A Meta-Analysis of the Protein Components in Rattlesnake Venom
Abstract
1. Introduction
2. Result
2.1. Venom Constituents in Crotalus Venom
2.1.1. Frequency of Protein Components in Crotalus Venom
2.1.2. Association between Various Venom Components in Crotalus Venom Using Presence/Absence Data
2.1.3. Association between Various Venom Components in Crotalus Venom Using the Relative Abundance of Protein Components
2.1.4. Hierarchical Clustering of Venom Components to Identify any Similarity or Dissimilarity with Phylogenetic Relationships
2.2. Venom Constituents in Sistrurus Venom
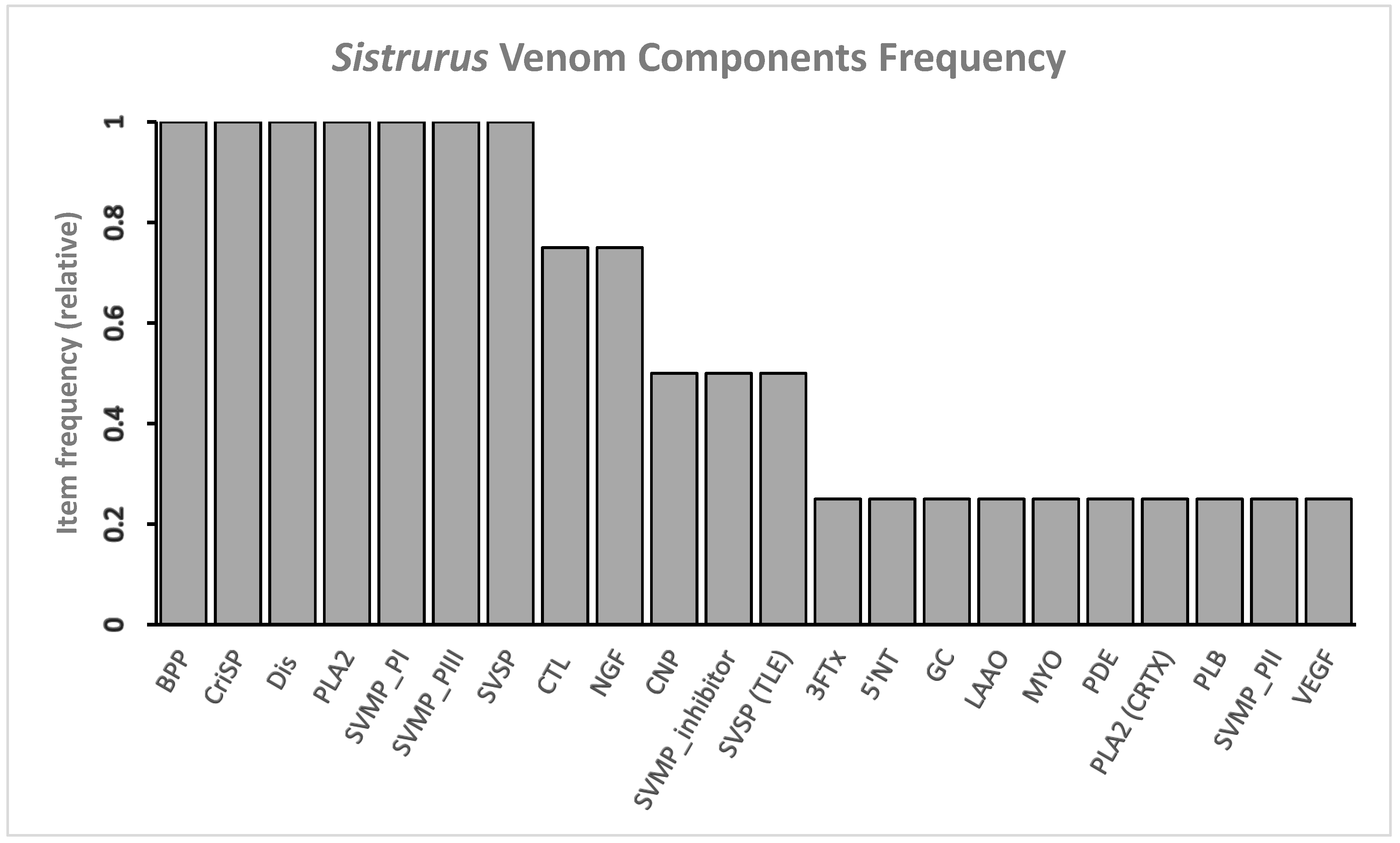
2.2.1. Frequency of Protein Components in Sistrurus Venom
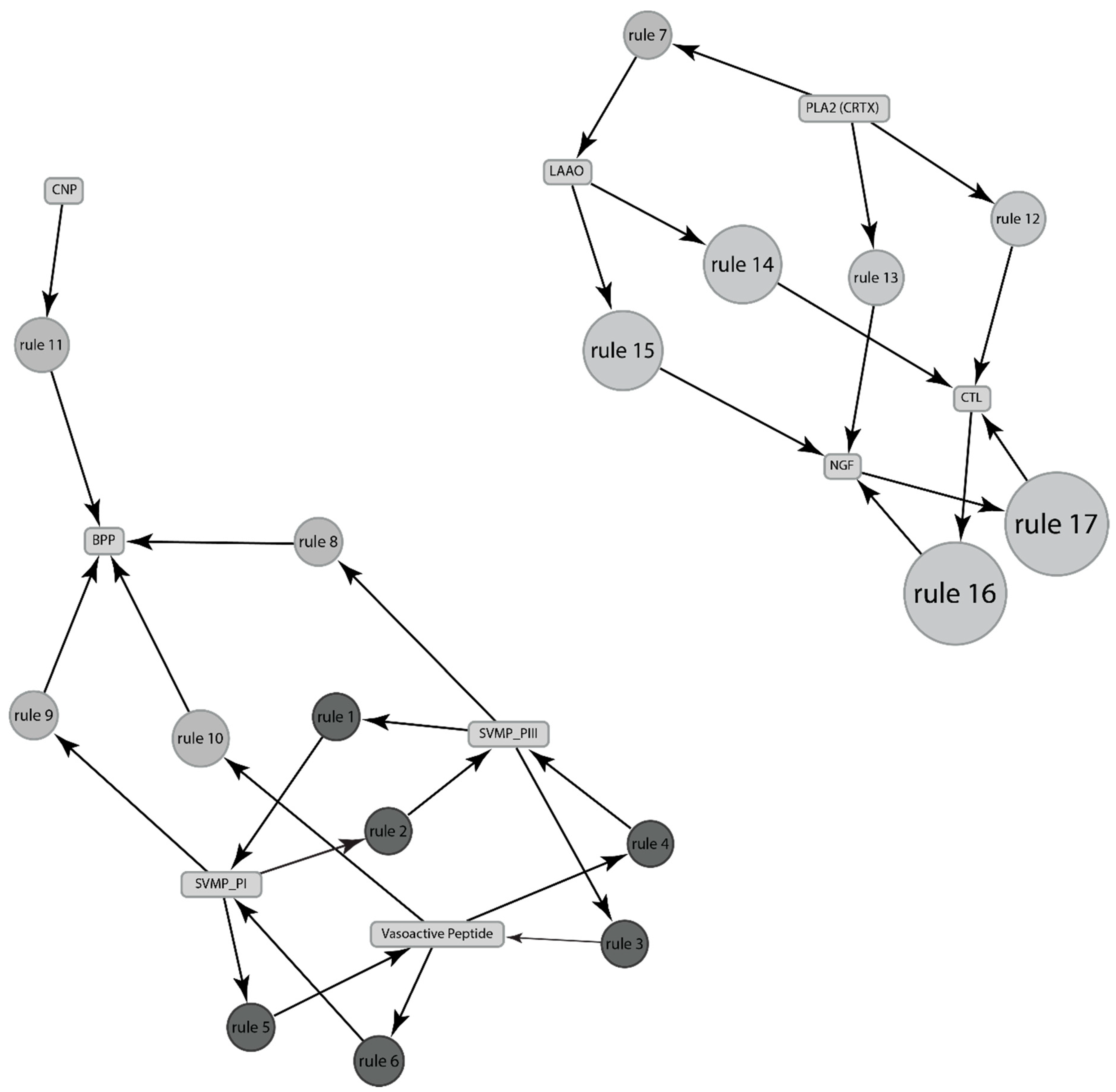
2.2.2. Association between Various Venom Components in Sistrurus Venom Using Presence/Absence Data
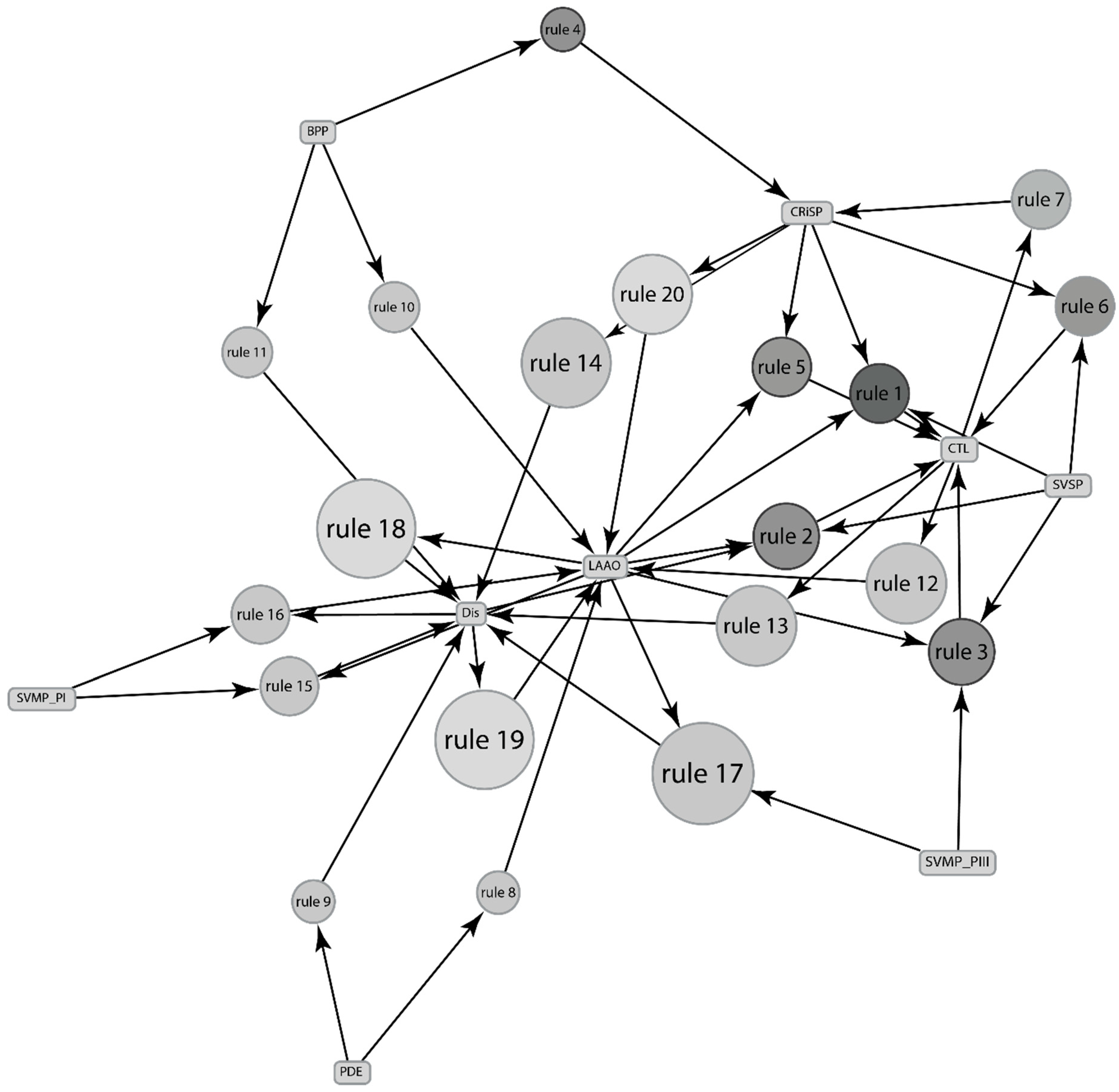
2.2.3. Association between Various Venom Components in Sistrurus Venom Using the Relative Abundance of Protein Components
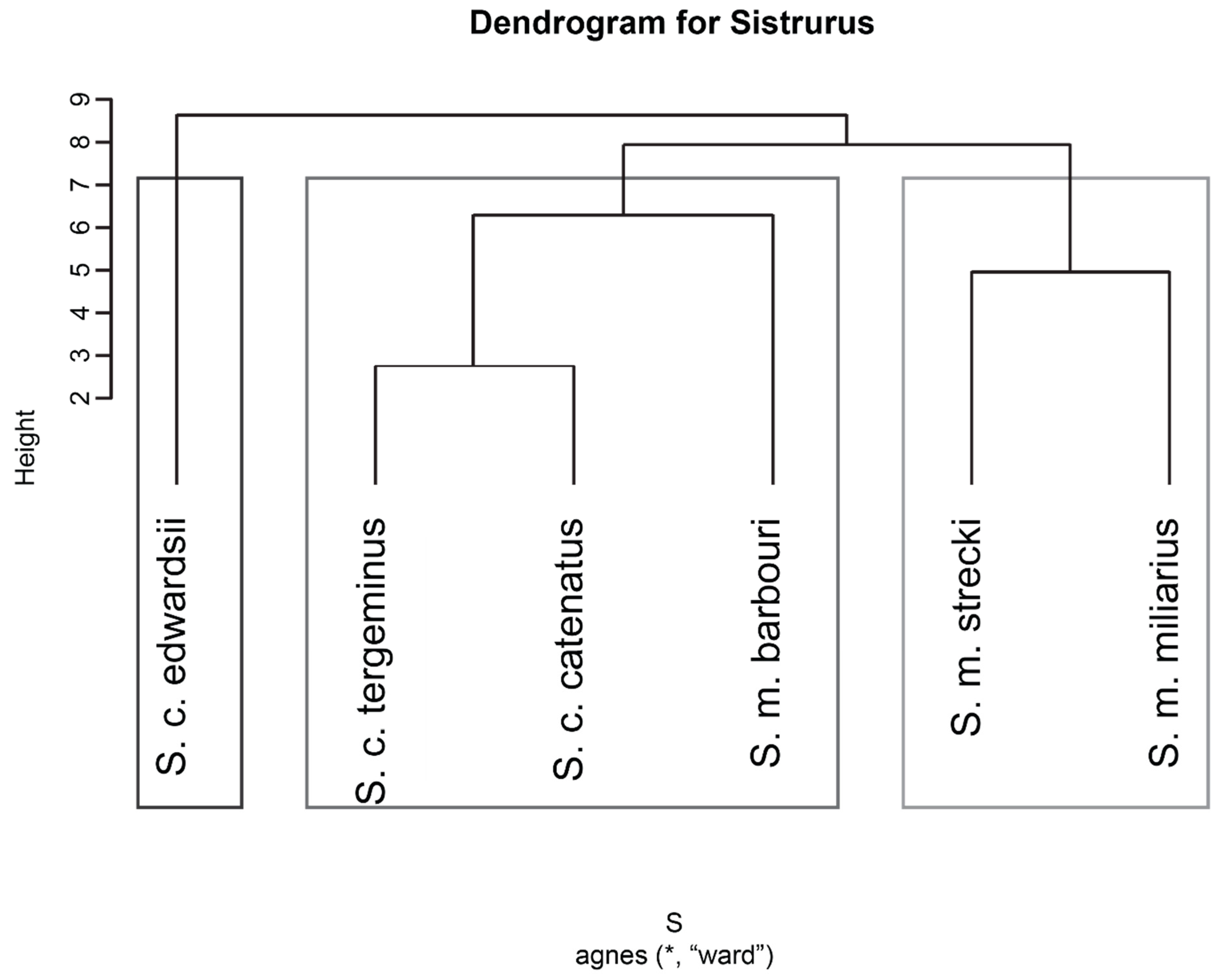
2.2.4. Hierarchical Clustering of Venom Components to Identify Similarities or Dissimilarities in Phylogenetic Relationships
3. Discussion
- (1)
- Two or more toxins interact with different targets on related biological pathways, resulting in synergistically increased toxicity;
- (2)
- Two or more toxins recognize and interact with the same target synergistically and produce the same effect, and is often called amplification;
- (3)
- One toxin (subunit) acts as a chaperone to potentiate another one. The chaperone may expose the active/functional site of the second toxin (subunit), or expose target sites, or increase affinity to target or modify the active surface of the other toxin (subunit). Such complexes usually dissociate after asserting their toxicity.
4. Conclusions
5. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3FTx | Three-finger toxin |
| 5′-NT | 5′-nucleotidase |
| Achase | Acetylcholinesterase |
| ANP | Natriuretic peptide type A |
| ATPase | Adenosine triphosphatase |
| BIP | Bradykinin inhibitory peptide |
| BNP | Natriuretic peptide type B |
| BPP | Bradykinin potentiate peptide |
| CTL | C-type Lectins |
| CNP | Natriuretic peptide type C |
| CysProt | Cysteine protease |
| CRiSP | Cysteine-rich secretory protein |
| CA | Crotapotin |
| CRTX | Crotoxin |
| Dis | Disintegrin |
| EGF | Epidermal growth factor |
| FGF | Fibroblast growth factor |
| GC | Guanylyl cyclase |
| Hya | Hyaluronidase |
| Kazal | Kazal-type inhibitor |
| Kun | Kunitz-type inhibitor |
| LAAO | L-amino acid oxidase |
| MTX | Mojave toxin |
| MYO | Myotoxin |
| NGF | Nerve growth factor |
| OHA | Ohanin |
| PDE | Phosphodiesterase |
| PDGF | Platelet-derived growth factor |
| PLA2 | Phospholipase A2 |
| PLB | Phospholipase B |
| PLD | Phospholipase D |
| SVMP | Snake venom metalloprotease |
| SVSP | Snake venom serine protease |
| TLE | Thrombin-like enzyme |
| VEGF | Vascular endothelial growth factor |
| WAP | Waparin |
| N/A | No information available |
| * | Dataset used for generating hierarchical clusters |
References
- Fry, B.G.; Wüster, W.; Kini, R.M.; Brusic, V.; Khan, A.; Venkataraman, D.; Rooney, A.P. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 2003, 57, 110–129. [Google Scholar] [CrossRef]
- Adade, C.M.; Carvalho, A.L.O.; Tomaz, M.A.; Costa, T.F.R.; Godinho, J.L.; Melo, P.A.; Lima, A.P.C.A.; Rodrigues, J.C.F.; Zingali, R.B.; Souto-Padrón, T. Crovirin, a snake venom cysteine-rich secretory protein (CRISP) with promising activity against Trypanosomes and Leishmania. PLoS Negl. Trop. Dis. 2014, 8, e3252. [Google Scholar] [CrossRef]
- Sanchez, E.E.; Galan, J.A.; Russell, W.K.; Soto, J.G.; Russell, D.H.; Perez, J.C. Isolation and characterization of two disintegrins inhibiting ADP-induced human platelet aggregation from the venom of Crotalus scutulatus scutulatus (Mohave Rattlesnake). Toxicol. Appl. Pharmacol. 2006, 212, 59–68. [Google Scholar] [CrossRef]
- Suntravat, M.; Jia, Y.; Lucena, S.E.; Sánchez, E.E.; Pérez, J.C. cDNA cloning of a snake venom metalloproteinase from the eastern diamondback rattlesnake (Crotalus adamanteus), and the expression of its disintegrin domain with anti-platelet effects. Toxicon Off. J. Int. Soc. Toxinol. 2013, 64, 43–54. [Google Scholar] [CrossRef]
- Suntravat, M.; Barret, H.S.; Jurica, C.A.; Lucena, S.E.; Perez, J.C.; Sánchez, E.E. Recombinant disintegrin (r-Cam-dis) from Crotalus adamanteus inhibits adhesion of human pancreatic cancer cell lines to laminin-1 and vitronectin. J. Venom Res. 2015, 6, 1–10. [Google Scholar] [PubMed]
- Galán, J.A.; Sánchez, E.E.; Rodríguez-Acosta, A.; Soto, J.G.; Bashir, S.; McLane, M.A.; Paquette-Straub, C.; Pérez, J.C. Inhibition of lung tumor colonization and cell migration with the disintegrin crotatroxin 2 isolated from the venom of Crotalus atrox. Toxicon Off. J. Int. Soc. Toxinol. 2008, 51, 1186–1196. [Google Scholar] [CrossRef]
- Mackessy, S.; Saviola, A.; Mukherjee, A. Venom toxins to drugs: Anti-thrombotic and anti-metastasis compounds from snake venoms. Toxicon 2018, 150, 320. [Google Scholar] [CrossRef]
- Arruda Macedo, J.K.; Fox, J.W.; de Souza Castro, M. Disintegrins from snake venoms and their applications in cancer research and therapy. Curr. Protein. Pept. Sci. 2015, 16, 532–548. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.; Lucena, S.; Aguilar, I.; Rodriguez-Acosta, A.; Salazar, A.M.; Sanchez, E.E.; Giron, M.E.; Carvajal, Z.; Arocha-Pinango, C.L.; Guerrero, B. Anti-platelet effect of cumanastatin 1, a disintegrin isolated from venom of South American Crotalus rattlesnake. Thromb. Res. 2009, 123, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Approaching the golden age of natural product pharmaceuticals from venom libraries: An overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications. Curr. Pharm. Des. 2007, 13, 2927–2934. [Google Scholar] [CrossRef] [PubMed]
- Lucena, S.E.; Romo, K.; Suntravat, M.; Sánchez, E.E. Anti-angiogenic activities of two recombinant disintegrins derived from the Mohave and Prairie rattlesnakes. Toxicon 2014, 78, 10–17. [Google Scholar] [CrossRef]
- Urra, F.A.; Araya-Maturana, R. Targeting Metastasis with Snake Toxins: Molecular Mechanisms. Toxins 2017, 9, 390. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A Review and Database of Snake Venom Proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef]
- Kumar, T.; Yang, P.; Lin, S.; Wu, C.; Lei, B.; Lo, S.; Tu, S.-C.; Yu, C. Cloning, Direct Expression, and Purification of a Snake Venom Cardiotoxin inEscherichia coli. Biochem. Biophys. Res. Commun. 1996, 219, 450–456. [Google Scholar] [CrossRef]
- Kumar, T.; Jayaraman, G.; Lee, C.S.; Arunkumar, A.; Sivaraman, T.; Samuel, D.; Yu, C. Snake venom cardiotoxins-structure, dynamics, function and folding. J. Biomol. Struct. Dyn. 1997, 15, 431–463. [Google Scholar] [CrossRef]
- Sivaraman, T.; Kumar, T.; Tu, Y.; Peng, H.; Yu, C. Structurally homologous toxins isolated from the Taiwan cobra (Naja naja atra) differ significantly in their structural stability. Arch. Biochem. Biophys. 1999, 363, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Daltry, J.C.; Wuster, W.; Thorpe, R.S. Diet and snake venom evolution. Nature 1996, 379, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Wickramaratna, J.C.; Hodgson, W.C.; Alewood, P.F.; Kini, R.; Ho, H.; Wüster, W. Electrospray liquid chromatography/mass spectrometry fingerprinting of Acanthophis (death adder) venoms: Taxonomic and toxinological implications. Rapid Commun. Mass Spectrom. 2002, 16, 600–608. [Google Scholar] [CrossRef]
