Production of plasminogen activator inhibitor-1 by human mast cells and its possible role in asthma. J. Immunol. 2000, 165, 3154–3161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weckroth, M.; Vaheri, A.; Virolainen, S.; Saarialho-Kere, U.; Jahkola, T.; Sirén, V. Epithelial tissue-type plasminogen activator expression, unlike that of urokinase, its receptor, and plasminogen activator inhibitor-1, is increased in chronic venous ulcers. Br. J. Dermatol. 2004, 151, 1189–1196. [Google Scholar] [CrossRef]
- Le Clainche, C.; Dwivedi, S.P.; Didry, D.; Carlier, M.F. Vinculin is a dually regulated actin filament barbed end-capping and side-binding protein. J. Biol. Chem. 2010, 285, 23420–23432. [Google Scholar] [CrossRef] [Green Version]
- von Essen, M.; Rahikainen, R.; Oksala, N.; Raitoharju, E.; Seppälä, I.; Mennander, A.; Sioris, T.; Kholová, I.; Klopp, N.; Illig, T.; et al. Talin and vinculin are downregulated in atherosclerotic plaque; Tampere Vascular Study. Atherosclerosis 2016, 255, 43–53. [Google Scholar] [CrossRef]
- Ezratty, E.J.; Partridge, M.A.; Gundersen, G.G. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 2005, 7, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.W.; Kim, Y.H.; Sohn, J.H.; Lee, H.; Kim, W.S. Focal adhesion kinase and Src expression in premalignant and malignant skin lesions. Exp. Dermatol. 2015, 24, 361–364. [Google Scholar] [CrossRef]
- Xia, H.; Nho, R.S.; Kahm, J.; Kleidon, J.; Henke, C.A. Focal adhesion kinase is upstream of phosphatidylinositol 3-kinase/Akt in regulating fibroblast survival in response to contraction of type I collagen matrices via a beta 1 integrin viability signaling pathway. J. Biol. Chem. 2004, 279, 33024–33034. [Google Scholar] [CrossRef] [Green Version]
- Shiwen, X.; Stratton, R.; Nikitorowicz-Buniak, J.; Ahmed-Abdi, B.; Ponticos, M.; Denton, C.; Abraham, D.; Takahashi, A.; Suki, B.; Layne, M.D.; et al. A Role of Myocardin Related Transcription Factor-A (MRTF-A) in Scleroderma Related Fibrosis. PLoS ONE 2015, 10, e0126015. [Google Scholar] [CrossRef]
- Haak, A.J.; Tsou, P.S.; Amin, M.A.; Ruth, J.H.; Campbell, P.; Fox, D.A.; Khanna, D.; Larsen, S.D.; Neubig, R.R. Targeting the myofibroblast genetic switch: Inhibitors of myocardin-related transcription factor/serum response factor-regulated gene transcription prevent fibrosis in a murine model of skin injury. J. Pharmacol. Exp. Ther. 2014, 349, 480–486. [Google Scholar] [CrossRef] [Green Version]
- Velasquez, L.S.; Sutherland, L.B.; Liu, Z.; Grinnell, F.; Kamm, K.E.; Schneider, J.W.; Olson, E.N.; Small, E.M. Activation of MRTF-A-dependent gene expression with a small molecule promotes myofibroblast differentiation and wound healing. Proc. Natl. Acad. Sci. USA 2013, 110, 16850–16855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, B.S. The role of microorganisms in atopic dermatitis. Clin. Exp. Immunol. 2006, 144, 1–9. [Google Scholar] [CrossRef]
- Begon, E.; Michel, L.; Flageul, B.; Beaudoin, I.; Jean-Louis, F.; Bachelez, H.; Dubertret, L.; Musette, P. Expression, subcellular localization and cytokinic modulation of Toll-like receptors (TLRs) in normal human keratinocytes: TLR2 up-regulation in psoriatic skin. Eur. J. Dermatol. EJD 2007, 17, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.E.; Lim, H.S.; Kim, Y.; Seo, C.S.; Yoo, S.R.; Shin, H.K.; Jeong, S.J. Traditional Herbal Formula Banhasasim-tang Exerts Anti-Inflammatory Effects in RAW 264.7 Macrophages and HaCaT Keratinocytes. Evid. Based Complement. Altern. Med. ECAM 2015, 2015, 728380. [Google Scholar] [CrossRef]
- Muthusamy, V.; Piva, T.J. The UV response of the skin: A review of the MAPK, NFkappaB and TNFalpha signal transduction pathways. Arch. Dermatol. Res. 2010, 302, 5–17. [Google Scholar] [CrossRef]
- Abdou, A.G.; Elwahed, M.G.; Serag El-Dien, M.M.; Eldien, D.S. Immunohistochemical expression of MCM2 in nonmelanoma epithelial skin cancers. Am. J. Dermatopathol. 2014, 36, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Gerbaud, P.; Guibourdenche, J.; Jarray, R.; Conti, M.; Palmic, P.; Leclerc-Mercier, S.; Bruneau, J.; Hermine, O.; Lepelletier, Y.; Raynaud, F. APN/CD13 is over-expressed by Psoriatic fibroblasts and is modulated by CGRP and IL-4 but not by retinoic acid treatment. J. Cell. Physiol. 2018, 233, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Kunii, R.; Nemoto, E.; Kanaya, S.; Tsubahara, T.; Shimauchi, H. Expression of CD13/aminopeptidase N on human gingival fibroblasts and up-regulation upon stimulation with interleukin-4 and interleukin-13. J. Periodontal Res. 2005, 40, 138–146. [Google Scholar] [CrossRef]
- Sorrell, J.M.; Brinon, L.; Baber, M.A.; Caplan, A.I. Cytokines and glucocorticoids differentially regulate APN/CD13 and DPPIV/CD26 enzyme activities in cultured human dermal fibroblasts. Arch. Dermatol. Res. 2003, 295, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.S.; Chi, H.; Lantry, L.E.; Stephens, R.E.; Ward, P.E. Substance P and neurokinin A metabolism by cultured human skeletal muscle myocytes and fibroblasts. Peptides 1996, 17, 1397–1403. [Google Scholar] [CrossRef]
- Bloom, D.A.; Burnett, J.W.; Alderslade, P. Partial purification of box jellyfish (Chironex fleckeri) nematocyst venom isolated at the beachside. Toxicon Off. J. Int. Soc. Toxinol. 1998, 36, 1075–1085. [Google Scholar] [CrossRef]
- Carrette, T.; Seymour, J. A rapid and repeatable method for venom extraction from cubozoan nematocysts. Toxicon Off. J. Int. Soc. Toxinol. 2004, 44, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Averbukh, L.A. Study using the probit-analysis method of the process of increasing resistance to dipin and bruneomycin in Fisher L-5178 lymphadenosis cells in experiments on animals. Antibiotiki 1976, 21, 169–174. [Google Scholar] [PubMed]
- Mortz, E.; Krogh, T.N.; Vorum, H.; Görg, A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 2001, 1, 1359–1363. [Google Scholar] [CrossRef]
- Shevchenko, A.; Jensen, O.N.; Podtelejnikov, A.V.; Sagliocco, F.; Wilm, M.; Vorm, O.; Mortensen, P.; Shevchenko, A.; Boucherie, H.; Mann, M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 1996, 93, 14440–14445. [Google Scholar] [CrossRef] [Green Version]






| S. No | Protein Name | String Interactions | KEGG Pathways |
|---|---|---|---|
| 1 | Elastin microfibril interface located protein 1 (EMILIN-1) |  | Regulation of actin cytoskeleton, focal adhesion, cell adhesion molecules, PI3K-AKT Signaling Pathway |
| 2 | Glucocorticoid |  | HIF signaling pathway, Thyroid hormone signaling pathway |
| 3 | Plasminogen |  | Complement and coagulation cascades |
| 4 | Vinculin |  | Focal adhesion, leukocyte trans-endothelial migration, regulation of actin cytoskeleton, adherens junction |
| 5 | Focal adhesion kinase 1 |  | Focal adhesion, bacterial invasion of epithelial cells, regulation of actin cytoskeleton |