- Assakura, M.T.; Salomao, M.G.; Puorto, G.; Mandelbaum, F.R. Hemorrhagic, fibrinogenolytic and edema-forming activities of the venom of the colubrid snake Philodryas olfersii (green snake). Toxicon 1992, 30, 427–438. [Google Scholar] [CrossRef]
- Jimenez Porras, J.M. Intraspecific variations in composition of venom of the jumping viper, bothrops nummifera. Toxicon 1964, 2, 187–195. [Google Scholar] [CrossRef]
- Glenn, J.L.; Straight, R.C.; Wolfe, M.C.; Hardy, D.L. Geographical variation in Crotalus scutulatus scutulatus (Mojave rattlesnake) venom properties. Toxicon 1983, 21, 119–130. [Google Scholar] [CrossRef]
- Yang, C.C.; Chang, L.S.; Wu, F.S. Venom constituents of Notechis scutatus scutatus (Australian tiger snake) from differing geographic regions. Toxicon 1991, 29, 1337–1344. [Google Scholar] [CrossRef]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wuster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Saravia, N.G.; Weigle, K.; Navas, C.; Segura, I.; Valderrama, L.; Valencia, A.Z.; Escorcia, B.; McMahon-Pratt, D. Heterogeneity, geographic distribution, and pathogenicity of serodemes of Leishmania viannia in Colombia. Am. J. Trop. Med. Hyg. 2002, 66, 738–744. [Google Scholar] [CrossRef]
- Creer, S.; Malhotra, A.; Thorpe, R.S.; Stocklin, R.S.; Favreau, P.S.; Hao Chou, W.S. Genetic and ecological correlates of intraspecific variation in pitviper venom composition detected using matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS) and isoelectric focusing. J. Mol. Evol. 2003, 56, 317–329. [Google Scholar] [CrossRef]
- Tonello, F.; Simonato, M.; Aita, A.; Pizzo, P.; Fernandez, J.; Lomonte, B.; Gutierrez, J.M.; Montecucco, C. A Lys49-PLA2 myotoxin of Bothrops asper triggers a rapid death of macrophages that involves autocrine purinergic receptor signaling. Cell Death Dis. 2012, 3, e343. [Google Scholar] [CrossRef]
- Fry, B. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery; Oxford University Press: New York, NY, USA, 2015. [Google Scholar]
- Mackessy, S.P.; Sixberry, N.M.; Heyborne, W.H.; Fritts, T. Venom of the Brown Treesnake, Boiga irregularis: Ontogenetic shifts and taxa-specific toxicity. Toxicon Off. J. Int. Soc. Toxinol. 2006, 47, 537–548. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Margres, M.J.; Calvin, K. Post-transcriptional Mechanisms Contribute Little to Phenotypic Variation in Snake Venoms. G3 Genes Genomes Genet. 2015, 5, 2375–2382. [Google Scholar] [CrossRef]
- Calvete, J.J.; Juarez, P.; Sanz, L. Snake venomics. Strategy and applications. J. Mass Spectrom. 2007, 42, 1405–1414. [Google Scholar] [CrossRef]
- Cintra, A.C.O.; De Toni, L.G.B.; Sartim, M.A.; Franco, J.J.; Caetano, R.C.; Murakami, M.T.; Sampaio, S.V. Batroxase, a new metalloproteinase from B. atrox snake venom with strong fibrinolytic activity. Toxicon 2012, 60, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W. A brief review of the scientific history of several lesser-known snake venom proteins: L-amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon 2013, 62, 75–82. [Google Scholar] [CrossRef]
- Kordis, D.; Gubensek, F. Adaptive evolution of animal toxin multigene families. Gene 2000, 261, 43–52. [Google Scholar] [CrossRef]
- Moura-da-Silva, A.M.; Paine, M.J.I.; Diniz, M.R.V.; Theakston, R.D.G.; Crampton, J.M. The molecular cloning of a phospholipase A 2 from Bothrops jararacussu snake venom: Evolution of venom group II phospholipase A 2’s may imply gene duplications. J. Mol. Evol. 2020, 41, 174–179. [Google Scholar] [CrossRef]
- Slowinski, J.B.; Knight, A.; Rooney, A.P. Inferring species trees from gene trees: A phylogenetic analysis of the Elapidae (Serpentes) based on the amino acid sequences of venom proteins. Mol. Phylogenet. Evol. 1997, 8, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Afifiyan, F.; Armugam, A.; Tan, C.H.; Gopalakrishnakone, P.; Jeyaseelan, K. Postsynaptic α-Neurotoxin Gene of the Spitting Cobra, Naja naja sputatrix: Structure, Organization, and Phylogenetic Analysis. Genome Res. 1999, 9, 259–266. [Google Scholar] [PubMed]
- Chang, L.; Lin, S.; Huang, H.; Hsiao, M. Genetic organization of alpha-bungarotoxins from Bungarus multicinctus (Taiwan banded krait): Evidence showing that the production of alpha-bungarotoxin isotoxins is not derived from edited mRNAs. Nucleic. Acids Res. 1999, 27, 3970–3975. [Google Scholar] [CrossRef]
- Kocholaty, W.F.; Ledford, E.B.; Daly, J.G.; Billings, T.A. Toxicity and some enzymatic properties and activities in the venoms of Crotalidae, Elapidae and Viperidae. Toxicon 1971, 9, 131–138. [Google Scholar] [CrossRef]
- Campbell, J.A.; Lamar, W.W.; Brodie, E.D. The Venomous Reptiles of the Western Hemisphere; Comstock Pub. Associates: Ithaca, NY, USA, 2004; Volume 2. [Google Scholar]
- Arnaud-Franco, G.; Cordero-Tapia, A.; Ortiz-Avila, V.; Moctezuma-Gonzalez, C.L.; Tejocote-Perez, M.; Carbajal-Saucedo, A. Comparison of biological and biochemical characteristics of venom from rattlesnakes in the southern Baja California Peninsula. Toxicon 2018, 148, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Borja, M.; Neri-Castro, E.; Castaneda-Gaytan, G.; Strickland, J.L.; Parkinson, C.L.; Castaneda-Gaytan, J.; Ponce-Lopez, R.; Lomonte, B.; Olvera-Rodriguez, A.; Alagon, A.; et al. Biological and Proteolytic Variation in the Venom of Crotalus scutulatus scutulatus from Mexico. Toxins 2018, 10, 35. [Google Scholar] [CrossRef]
- Mackessy, S.P. Evolutionary trends in venom composition in the western rattlesnakes (Crotalus viridis sensu lato): Toxicity vs. tenderizers. Toxicon 2010, 55, 1463–1474. [Google Scholar] [CrossRef]
- Findrik, Z.; Vasić-Rački, Ð.; Primožič, M.; Habulin, M.; Knez, Ž. Enzymatic activity of L-amino acid oxidase from snake venom Crotalus adamanteus in supercritical CO2. Biocatal. Biotransform. 2005, 23, 315–321. [Google Scholar] [CrossRef]
- Calvete, J.J.; Fasoli, E.; Sanz, L.; Boschetti, E.; Righetti, P.G. Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res. 2009, 8, 3055–3067. [Google Scholar] [CrossRef] [PubMed]
- Boldrini-Franca, J.; Correa-Netto, C.; Silva, M.M.; Rodrigues, R.S.; De La Torre, P.; Perez, A.; Soares, A.M.; Zingali, R.B.; Nogueira, R.A.; Rodrigues, V.M.; et al. Snake venomics and antivenomics of Crotalus durissus subspecies from Brazil: Assessment of geographic variation and its implication on snakebite management. J. Proteom. 2010, 73, 1758–1776. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.; Burin, S.M.; Menaldo, D.L.; de Castro, F.A.; Sampaio, S.V. Snake venom L-amino acid oxidases: An overview on their antitumor effects. J. Venom. Anim. Toxins Incl. Trop. Dis. 2014, 20, 23. [Google Scholar] [CrossRef]
- Mackessy, S.P. Fractionation of red diamond rattlesnake (Crotalus ruber ruber) venom: Protease, phosphodiesterase, L-amino acid oxidase activities and effects of metal ions and inhibitors on protease activity. Toxicon 1985, 23, 337–340. [Google Scholar] [CrossRef]
- Mackessy, S.P.; Leroy, J.; Mociño-Deloya, E.; Setser, K.; Bryson, R.W.; Saviola, A.J. Venom Ontogeny in the Mexican Lance-Headed Rattlesnake (Crotalus polystictus). Toxins 2018, 10, 271. [Google Scholar] [CrossRef]
- Saviola, A.J.; Gandara, A.J.; Bryson, R.W., Jr.; Mackessy, S.P. Venom phenotypes of the Rock Rattlesnake (Crotalus lepidus) and the Ridge-nosed Rattlesnake (Crotalus willardi) from Mexico and the United States. Toxicon 2017, 138, 119–129. [Google Scholar] [CrossRef]
- Komori, Y.; Sakai, K.; Masuda, K.; Nikai, T. Isolation and Biochemical Characterization of Rubelase, a Non-Hemorrhagic Elastase from Crotalus ruber ruber (Red Rattlesnake) Venom. Toxins 2011, 3, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P. Characterization of the major metalloprotease isolated from the venom of the northern pacific rattlesnake, Crotalus viridis oreganus. Toxicon 1996, 34, 1277–1285. [Google Scholar] [CrossRef]
- Smith, C.F.; Mackessy, S.P. The effects of hybridization on divergent venom phenotypes: Characterization of venom from Crotalus scutulatus scutulatus x Crotalus oreganus helleri hybrids. Toxicon 2016, 120, 110–123. [Google Scholar] [CrossRef]
- Boldrini-Franca, J.; Pinheiro-Junior, E.L.; Arantes, E.C. Functional and biological insights of rCollinein-1, a recombinant serine protease from Crotalus durissus collilineatus. J. Venom. Anim. Toxins Incl. Trop. Dis. 2019, 25, e147118. [Google Scholar] [CrossRef]
- Calvete, J.J.; Sanz, L.; Cid, P.; de la Torre, P.; Flores-Diaz, M.; Dos Santos, M.C.; Borges, A.; Bremo, A.; Angulo, Y.; Lomonte, B.; et al. Snake venomics of the Central American rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J. Proteome Res. 2010, 9, 528–544. [Google Scholar] [CrossRef]
- Calvete, J.J.; Perez, A.; Lomonte, B.; Sanchez, E.E.; Sanz, L. Snake venomics of Crotalus tigris: The minimalist toxin arsenal of the deadliest Nearctic rattlesnake venom. Evolutionary Clues for generating a pan-specific antivenom against crotalid type II venoms [corrected]. J. Proteome Res. 2012, 11, 1382–1390. [Google Scholar] [CrossRef]
- Almeida, J.R.; Resende, L.M.; Silva, A.G.; Ribeiro, R.I.; Stabeli, R.G.; Soares, A.M.; Calderon, L.A.; Marangoni, S.; Da Silva, S.L. Biochemical and functional studies of ColTx-I, a new myotoxic phospholipase A2 isolated from Crotalus oreganus lutosus (Great Basin rattlesnake) snake venom. Toxicon 2016, 117, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.R.; Lancellotti, M.; Soares, A.M.; Calderon, L.A.; Ramirez, D.; Gonzalez, W.; Marangoni, S.; Da Silva, S.L. CoaTx-II, a new dimeric Lys49 phospholipase A2 from Crotalus oreganus abyssus snake venom with bactericidal potential: Insights into its structure and biological roles. Toxicon 2016, 120, 147–158. [Google Scholar] [CrossRef]
- Macías-Rodríguez, E.F.; Díaz-Cárdenas, C.O.; Gatica-Colima, A.B.; Plenge-Tellechea, L.F. Seasonal variation in protein content and PLA2 activity of Crotalus molossus molossus venom from captive and wild specimens. Acta Univ. 2014, 24, 38–47. [Google Scholar] [CrossRef]
- Rivas, E.; Neri, E.; Benard, M.I.; Hernánez-Dávila, A.; Zamudio, F.; Alagón, A. General characterization of the venoms from two species of rattlesnakes and an intergrade population (C. lepidus x aquilus ) from Aguascalientes and Zacatecas, Mexico. Toxicon 2017, 138. [Google Scholar] [CrossRef] [PubMed]
- Griffin, P.R.; Aird, S.D. A new small myotoxin from the venom of the prairie rattlesnake (Crotalus viridis viridis). FEBS Lett. 1990, 274, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Ownby, C.L.; Colberg, T.R.; White, S.P. Isolation, characterization and crystallization of a phospholipase A2 myotoxin from the venom of the prairie rattlesnake (Crotalus viridis viridis). Toxicon 1997, 35, 111–124. [Google Scholar] [CrossRef]
- Saviola, A.J.; Pla, D.; Sanz, L.; Castoe, T.A.; Calvete, J.J.; Mackessy, S.P. Comparative venomics of the Prairie Rattlesnake (Crotalus viridis viridis) from Colorado: Identification of a novel pattern of ontogenetic changes in venom composition and assessment of the immunoreactivity of the commercial antivenom CroFab(R). J. Proteom. 2015, 121, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Strickland, J.L.; Mason, A.J.; Rokyta, D.R.; Parkinson, C.L. Phenotypic Variation in Mojave Rattlesnake (Crotalus scutulatus) Venom Is Driven by Four Toxin Families. Toxins 2018, 10, 135. [Google Scholar] [CrossRef]
- Evangelista, J.S.A.M.; Martins, A.M.C.; Nascimento, N.R.F.; Sousa, C.M.; Alves, R.S.; Toyama, D.O.; Toyama, M.H.; Evangelista, J.J.F.; Menezes, D.B.d.; Fonteles, M.C.; et al. Renal and vascular effects of the natriuretic peptide isolated from Crotalus durissus cascavella venom. Toxicon Off. J. Int. Soc. Toxinol. 2008, 52, 737–744. [Google Scholar] [CrossRef]
- Gomes, C.L.; Konno, K.; Conceição, I.M.; Ianzer, D.; Yamanouye, N.; Prezoto, B.C.; Assakura, M.T.; Rádis-Baptista, G.; Yamane, T.; Santos, R.A.; et al. Identification of novel bradykinin-potentiating peptides (BPPs) in the venom gland of a rattlesnake allowed the evaluation of the structure-function relationship of BPPs. Biochem. Pharmacol. 2007, 74, 1350–1360. [Google Scholar] [CrossRef]
- Carey, C.M.; Bueno, R.; Gutierrez, D.A.; Petro, C.; Lucena, S.E.; Sanchez, E.E.; Soto, J.G. Recombinant rubistatin (r-Rub), an MVD disintegrin, inhibits cell migration and proliferation, and is a strong apoptotic inducer of the human melanoma cell line SK-Mel-28. Toxicon 2012, 59, 241–248. [Google Scholar] [CrossRef]
- Saviola, A.J.; Modahl, C.M.; Mackessy, S.P. Disintegrins of Crotalus simus tzabcan venom: Isolation, characterization and evaluation of the cytotoxic and anti-adhesion activities of tzabcanin, a new RGD disintegrin. Biochimie 2015, 116, 92–102. [Google Scholar] [CrossRef]
- Saviola, A.J.; Burns, P.D.; Mukherjee, A.K.; Mackessy, S.P. The disintegrin tzabcanin inhibits adhesion and migration in melanoma and lung cancer cells. Int. J. Biol. Macromol. 2016, 88, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Roldán-Padrón, O.; Castro-Guillén, J.L.; García-Arredondo, J.A.; Cruz-Pérez, M.S.; Díaz-Peña, L.F.; Saldaña, C.; Blanco-Labra, A.; García-Gasca, T. Snake Venom Hemotoxic Enzymes: Biochemical Comparison between Crotalus Species from Central Mexico. Molecules 2019, 24, 1489. [Google Scholar] [CrossRef]
- Gibbs, H.L.; Mackessy, S.P. Functional basis of a molecular adaptation: Prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon 2009, 53, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.L.; Sanz, L.; Sovic, M.G.; Calvete, J.J. PMC3691181; Phylogeny-based comparative analysis of venom proteome variation in a clade of rattlesnakes (Sistrurus sp.). PLoS ONE 2013, 8, e67220. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wagstaff, S.C.; Wüster, W.; Cook, D.A.; Bolton, F.M.; King, S.I.; Pla, D.; Sanz, L.; Calvete, J.J.; Harrison, R.A. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl. Acad. Sci. 2014, 111, 9205–9210. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Huang, C. Synergistic Strategies of Predominant Toxins in Snake Venoms. Toxicol. Lett. 2018, 287. [Google Scholar] [CrossRef]
- Groten, J.P.; Feron, V.J.; Sühnel, J. Toxicology of Simple and Complex Mixtures. Trends Pharmacol. Sci. 2001, 22. [Google Scholar] [CrossRef]