| 6 | Myocardin Related Transcription Factor-A |  | Platelet activation, focal adhesion, tight junction, RAP1 signaling pathway |
| 7 | Toll-like receptor 4 |  | Toll-like receptor signaling pathway, NF- kappa B signaling pathway, pertussis |
| 8 | Aminopeptidase N |  | Metabolic pathways, glutathione metabolism, hematopoietic cell lineage |
| 9 | DNA replication licensing factor MCM2 |  | Cell cycle, DNA replication |
| Spot No | Accession Number 1 | Protein Name | Uniprot ID | Theoretical MW/Pi 2 | Gene | Matched Peptide 3 | MOWSE Score | Biological Process |
|---|---|---|---|---|---|---|---|---|
| Proteins down-regulated by NnV | ||||||||
| 2319 | P18206 | Vinculin | VINC_HUMAN | 123,800/5.5 | VCL | 36.80% | 1.64 × 1014 | epithelial cell-cell adhesion, cell-matrix adhesion |
| 1901 | Q9Y6U3 | Adseverin | ADSV_HUMAN | 80,490/5.5 | SCIN | 27.30% | 6.09 × 106 | regulation of chondrocyte differentiation, negative regulation of cell population proliferation |
| 1417 | O95259 | Potassium voltage-gated channel subfamily H member 1 | KCNH1_HUMAN | 111,424/7.5 | KCNH1 | 20.10% | 20.10% | regulation of cell proliferation, myoblast fusion, potassium ion transport |
| 1989 | Q9NQ38 | Serine protease inhibitor Kazal-type 5 | ISK5_HUMAN | 120,716/8.5 | SPINK5 | 19.3% | 2.09 × 107 | extracellular matrix organization, epidermal cell differentiation, regulation of T cell differentiation |
| 1956 | P14210 | Hepatocyte growth factor | HGF_HUMAN | 83,135/8.2 | HGF | 28.2% | 2.65 × 107 | activation of MAPK activity, epithelial to mesenchymal transition, mitotic cell cycle |
| 1937 | Q6ZMZ3 | Nesprin-3 | SYNE3_HUMAN | 112,217/5.9 | SYNE3 | 18.60% | 9.88 × 107 | cytoskeletal anchoring at the nuclear membrane, nuclear migration, regulation of cell shape |
| 1937 | P00747 | Plasminogen | PLMN_HUMAN | 90,570/7.0 | PLG | 19.30% | 2.15 × 106 | blood coagulation, fibrinolysis, proteolysis, platelet degranulation |
| 1649 | P16234 | Platelet-derived growth factor receptor alpha | PGFRA_HUMAN | 122,671/5.1 | PDGFRA | 26.40% | 5.01 × 108 | double-strand break repair, signal transduction, histone deacetylation |
| 816 | O43707 | Alpha-actinin-4 | ACTN4_HUMAN | 104,855/5.3 | ACTN4 | 31.60% | 4.53 × 109 | actin filament bundle assembly, positive regulation of NIK/NF-kappa B signaling |
| 1052 | P54762 | Ephrin type-B receptor 1 | EPHB1_HUMAN | 109,886/6.0 | EPHB1 | 29.4% | 9.78 × 108 | cell differentiation, cell-substrate adhesion, angiogenesis |
| 1915 | O00203 | Ubiquitin carboxyl-terminal hydrolase 36 | UBP36_HUMAN | 122,653/9.7 | USP36 | 14.90% | 1.72 × 106 | negative regulation of macroautophagy, regulation of protein stability, nucleolus organization |
| 1793 | Q9UBK2 | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | PRGC1_HUMAN | 91,028/5.7 | PPARGC1A | 19.00% | 3.55 × 107 | regulation of transcription, DNA-templated, response to reactive oxygen species, response to ischemia |
| 2057 | Q5T0W9 | Protein FAM83B | TMF1_HUMAN | 114,800/9.0 | FAM83B | 28.8% | 3.02 × 1012 | cell proliferation, epidermal growth factor receptor signaling pathway |
| 1903 | Q6ZN19 | Zinc finger protein 841 | ZN841_HUMAN | 93,149/9.5 | ZNF841 | 23.1% | 2.49 × 106 | transcription, transcription regulation |
| 1420 | Q6ZN30 | Zinc finger protein basonuclin-2 | BNC2_HUMAN | 122,331/6.1 | BNC2 | 27.00% | 1.71 × 101 | endochondral bone growth, mesenchyme development |