- Buschek, S.; Ignjatovic, V.; Summerhayes, R.; Lowe, R. The effect of different snake venoms and anti-venoms on thrombin clotting time in human plasma. Thromb Res. 2010, 125, e149–e152. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.N.; Lomonte, B.; del Carmen Gutierrez, M.; Alagon, A.; Gutierrez, J.M. Intraspecies variation in the venom of the rattlesnake Crotalus simus from Mexico: Different expression of crotoxin results in highly variable toxicity in the venoms of three subspecies. J. Proteom. 2013, 87, 103–121. [Google Scholar] [CrossRef]
- Viala, V.L.; Hildebrand, D.; Fucase, T.M.; Sciani, J.M.; Prezotto-Neto, J.P.; Riedner, M.; Sanches, L.; Nishimura, P.J.; Oguiura, N.; Pimenta, D.C.; et al. Proteomic analysis of the rare Uracoan rattlesnake Crotalus vegrandis venom: Evidence of a broad arsenal of toxins. Toxicon 2015, 107, 234–251. [Google Scholar] [CrossRef]
- Margres, M.J.; McGivern, J.J.; Wray, K.P.; Seavy, M.; Calvin, K.; Rokyta, D.R. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). J. Proteom. 2014, 96, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.R.; Lemmon, A.R.; Margres, M.J.; Aronow, K. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genom. 2012, 13, 312. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Wray, K.P.; Lemmon, A.R.; Lemmon, E.M.; Caudle, S.B. A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon Off. J. Int. Soc. Toxinol. 2011, 57, 657–671. [Google Scholar] [CrossRef]
- Vincent, M.; Curti, B. On the Reaction Mechanism of Crotalus adamanteus l-Amino Acid Oxidase. J. Biol. Chem. 1967, 242, 1259–1264. [Google Scholar]
- Heinrikson, R.L.; Krueger, E.T.; Keim, P.S. Amino acid sequence of phospholipase A2-alpha from the venom of Crotalus adamanteus. A new classification of phospholipases A2 based upon structural determinants. J. Biol. Chem. 1977, 252, 4913–4921. [Google Scholar] [CrossRef]
- Jia, Y.; Olvera, P.; Rangel, F.; Mendez, B.; Reddy, S. Rapid Identification of Phospholipase A2 Transcripts from Snake Venoms. Toxins 2019, 11, 69. [Google Scholar] [CrossRef]
- Wellner, D.; Meister, A. Crystalline L-amino acid oxidase of Crotalus adamanteus. J. Biol. Chem. 1960, 235, 2013–2018. [Google Scholar] [CrossRef]
- Samy, R.P.; Kandasamy, M.; Gopalakrishnakone, P.; Stiles, B.G.; Rowan, E.G.; Becker, D.; Shanmugam, M.K.; Sethi, G.; Chow, V.T.K. Wound healing activity and mechanisms of action of an antibacterial protein from the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). PLoS ONE 2014, 9, e80199. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Margres, M.J.; Ward, M.J.; Sanchez, E.E. The genetics of venom ontogeny in the eastern diamondback rattlesnake (Crotalus adamanteus). PeerJ 2017, 5, e3249. [Google Scholar] [CrossRef] [PubMed]
- Gomis-Rüth, F.X.; Kress, L.F.; Kellermann, J.; Mayr, I.; Lee, X.; Huber, R.; Bode, W. Refined 2·0 Å X-ray Crystal Structure of the Snake Venom Zinc-endopeptidase Adamalysin II: Primary and Tertiary Structure Determination, Refinement, Molecular Structure and Comparison with Astacin, Collagenase and Thermolysin. J. Mol. Biol. 1994, 239, 513–544. [Google Scholar] [CrossRef]
- Suntravat, M.; Langlais, P.R.; Sánchez, E.E.; Nielsen, V.G. CatroxMP-II: A heme-modulated fibrinogenolytic metalloproteinase isolated from Crotalus atrox venom. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 2018, 31, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Sultan, M.T.; Li, H.-M.; Lee, Y.Z.; Lim, S.S.; Song, D.-K. Identification of Lys49-PLA2 from crude venom of Crotalus atrox as a human neutrophil-calcium modulating protein. Korean J. Physiol. Pharmacol. 2016, 20, 177–183. [Google Scholar] [CrossRef]
- Igarashi, T.; Araki, S.; Mori, H.; Takeda, S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007, 581, 2416–2422. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M.T. Timeline of key events in snake venom metalloproteinase research. J. Proteom. 2009, 72, 200–209. [Google Scholar] [CrossRef]
- Torii, S.; Naito, M.; Tsuruo, T. Apoxin I, a novel apoptosis-inducing factor with L-amino acid oxidase activity purified from Western diamondback rattlesnake venom. J. Biol. Chem. 1997, 272, 9539–9542. [Google Scholar] [CrossRef]
- Kikushima, E.; Nakamura, S.; Oshima, Y.; Shibuya, T.; Miao, J.Y.; Hayashi, H.; Nikai, T.; Araki, S. Hemorrhagic activity of the vascular apoptosis-inducing proteins VAP1 and VAP2 from Crotalus atrox. Toxicon Off. J. Int. Soc. Toxinol. 2008, 52, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, J.B.; Fox, J.W. Proteolytic specificity and cobalt exchange of hemorrhagic toxin e, a zinc protease isolated from the venom of the western diamondback rattlesnake (Crotalus atrox). Biochemistry 1983, 22, 3770–3778. [Google Scholar] [CrossRef]
- Bjarnason, J.B.; Barish, A.; Direnzo, G.S.; Campbell, R.; Fox, J.W. Kallikrein-like Enzymes From Crotalus Atrox Venom. J. Biol. Chem. 1983, 258, 12566–12573. [Google Scholar] [CrossRef]
- Segura, A.; Herrera, M.; Mares, F.R.; Jaime, C.; Sanchez, A.; Vargas, M.; Villalta, M.; Gomez, A.; Gutierrez, J.M.; Leon, G. Proteomic, toxicological and immunogenic characterization of Mexican west -coast rattlesnake (Crotalus basiliscus) venom and its immunological relatedness with the venom of Central American rattlesnake (Crotalus simus). J. Proteom. 2017, 158, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Datta, G.; Dong, A.; Witt, J.; Tu, A.T. Biochemical characterization of basilase, a fibrinolytic enzyme from Crotalus basiliscus basiliscus. Arch. Biochem. Biophys. 1995, 317, 365–373. [Google Scholar] [CrossRef]
- Scarborough, R.M.; Rose, J.W.; Naughton, M.A.; Phillips, D.R.; Nannizzi, L.; Arfsten, A.; Campbell, A.M.; Charo, I.F. Characterization of the Integrin Specificities of Disintegrins Isolated From American Pit Viper Venoms. J. Biol. Chem. 1993, 268, 1058–1065. [Google Scholar] [CrossRef]
- Retzios, A.D.; Markland, J.F.S. Purification, characterization, and fibrinogen cleavage sites of three fibrinolytic enzymes from the venom of Crotalus basiliscus basiliscus. Biochemistry 1992, 31, 4547–4557. [Google Scholar] [CrossRef] [PubMed]
- Rautsaw, R.M.; Hofmann, E.P.; Margres, M.J.; Holding, M.L.; Strickland, J.L.; Mason, A.J.; Rokyta, D.R.; Parkinson, C.L. Intraspecific sequence and gene expression variation contribute little to venom diversity in sidewinder rattlesnakes (Crotalus cerastes). Proc. Biol. Sci. 2019, 286. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, E.P.; Rautsaw, R.M.; Strickland, J.L.; Holding, M.L.; Hogan, M.P.; Mason, A.J.; Rokyta, D.R.; Parkinson, C.L. Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Sci. Rep. 2018, 8, 15534. [Google Scholar] [CrossRef] [PubMed]
- Bosak, A.R.; Ruha, A.M.; Graeme, K.A. A case of neurotoxicity following envenomation by the Sidewinder rattlesnake, Crotalus cerastes. J. Med. Toxicol. 2014, 10, 229–231. [Google Scholar] [CrossRef]
- Bandeira, I.C.J.; Bandeira-Lima, D.; Mello, C.P.; Pereira, T.P.; De Menezes, R.; Sampaio, T.L.; Falcao, C.B.; Radis-Baptista, G.; Martins, A.M.C. Antichagasic effect of crotalicidin, a cathelicidin-like vipericidin, found in Crotalus durissus terrificus rattlesnake’s venom gland. Parasitology 2018, 145, 1059–1064. [Google Scholar] [CrossRef]
- Wiezel, G.A.; Shibao, P.Y.T.; Cologna, C.T.; Morandi Filho, R.; Ueira-Vieira, C.; De Pauw, E.; Quinton, L.; Arantes, E.C. In-Depth Venome of the Brazilian Rattlesnake Crotalus durissus terrificus: An Integrative Approach Combining Its Venom Gland Transcriptome and Venom Proteome. J. Proteome Res. 2018, 17, 3941–3958. [Google Scholar] [CrossRef] [PubMed]
- Bordon, K.C.; Perino, M.G.; Giglio, J.R.; Arantes, E.C. Isolation, enzymatic characterization and antiedematogenic activity of the first reported rattlesnake hyaluronidase from Crotalus durissus terrificus venom. Biochimie 2012, 94, 2740–2748. [Google Scholar] [CrossRef]
- de Oliveira, S.A.; Magalhaes, M.R.; de Oliveira, L.P.; da Cunha, L.C. Identification of antinociceptive fraction of snake venom from Crotalus durissus collilineatus crotamine-negative and its acute toxicity evaluation. Toxicon 2016, 122, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Lopes, D.M.; Junior, N.E.; Costa, P.P.; Martins, P.L.; Santos, C.F.; Carvalho, E.D.; Carvalho, M.D.; Pimenta, D.C.; Cardi, B.A.; Fonteles, M.C.; et al. A new structurally atypical bradykinin-potentiating peptide isolated from Crotalus durissus cascavella venom (South American rattlesnake). Toxicon 2014, 90, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Pereañez, J.A.; Núñez, V.; Huancahuire-Vega, S.; Marangoni, S.; Ponce-Soto, L.A. Biochemical and Biological Characterization of a PLA2 From Crotoxin Complex of Crotalus Durissus Cumanensis. Toxicon Off. J. Int. Soc. Toxinol. 2009, 53. [Google Scholar] [CrossRef]
- Marcussi, S.; Santos, P.R.S.; Menaldo, D.L.; Silveira, L.B.; Santos-Filho, N.; Mazzi, M.V.; da Silva, S.L.; Stábeli, R.G.; Antunes, L.M.G.; Soares, A.M. Evaluation of the genotoxicity of Crotalus durissus terrificus snake venom and its isolated toxins on human lymphocytes. Mutat. Res. 2011, 724, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, J.F.; Pereira, C.M.; Bittar, C.; Batista, M.N.; Campos, G.R.F.; da Silva, S.; Cintra, A.C.O.; Zothner, C.; Harris, M.; Sampaio, S.V.; et al. Multiple effects of toxins isolated from Crotalus durissus terrificus on the hepatitis C virus life cycle. PLoS ONE 2017, 12, e0187857. [Google Scholar] [CrossRef]
- Cavalcante, W.L.; Ponce-Soto, L.A.; Marangoni, S.; Gallacci, M. Neuromuscular effects of venoms and crotoxin-like proteins from Crotalus durissus ruruima and Crotalus durissus cumanensis. Toxicon 2015, 96, 46–49. [Google Scholar] [CrossRef]
- Cavalcante, W.L.G.; Noronha-Matos, J.B.; Timoteo, M.A.; Fontes, M.R.M.; Gallacci, M.; Correia-de-Sa, P. Neuromuscular paralysis by the basic phospholipase A2 subunit of crotoxin from Crotalus durissus terrificus snake venom needs its acid chaperone to concurrently inhibit acetylcholine release and produce muscle blockage. Toxicol Appl. Pharmacol. 2017, 334, 8–17. [Google Scholar] [CrossRef]
- Costa, C.R.C.; Belchor, M.N.; Rodrigues, C.F.B.; Toyama, D.O.; de Oliveira, M.A.; Novaes, D.P.; Toyama, M.H. Edema Induced by a Crotalus durissus terrificus Venom Serine Protease (Cdtsp 2) Involves the PAR Pathway and PKC and PLC Activation. Int. J. Mol. Sci. 2018, 19, 2405. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, S.A.M.; Magalhães, M.R.; Salazar, V.C.R.; Valadares, M.C.; da Cunha, L.C. Identification of crotamine in the venom of Crotalus durissus collilineatus by three different methods. Toxicon 2015, 95, 46–51. [Google Scholar] [CrossRef]
- de Oliveira, L.A.; Ferreira, R.S., Jr.; Barraviera, B.; de Carvalho, F.C.T.; de Barros, L.C.; Dos Santos, L.D.; Pimenta, D.C. Crotalus durissus terrificus crotapotin naturally displays preferred positions for amino acid substitutions. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 46. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.P.; Favoretto, B.C.; Clissa, P.B.; Sampaio, S.C.; Faquim-Mauro, E.L. Crotoxin Isolated from Crotalus durissus terrificus Venom Modulates the Functional Activity of Dendritic Cells via Formyl Peptide Receptors. J. Immunol. Res. 2018, 2018, 7873257. [Google Scholar] [CrossRef]
- Muller, V.D.; Soares, R.O.; dos Santos, N.N., Jr.; Trabuco, A.C.; Cintra, A.C.; Figueiredo, L.T.; Caliri, A.; Sampaio, S.V.; Aquino, V.H. Phospholipase A2 isolated from the venom of Crotalus durissus terrificus inactivates dengue virus and other enveloped viruses by disrupting the viral envelope. PLoS ONE 2014, 9, e112351. [Google Scholar] [CrossRef]
- Vargas, L.J.; Quintana, J.C.; Pereanez, J.A.; Nunez, V.; Sanz, L.; Calvete, J. Cloning and characterization of an antibacterial L-amino acid oxidase from Crotalus durissus cumanensis venom. Toxicon 2013, 64, 1–11. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Wray, K.P.; Margres, M.J. The genesis of an exceptionally lethal venom in the timber rattlesnake (Crotalus horridus) revealed through comparative venom-gland transcriptomics. BMC Genom. 2013, 14, 394. [Google Scholar] [CrossRef]
- Galán, J.A.; Sánchez, E.E.; Bashir, S.A.; Pérez, J.C. Characterization and identification of disintegrins in Crotalus horridus venom by liquid chromatography and tandem matrix-assisted laser desorption ionization—Quadrupole ion trap time-of-flight (MALDI-QIT-TOF) mass spectrometry. Can. J. Chem. 2005, 83, 1124–1131. [Google Scholar] [CrossRef]
- Borja, M.; Galan, J.A.; Cantu, E., Jr.; Zugasti-Cruz, A.; Rodriguez-Acosta, A.; Lazcano, D.; Lucena, S.; Suntravat, M.; Sanchez, Y.E.E. Morulustatin, A Disintegrin that Inhibits ADP-Induced Platelet Aggregation, Isolated from the Mexican Tamaulipan Rock Rattlesnake (Crotalus lepidus morulus). Rev. Cient. 2016, 26, 86–94. [Google Scholar]
- Martinez-Romero, G.; Rucavado, A.; Lazcano, D.; Gutierrez, J.M.; Borja, M.; Lomonte, B.; Garza-Garcia, Y.; Zugasti-Cruz, A. Comparison of venom composition and biological activities of the subspecies Crotalus lepidus lepidus, Crotalus lepidus klauberi and Crotalus lepidus morulus from Mexico. Toxicon 2013, 71, 84–95. [Google Scholar] [CrossRef]
- Borja, M.; Lazcano, D.; Martínez-Romero, G.; Morlett, J.; Sánchez, E.; Cepeda-Nieto, A.C.; Garza-García, Y.; Zugasti-Cruz, A. Intra-specific Variation in the Protein Composition and Proteolytic Activity of Venom of Crotalus lepidus morulus from the Northeast of Mexico. Copeia 2013, 2013, 707–716. [Google Scholar] [CrossRef]
- Holzer, M.; Mackessy, S.P. An aqueous endpoint assay of snake venom phospholipase A2. Toxicon 1996, 34, 1149–1155. [Google Scholar] [CrossRef]
- Tan, K.K.; Ler, S.G.; Gunaratne, J.; Bay, B.H.; Ponnampalam, G. In Vitro Cytotoxicity of L-amino Acid Oxidase From the Venom of Crotalus Mitchellii Pyrrhus. Toxicon Off. J. Int. Soc. Toxinol. 2017, 139. [Google Scholar] [CrossRef]
- Melendez-Martinez, D.; Macias-Rodriguez, E.; Vazquez-Briones, R.; Lopez-Vera, E.; Sandra Cruz-Perez, M.; Vargas-Caraveo, A.; Gatica-Colima, A.; Fernando Plenge-Tellechea, L. In vitro hemotoxic, alpha-neurotoxic and vasculotoxic effects of the Mexican black-tailed rattlesnake (Crotalus molossus nigrescens) venom. J. Venom Res. 2017, 8, 1–8. [Google Scholar]
- Rael, E.D.; Martinez, M.; Molina, O. Isolation of a fibrinolytic protease, M4, from venom of Crotalus molossus molossus (northern blacktail rattlesnake). Haemostasis 1992, 22, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Borja, M.; Neri-Castro, E.; Perez-Morales, R.; Strickland, J.L.; Ponce-Lopez, R.; Parkinson, C.L.; Espinosa-Fematt, J.; Saenz-Mata, J.; Flores-Martinez, E.; Alagon, A.; et al. Ontogenetic Change in the Venom of Mexican Black-Tailed Rattlesnakes (Crotalus molossus nigrescens). Toxins 2018, 10, 501. [Google Scholar] [CrossRef]