| 1424 | Q9Y6C2 | Elastin microfibril interface located protein 1 (EMILIN-1) | EMIL1_HUMAN | 106,696/5.1 | EMILIN1 | 20.5% | 3.88 × 107 | cell-matrix adhesion, negative regulation of collagen biosynthetic process |
| 1792 | Q9UMS6 | Synaptopodin-2 | SYNP2_HUMAN | 117,515/8.8 | SYNPO2 | 27.3% | 9.57 × 108 | positive regulation of cell migration, chaperone-mediated autophagy |
| 1423 | Q05397 | Focal adhesion kinase 1 | FAK1_HUMAN | 119,234/6.2 | PTK2 | 20.80% | 5.88 × 106 | positive regulation of cell migration, chaperone-mediated autophagy |
| 1266 | Q12866 | Tyrosine-protein kinase Mer | MERTK_HUMAN | 110,250/5.5 | MERTK | 16.90% | 2.50 × 108 | apoptotic cell clearance, cell differentiation, phagocytosis |
| 1933 | Q96NW4 | Mismatch repair endonuclease PMS2 | ANR27_HUMAN | 116,985/6.4 | ANKRD27 | 33.90% | 2.92 × 1012 | mismatch repair |
| 822 | P04150 | Glucocorticoid receptor | GCR_HUMAN | 85,660/6.0 | NR3C1 | 32.60% | 7.99 × 1011 | cellular response to glucocorticoid stimulus, signal transduction |
| 2233 | Q96M96 | FYVE, RhoGEF and PH domain-containing protein 4 | FGD4_HUMAN | 86,627/5.8 | FGD4 | 16.10% | 9.69 × 106 | actin cytoskeleton organization, regulation of cell shape, cytoskeleton organization |
| 1944 | Q13342 | Nuclear body protein SP140 | SP140_HUMAN | 98,224/5.2 | SP140 | 35.60% | 1.39 × 109 | defense response |
| 1166 | O14730 | Serine/threonine-protein kinase RIO3 | RIOK3_HUMAN | 59,094/5.5 | RIOK3 | 26.40% | 4.31 × 106 | positive regulation of interferon-beta production, cellular response to dsDNA, innate immune response |
| 1553 | Q13563 | Polycystin-2 | PKD2_HUMAN | 109,692/5.5 | PKD2 | 19.90% | 1.20 × 108 | cellular response to osmotic stress, regulation of cell proliferation, positive regulation of cell cycle arrest |
| 1421 | O43182 | Rho GTPase-activating protein 6 | RHG06_HUMAN | 105,948/7.0 | ARHGAP6 | 27.60% | 9.16 × 107 | inflammatory response, transmembrane transport |
| Proteins up-regulated by NnV | ||||||||
| 2250 | O75330 | Hyaluronan mediated motility receptor | ZN33B_HUMAN | 84,101/5.7 | HMMR | 35.5% | 5.68 × 109 | Hyaluronan catabolic process, regulation of G2/M transition of mitotic cell cycle |
| 1562 | P15144 | Aminopeptidase N | AMPN_HUMAN | 109,541/5.3 | ANPEP | 34.20% | 5.11 × 1011 | cell differentiation, proteolysis |
| 1978 | Q86VD1 | MORC family CW-type zinc finger protein 1 | MORC1_HUMAN | 112,882/8.1 | MORC1 | 20.80% | 8.49 × 106 | negative regulation of transposition, cell differentiation, multicellular organism development |
| 2020 | P51784 | Ubiquitin carboxyl-terminal hydrolase 11 | UBP11_HUMAN | 109,818/5.3 | USP11 | 23.90% | 1.19 × 1010 | ubiquitin-dependent protein catabolic process, protein deubiquitination |
| 1378 | O00206 | Toll-like receptor 4 | TLR4_HUMAN | 95,681/5.9 | TLR4 | 31.90% | 6.08 × 108 | activation of MAPK activity, B cell proliferation involved in immune response |
| 2080 | Q8N392 | Rho GTPase-activating protein 18 | RHG18_HUMAN | 96,255/7.3 | ARHGAP18 | 21.40% | 8.53 × 106 | actin filament organization, phagocytosis, engulfment |
| 2514 | Q86UV5 | Ubiquitin carboxyl-terminal hydrolase 48 | UBP48_HUMAN | 119,033/5.7 | USP48 | 21.50% | 8.78 × 107 | protein deubiquitination, ubiquitin-dependent protein catabolic process |
| 1696 | Q969V6 | MKL/myocardin-like protein 1 | MKL1_HUMAN | 98,920/5.6 | Q969V6 | 28.20% | 1.57 × 107 | actin cytoskeleton organization, smooth muscle cell differentiation |