- Chen, T.; Rael, E.D. Purification of M5, a fibrinolytic proteinase from Crotalus molossus molossus venom that attacks complement. Int. J. Biochem. Cell Biol. 1997, 29, 789–799. [Google Scholar] [CrossRef]
- Sanchez, E.E.; Soliz, L.A.; Ramirez, M.S.; Perez, J.C. Partial characterization of a basic protein from Crotalus molossus molossus (northern blacktail rattlesnake) venom and production of a monoclonal antibody. Toxicon 2001, 39, 523–537. [Google Scholar] [CrossRef]
- Sanchez, E.E.; Gonzalez, R.; Lucena, S.; Garcia, S.; Finol, H.J.; Suntravat, M.; Giron, M.E.; Fernandez, I.; Rodriguez-Acosta, A. Crotamine-like from Southern Pacific rattlesnake (Crotalus oreganus helleri) Venom acts on human leukemia (K-562) cell lines and produces ultrastructural changes on mice adrenal gland. Ultrastruct. Pathol. 2018, 42, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Martins, W.; Baldasso, P.A.; Honorio, K.M.; Maltarollo, V.G.; Ribeiro, R.I.; Carvalho, B.M.; Soares, A.M.; Calderon, L.A.; Stabeli, R.G.; Caballol, M.A.; et al. A novel phospholipase A2 (D49) from the venom of the Crotalus oreganus abyssus (North American Grand canyon rattlesnake). BioMed Res. Int. 2014, 2014, 654170. [Google Scholar] [CrossRef]
- Da Silva, S.L.; Dias-Junior, C.A.; Baldasso, P.A.; Damico, D.C.S.; Carvalho, B.M.A.; Garanto, A.; Acosta, G.; Oliveira, E.; Albericio, F.; Soares, A.M.; et al. Vascular effects and electrolyte homeostasis of the natriuretic peptide isolated from Crotalus oreganus abyssus (North American Grand Canyon rattlesnake) venom. Peptides 2012, 36, 206–212. [Google Scholar] [CrossRef]
- Salazar, A.M.; Guerrero, B.; Cantu, B.; Cantu, E.; Rodríguez-Acosta, A.; Pérez, J.C.; Galán, J.A.; Tao, A.; Sánchez, E.E. Venom Variation in Hemostasis of the Southern Pacific Rattlesnake (Crotalus Oreganus Helleri): Isolation of Hellerase. Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP 2009, 149. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Undheim, E.A.; Scheib, H.; Gren, E.C.; Cochran, C.; Person, C.E.; Koludarov, I.; Kelln, W.; Hayes, W.K.; King, G.F.; et al. Intraspecific Venom Variation in the Medically Significant Southern Pacific Rattlesnake (Crotalus Oreganus Helleri): Biodiscovery, Clinical and Evolutionary Implications. J. Proteom. 2014, 99. [Google Scholar] [CrossRef]
- Almeida, J.R.; Mendes, B.; Lancellotti, M.; Marangoni, S.; Vale, N.; Passos, O.; Ramos, M.J.; Fernandes, P.A.; Gomes, P.; Da Silva, S.L. A novel synthetic peptide inspired on Lys49 phospholipase A2 from Crotalus oreganus abyssus snake venom active against multidrug-resistant clinical isolates. Eur. J. Med. Chem. 2018, 149, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Marquez, J.; Parra, V.; Suntravat, M.; Sanchez, E.E. Purification and characterization of cysteine rich-secretory proteins (CRiSPs) from the venom of the Southern Pacific rattlesnake (Crotalus oreganus helleri): Their role on blood and lymphatic endothelial cell permeability. Toxicon 2018, 150, 315–334. [Google Scholar] [CrossRef]
- Hamako, J.; Suzuki, Y.; Hayashi, N.; Kimura, M.; Ozeki, Y.; Hashimoto, K.; Matsui, T. Amino acid sequence and characterization of C-type lectin purified from the snake venom of Crotalus ruber. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2007, 146, 299–306. [Google Scholar] [CrossRef]
- Mori, N.; Sugihara, H. Comparative study of two arginine ester hydrolases, E-I and E-II from the venom of Crotalus ruber ruber (red rattlesnake). Comp. Biochem. Physiol. B 1989, 92, 537–547. [Google Scholar] [CrossRef]
- Straight, R.C.; Glenn, J.L.; Wolt, T.B.; Wolfe, M.C. North-south regional variation in phospholipase A activity in the venom of Crotalus ruber. Comp. Biochem. Physiol. B 1992, 103, 635–639. [Google Scholar] [CrossRef]
- Mori, N.; Nikai, T.; Sugihara, H.; Tu, A.T. Biochemical characterization of hemorrhagic toxins with fibrinogenase activity isolated from Crotalus ruber ruber venom. Arch. Biochem. Biophys 1987, 253, 108–121. [Google Scholar] [CrossRef]
- Mori, N.; Nikai, T.; Sugihara, H. Phosphodiesterase from the venom of Crotalus ruber ruber. Int. J. Biochem. 1987, 19, 115–119. [Google Scholar] [CrossRef]
- Mori, N.; Sugihara, H. Characterization of kallikrein-like enzyme from Crotalus ruber ruber (red rattlesnake) venom. Int. J. Biochem. 1989, 21, 83–90. [Google Scholar] [CrossRef]
- Sanchez, E.E.; Galan, J.A.; Powell, R.L.; Reyes, S.R.; Soto, J.G.; Russell, W.K.; Russell, D.H.; Perez, J.C. Disintegrin, hemorrhagic, and proteolytic activities of Mohave rattlesnake, Crotalus scutulatus scutulatus venoms lacking Mojave toxin. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2005, 141, 124–132. [Google Scholar] [CrossRef]
- Borja, M.; Castañeda, G.; Espinosa, J.; Neri, E.; Carbajal, A.; Clement, H.; García, O.; Alagon, A. Mojave Rattlesnake (Crotalus scutulatus scutulatus) with Type B Venom from Mexico. Copeia 2014, 2014, 7–13. [Google Scholar] [CrossRef]
- Massey, D.J.; Calvete, J.J.; Sanchez, E.E.; Sanz, L.; Richards, K.; Curtis, R.; Boesen, K. Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from Southern Arizona. J. Proteom. 2012, 75, 2576–2587. [Google Scholar] [CrossRef] [PubMed]
- Hinson, M.W.; Childs, C.; Johnson, B.D.; Sifford, D.H. Concanavalin A-Binding Enzymes of Crotalus scutulatus scutulatus Venom. J. Ark. Acad. Sci. 1985, 39, 50–54. [Google Scholar]
- Ho, C.L.; Lee, C.Y. Presynaptic actions of Mojave toxin isolated from Mojave rattlesnake (crotalus scutulatus) venom. Toxicon 1981, 19, 889–892. [Google Scholar] [CrossRef]
- Modahl, C.M.; Mackessy, S.P. Full-Length Venom Protein cDNA Sequences from Venom-Derived mRNA: Exploring Compositional Variation and Adaptive Multigene Evolution. PLoS Negl. Trop. Dis. 2016, 10, e0004587. [Google Scholar] [CrossRef]
- Durban, J.; Sanz, L.; Trevisan-Silva, D.; Neri-Castro, E.; Alagon, A.; Calvete, J.J. Integrated Venomics and Venom Gland Transcriptome Analysis of Juvenile and Adult Mexican Rattlesnakes Crotalus simus, C. tzabcan, and C. culminatus Revealed miRNA-modulated Ontogenetic Shifts. J. Proteome Res. 2017, 16, 3370–3390. [Google Scholar] [CrossRef]
- Nielsen, V.G.; Boyer, L.V.; Redford, D.T.; Ford, P. Thrombelastographic characterization of the thrombin-like activity of Crotalus simus and Bothrops asper venoms. Blood Coagul. Fibrinolysis 2017, 28, 211–217. [Google Scholar] [CrossRef]
- Weinstein, S.A.; Smith, L.A. Preliminary fractionation of tiger rattlesnake (Crotalus tigris) venom. Toxicon 1990, 28, 1447–1455. [Google Scholar] [CrossRef]
- Minton, S.A.; Weinstein, S.A. Protease activity and lethal toxicity of venoms from some little known rattlesnakes. Toxicon 1984, 22, 828–830. [Google Scholar] [CrossRef]
- Powell, R.L.; Lieb, C.S.; Rael, E.D. Identification of a neurotoxic venom component in the Tiger rattlesnake, Crotalus tigris. J. Herpetol. 2004, 38, 149–152. [Google Scholar] [CrossRef]
- Kaiser, I.I.; Aird, S.D. A crotoxin homolog from the venom of the Uracoan rattlesnake (Crotalus vegrandis). Toxicon 1987, 25, 1113–1120. [Google Scholar] [CrossRef]
- Scannone, H.R.; Rodriguez, O.G.; Lancini, A.R. Enzymatic activities and other characteristics of Crotalus vegrandis snake venom. In Toxins: Animals, Plants and Microbials; Rosenberg, P., Ed.; Pergamon Press: New York, NY, USA, 1978; pp. 223–229. [Google Scholar] [CrossRef]
- Moore, S.W.; Bhat, V.K.; Flatt, P.R.; Gault, V.A.; McClean, S. Isolation and characterisation of insulin-releasing compounds from Crotalus adamanteus, Crotalus vegrandis and Bitis nasicornis venom. Toxicon 2015, 101, 48–54. [Google Scholar] [CrossRef]
- Giron, M.; Pinto, A.; Finol, H.J.; Aguilar, I.; Rodriguez-Acosta, A. Kidney structural and ultrastructural pathological changes induced by uracoan rattlesnake (Crotalus vegrandis Klauber 1941) venom. J. Submicrosc. Cytol. Pathol. 2002, 34, 447–459. [Google Scholar] [PubMed]
- Aguilar, I.; Giron, M.E.; Rodriguez-Acosta, A. Purification and characterisation of a haemorrhagic fraction from the venom of the Uracoan rattlesnake Crotalus vegrandis. Biochim. Biophys. Acta 2001, 1548, 57–65. [Google Scholar] [CrossRef]
- Adade, C.; Fernandes Anne Cristine, S.; Carvalho Ana Lúcia, O.; Zingali, R.; Souto-Padrón, T. 44. Leishmanicidal Effects of a Phospholipase A2 Isolated from Crotalus viridis viridis Snake Venom. Toxicon 2012, 60, 117. [Google Scholar] [CrossRef]
- Adade, C.M.; Cons, B.L.; Melo, P.A.; Souto-Padron, T. Effect of Crotalus viridis viridis snake venom on the ultrastructure and intracellular survival of Trypanosoma cruzi. Parasitology 2011, 138, 46–58. [Google Scholar] [CrossRef]
- Zancolli, G.; Baker, T.G.; Barlow, A.; Bradley, R.K.; Calvete, J.J.; Carter, K.C.; de Jager, K.; Owens, J.B.; Price, J.F.; Sanz, L.; et al. Is Hybridization a Source of Adaptive Venom Variation in Rattlesnakes? A Test, Using a Crotalus scutulatus × viridis Hybrid Zone in Southwestern New Mexico. Toxins 2016, 8, 188. [Google Scholar] [CrossRef]
- Nget-Hong, T.; Ponnudurai, G. A Comparative Study of the Biological Activities of Rattlesnake (Genera Crotalus and Sistrurus) Venoms. Comp. Biochem. Physiol. C Comp. Pharmacol. Toxicol. 1991, 98. [Google Scholar] [CrossRef]
- Borgelt, C. Frequent item set mining. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2012, 2, 437–456. [Google Scholar] [CrossRef]
- Agrawal, R.; Imieliński, T.; Swami, A. Mining association rules between sets of items in large databases. In Proceedings of the 1993 ACM SIGMOD international conference on Management of data, Washington, DC, USA, 26–28 May 1993; pp. 207–216. [Google Scholar]
- Hahsler, M.; Chelluboina, S. Visualizing association rules: Introduction to the R-extension package arulesViz. R Proj. Modul. 2011, 223–238. [Google Scholar]
- Hornik, K.; Grün, B.; Hahsler, M. arules-A computational environment for mining association rules and frequent item sets. J. Stat. Softw. 2005, 14, 1–25. [Google Scholar]
- Pahari, S.; Mackessy, S.P.; Kini, R.M. The venom gland transcriptome of the Desert Massasauga rattlesnake (Sistrurus catenatus edwardsii): Towards an understanding of venom composition among advanced snakes (Superfamily Colubroidea). BMC Mol. Biol. 2007, 8, 115. [Google Scholar] [CrossRef] [PubMed]
- Doley, R.; Pahari, S.; Mackessy, S.P.; Kini, R.M. Accelerated exchange of exon segments in Viperid three-finger toxin genes (Sistrurus catenatus edwardsii; Desert Massasauga). BMC Evol. Biol. 2008, 8, 196. [Google Scholar] [CrossRef]
- Lisle Gibbs, H.; Chiucchi, J.E. Deconstructing a complex molecular phenotype: Population-level variation in individual venom proteins in Eastern Massasauga Rattlesnakes (Sistrurus c. catenatus). J. Mol. Evol. 2011, 72, 383–397. [Google Scholar] [CrossRef]
- Sunagar, K.; Jackson, T.N.; Undheim, E.A.; Ali, S.A.; Antunes, A.; Fry, B.G. Three-fingered RAVERs: Rapid Accumulation of Variations in Exposed Residues of snake venom toxins. Toxins 2013, 5, 2172–2208. [Google Scholar] [CrossRef]
- Chapeaurouge, A.; Reza, M.A.; Mackessy, S.P.; Carvalho, P.C.; Valente, R.H.; Teixeira-Ferreira, A.; Perales, J.; Lin, Q.; Kini, R.M. Interrogating the Venom of the Viperid Snake Sistrurus catenatus edwardsii by a Combined Approach of Electrospray and MALDI Mass Spectrometry. PLoS ONE 2015, 10, e0092091. [Google Scholar] [CrossRef]
- Gibbs, H.L.; Sanz, L.; Calvete, J.J. Snake population venomics: Proteomics-based analyses of individual variation reveals significant gene regulation effects on venom protein expression in Sistrurus rattlesnakes. J. Mol. Evol. 2009, 68, 113–125. [Google Scholar] [CrossRef]
- Juarez, P.; Sanz, L.; Calvete, J.J. Snake venomics: Characterization of protein families in Sistrurus barbouri venom by cysteine mapping, N-terminal sequencing, and tandem mass spectrometry analysis. Proteomics 2004, 4, 327–338. [Google Scholar] [CrossRef]
- Gibbs, H.L.; Sanz, L.; Chiucchi, J.E.; Farrell, T.M.; Calvete, J.J. Proteomic analysis of ontogenetic and diet-related changes in venom composition of juvenile and adult Dusky Pigmy rattlesnakes (Sistrurus miliarius barbouri). J. Proteom. 2011, 74, 2169–2179. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.; Gibbs, H.L.; Mackessy, S.P.; Calvete, J.J. Venom Proteomes of Closely Related Sistrurus Rattlesnakes with Divergent Diets. J. Proteome Res. 2006, 5, 2098–2112. [Google Scholar] [CrossRef]
- Carstairs, S.D.; Kreshak, A.A.; Tanen, D.A. Crotaline Fab antivenom reverses platelet dysfunction induced by Crotalus scutulatus venom: An in vitro study. Acad. Emerg. Med. 2013, 20, 522–525. [Google Scholar] [CrossRef]
- Strydom, D.J.; Botes, D.P. Snake Venom Toxins. I. Preliminary Studies on the Separation of Toxins of Elapidae Venoms. Toxicon Off. J. Int. Soc. Toxinol. 1970, 8. [Google Scholar] [CrossRef]
- Laustsen, A.H. Toxin synergism in snake venoms. Toxin Rev. 2016, 35, 165–170. [Google Scholar] [CrossRef]
- Lomeo, R.D.S.; Gonçalves, A.P.D.F.; Silva, C.N.D.; de Paula, A.T.; Costa Santos, D.O.; Fortes-Dias, C.; Gomes, D.A.; de Lima, M.E. Crotoxin from Crotalus durissus terrificus snake venom induces the release of glutamate from cerebrocortical synaptosomes via N and P/Q calcium channels. Toxicon 2014, 85, 5–16. [Google Scholar] [CrossRef]
- Faure, G.; Xu, H.; Saul, F.A. Crystal structure of crotoxin reveals key residues involved in the stability and toxicity of this potent heterodimeric β-neurotoxin. J. Mol. Biol. 2011, 412, 176–191. [Google Scholar] [CrossRef]
- Gay, C.C.; Leiva, L.C.; Maruñak, S.; Teibler, P.; De Pérez, O.A. Proteolytic, Edematogenic and Myotoxic Activities of a Hemorrhagic Metalloproteinase Isolated From Bothrops Alternatus Venom. Toxicon Off. J. Int. Soc. Toxinol. 2005, 46. [Google Scholar] [CrossRef]
- Denegri, M.E.G.; Acosta, O.C.; Huancahuire-Vega, S.; Martins-de-Souza, D.; Marangoni, S.; Maruñak, S.L.; Teibler, G.P.; Leiva, L.C.; Ponce-Soto, L.A. Isolation and Functional Characterization of a New Acidic PLA(2) Ba SpII RP4 of the Bothrops Alternatus Snake Venom From Argentina. Toxicon Off. J. Int. Soc. Toxinol. 2010, 56. [Google Scholar] [CrossRef]
- Bustillo, S.; Gay, C.C.; Denegri, M.E.G.; Ponce-Soto, L.A.; de Kier Joffé, E.B.; Acosta, O.; Leiva, L.C. Synergism Between Baltergin Metalloproteinase and Ba SPII RP4 PLA2 from Bothrops Alternatus Venom on Skeletal Muscle (C2C12) Cells. Toxicon Off. J. Int. Soc. Toxinol. 2012, 59. [Google Scholar] [CrossRef]
- Bustillo, S.; García-Denegri, M.E.; Gay, C.; Van de Velde, A.C.; Acosta, O.; Angulo, Y.; Lomonte, B.; Gutiérrez, J.M.; Leiva, L. Phospholipase A(2) Enhances the Endothelial Cell Detachment Effect of a Snake Venom Metalloproteinase in the Absence of Catalysis. Chem. Biol. Interact. 2015, 240. [Google Scholar] [CrossRef]