| 1929 | Q9Y4L1 | Hypoxia up-regulated protein 1 | HYOU1_HUMAN | 111,336/5.2 | Q9Y4L1 | 31.20% | 1.45 × 1010 | cellular response to Hypoxia, response to ischemia, receptor-mediated endocytosis |
| 1590 | Q14587 | Heat shock protein 105 kDa | ZN268_HUMAN | 96,866/5.3 | ZNF268 | 23.00% | 5.01 × 107 | receptor-mediated endocytosis, regulation of cellular response to heat, positive regulation of NK T cell activation |
| 1198 | Q96FS4 | Signal-induced proliferation-associated protein 1 | SIPA1_HUMAN | 112,150/6.2 | SIPA1 | 15.90% | 1.91 × 106 | cell proliferation, negative regulation of cell growth, negative regulation of cell cycle |
| 1555 | Q5T7N2 | LINE-1 type transposase domain-containing protein 1 | LITD1_HUMAN | 98,850/4.9 | L1TD1 | 18.20% | 1.50 × 106 | transposition, RNA-mediated |
| 1886 | P15918 | V(D)J recombination-activating protein 1 | RAG1_HUMAN | 119,098/8.9 | RAG1 | 15.8% | 1.42 × 106 | B cell differentiation, T cell homeostasis, negative regulation of thymocyte apoptotic process |
| 710 | A6NI28 | Rho GTPase-activating protein 42 | RHG42_HUMAN | 98,570/8.2 | ARHGAP42 | 26.9% | 2.94 × 107 | activation of GTPase activity, negative regulation of vascular smooth muscle contraction |
| 2199 | P21709 | Ephrin type-A receptor 1 | EPHA1_HUMAN | 108,128/6.2 | EPHA1 | 22.0% | 3.45 × 109 | negative regulation of cell migration, cell surface receptor signaling pathway |
| 1389 | Q93033 | Immunoglobulin superfamily member 2 | IGSF2_HUMAN | 115,109/6.5 | CD101 | 22.9% | 1.59 × 1012 | cell surface receptor signaling pathway, positive regulation of myeloid leukocyte differentiation |
| 2305 | Q4V348 | Zinc finger protein 658B | Z658B_HUMAN | 94,332/8.9 | ZNF658B | 25.40% | 1.51 × 107 | bile acid biosynthetic process |
| 1122 | Q02156 | Protein kinase C epsilon type | KPCE_HUMAN | 83,675/6.7 | PRKCE | 30.10% | 1.18 × 108 | apoptotic process, apoptotic process, |
| 1491 | P49736 | DNA replication licensing factor MCM2 | MCM2_HUMAN | 101,897/5.3 | MCM2 | 26.0% | 9.15 × 108 | apoptotic process, apoptotic process, DNA replication initiation, nucleosome assembly |
| 1827 | P35527 | Keratin, type I cytoskeletal 9 | K1C9_HUMAN | 62,065/5.1 | KRT9 | 20.50% | 1.04 × 106 | intermediate filament organization, cornification |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choudhary, I.; Hwang, D.; Chae, J.; Yoon, W.; Kang, C.; Kim, E. Proteomic Changes during the Dermal Toxicity Induced by Nemopilema nomurai Jellyfish Venom in HaCaT Human Keratinocyte. Toxins 2021, 13, 311. https://doi.org/10.3390/toxins13050311
Choudhary I, Hwang D, Chae J, Yoon W, Kang C, Kim E. Proteomic Changes during the Dermal Toxicity Induced by Nemopilema nomurai Jellyfish Venom in HaCaT Human Keratinocyte. Toxins. 2021; 13(5):311. https://doi.org/10.3390/toxins13050311
Chicago/Turabian StyleChoudhary, Indu, Duhyeon Hwang, Jinho Chae, Wonduk Yoon, Changkeun Kang, and Euikyung Kim. 2021. "Proteomic Changes during the Dermal Toxicity Induced by Nemopilema nomurai Jellyfish Venom in HaCaT Human Keratinocyte" Toxins 13, no. 5: 311. https://doi.org/10.3390/toxins13050311
APA StyleChoudhary, I., Hwang, D., Chae, J., Yoon, W., Kang, C., & Kim, E. (2021). Proteomic Changes during the Dermal Toxicity Induced by Nemopilema nomurai Jellyfish Venom in HaCaT Human Keratinocyte. Toxins, 13(5), 311. https://doi.org/10.3390/toxins13050311