- Šribar, J.; Oberčkal, J.; Križaj, I. Understanding the Molecular Mechanism Underlying the Presynaptic Toxicity of Secreted Phospholipases A(2): An Update. Toxicon Off. J. Int. Soc. Toxinol. 2014, 89. [Google Scholar] [CrossRef]
- Higuchi, S.; Murayama, N.; Saguchi, K.; Ohi, H.; Fujita, Y.; Camargo, A.C.; Ogawa, T.; Deshimaru, M.; Ohno, M. Bradykinin-potentiating peptides and C-type natriuretic peptides from snake venom. Immunopharmacology 1999, 44, 129–135. [Google Scholar] [CrossRef]
- Murayama, N.; Hayashi, M.A.; Ohi, H.; Ferreira, L.A.; Hermann, V.V.; Saito, H.; Fujita, Y.; Higuchi, S.; Fernandes, B.L.; Yamane, T.; et al. Cloning and sequence analysis of a Bothrops jararaca cDNA encoding a precursor of seven bradykinin-potentiating peptides and a C-type natriuretic peptide. Proc. Natl. Acad. Sci. USA 1997, 94, 1189–1193. [Google Scholar] [CrossRef]
- Rebello Horta, C.C.; Chatzaki, M.; Rezende, B.A.; Magalhaes Bde, F.; Duarte, C.G.; Felicori, L.F.; Ribeiro Oliveira-Mendes, B.B.; do Carmo, A.O.; Chavez-Olortegui, C.; Kalapothakis, E. Cardiovascular-Active Venom Toxins: An Overview. Curr. Med. Chem. 2016, 23, 603–622. [Google Scholar] [CrossRef]
- Cushman, D.W.; Ondetti, M.A. History of the design of captopril and related inhibitors of angiotensin converting enzyme. Hypertension 1991, 17, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, W.; Ma, B.; Huang, K.; Song, M.; Zhang, N.; Zhang, Y.; Wang, Y.; Dai, Y.; Luo, Y. Isolation and Characterisation of a Kallikrein-Like Enzyme from Agkistrodon Halys Pallas Snake Venom. J. Sci. Food Agric. 2012, 92. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.A.; Camargo, A.C. The Bradykinin-potentiating Peptides from Venom Gland and Brain of Bothrops Jararaca Contain Highly Site Specific Inhibitors of the Somatic Angiotensin-Converting Enzyme. Toxicon Off. J. Int. Soc. Toxinol. 2005, 45. [Google Scholar] [CrossRef]
- Budnitskaya, P.; Gapanhuk, E.; Henriques, O.B. Comparative Action of Various Kininogenases on Crude Horse Plasma Substrates. Biochem. Pharmacol. 1970, 19. [Google Scholar] [CrossRef]
- Felicori, L.F.; Souza, C.T.; Velarde, D.T.; Magalhaes, A.; Almeida, A.P.; Figueiredo, S.; Richardson, M.; Diniz, C.R.; Sanchez, E.F. Kallikrein-like Proteinase from Bushmaster Snake Venom. Protein Expr. Purif. 2003, 30. [Google Scholar] [CrossRef]
- Soto, J.G.; White, S.A.; Reyes, S.R.; Regalado, R.; Sanchez, E.E.; Perez, J.C. Molecular evolution of PIII-SVMP and RGD disintegrin genes from the genus Crotalus. Gene 2007, 389, 66–72. [Google Scholar] [CrossRef]
- Calvete, J.J.; Moreno-Murciano, M.P.; Theakston, R.D.; Kisiel, D.G.; Marcinkiewicz, C. Snake venom disintegrins: Novel dimeric disintegrins and structural diversification by disulphide bond engineering. Biochem. J. 2003, 372, 725–734. [Google Scholar] [CrossRef]
- Markland, F.S.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Baramova, E.N.; Shannon, J.D.; Bjarnason, J.B.; Gonias, S.L.; Fox, J.W. Interaction of Hemorrhagic Metalloproteinases With Human Alpha 2-macroglobulin. Biochemistry 1990, 29. [Google Scholar] [CrossRef]
- Calvete, J.J.; Marcinkiewicz, C.; Monleon, D.; Esteve, V.; Celda, B.; Juarez, P.; Sanz, L. Snake venom disintegrins: Evolution of structure and function. Toxicon 2005, 45, 1063–1074. [Google Scholar] [CrossRef]
- Yee, K.T.; Pitts, M.; Tongyoo, P.; Rojnuckarin, P.; Wilkinson, M.C. Snake Venom Metalloproteinases and Their Peptide Inhibitors from Myanmar Russell’s Viper Venom. Toxins 2017, 9, 15. [Google Scholar] [CrossRef]
- Sunagar, K.; Fry, B.G.; Jackson, T.N.W.; Casewell, N.R.; Undheim, E.A.B.; Vidal, N.; Ali, S.A.; King, G.F.; Vasudevan, K.; Vasconcelos, V.; et al. Molecular Evolution of Vertebrate Neurotrophins: Co-Option of the Highly Conserved Nerve Growth Factor Gene into the Advanced Snake Venom Arsenalf. PLoS ONE 2013, 8, e81827. [Google Scholar] [CrossRef]
- Wijeyewickrema, L.C.; Gardiner, E.E.; Gladigau, E.L.; Berndt, M.C.; Andrews, R.K. Nerve growth factor inhibits metalloproteinase-disintegrins and blocks ectodomain shedding of platelet glycoprotein VI. J. Biol. Chem. 2010, 285, 11793–11799. [Google Scholar] [CrossRef]
- Frade, J.M.; Barde, Y.A. Genetic Evidence for Cell Death Mediated by Nerve Growth Factor and the Neurotrophin Receptor p75 in the Developing Mouse Retina and Spinal Cord. Development 1999, 126, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Frade, J.M.; Rodríguez-Tébar, A.; Barde, Y.A. Induction of Cell Death by Endogenous Nerve Growth Factor Through Its p75 Receptor. Nature 1996, 383. [Google Scholar] [CrossRef]
- Girish, K.S.; Jagadeesha, D.K.; Rajeev, K.B.; Kemparaju, K. Snake Venom Hyaluronidase: An Evidence for Isoforms and Extracellular Matrix Degradation. Mol. Cell. Biochem. 2002, 240. [Google Scholar] [CrossRef] [PubMed]
- Payne, V.; Kam, P.C.A. Mast Cell Tryptase: A Review of Its Physiology and Clinical Significance. Anaesthesia 2004, 59. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Velasco, J.; Meik, J.M.; Smith, E.N.; Castoe, T.A. Phylogenetic relationships of the enigmatic longtailed rattlesnakes (Crotalus ericsmithi, C. lannomi, and C. stejnegeri). Mol. Phylogenet Evol. 2013, 69, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Nunes Edos, S.; de Souza, M.A.; Vaz, A.F.; Santana, G.M.; Gomes, F.S.; Coelho, L.C.; Paiva, P.M.; da Silva, R.M.; Silva-Lucca, R.A.; Oliva, M.L.; et al. Purification of a lectin with antibacterial activity from Bothrops leucurus snake venom. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 159, 57–63. [Google Scholar] [CrossRef]
- Niewiarowski, S.; McLane, M.A.; Kloczewiak, M.; Stewart, G.J. Disintegrins and Other Naturally Occurring Antagonists of Platelet Fibrinogen Receptors. Semin. Hematol. 1994, 31, 289–300. [Google Scholar]
- Pinheiro, E.; Bogen, D.L.; Hoxha, D.; Ciolino, J.D.; Wisner, K.L. Sertraline and breastfeeding: Review and meta-analysis. Arch. Women’s Ment. Health 2015, 18. [Google Scholar] [CrossRef]




| Species | Venom Components | Reference |
|---|---|---|
| C. adamanteus | 5′NT, BPP, carboxypeptidase (E-Like), CNP, CRiSP, CTL, dipeptidase, Dis, EF-hand protein, EGF, GC, Hya, Kun, LAAO, MYO, NGF, PDE, PLA2, PLB, SVMP-P I/II/III, SVSP, VEGF, vespryn | [75,76,77,78,79,80,81,82,83,84,85,86,87] |
| C. aquilus | Hya, PLA2, SVMP P-III, SVSP (TLE) | [59,69] |
| C. atrox | BIPs, BPPs, CNP, CRiSP, Dis, Hya, LAAO, CTL, PLA2, SVMP P-I/III, SVSP, VEGF | [6,44,80,88,89,90,91,92,93,94,95] |
| C. basiliscus | BPP, CRISP, CTL, Dis, LAAO, PLA2 (CRTX, non-CRTX), SVMP P-I/II/III, SVMP inhibitor, SVSP | [96,97,98,99] |
| C. catalinesis | SVSP, SVMP P-III, PLA2 | [40] |
| C. cerastes | 3FTx, 5′NT, BPP, CRiSP, CTL, Dis, ficolin, Hya, Kun, LAAO, MYO, NGF, PDE, PLA2, SVMP P-II/III, SVSP, VEGF, vespryn, WAP | [98,100,101,102] |
| C. durissus | 3FTx, achase, aminopeptidase, angiogenin, BPP, carboxypeptidase, CNP, CRiSP, CTL, CysProt inhibitor, CysProt, dipeptidyl peptidase, Dis, FGF, fraction 5, Hya, Kazal, Kun, LAAO, lipase, MYO, NGF, PDGF, PLA2 (non-CRTX, CRTX), PLB, PLD, Serpin-like, SVMP inhibitor, SVMP P-III, SVSP, VEGF, vespryn, WAP | [45,64,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118] |
| C. enyo | SVSP, SVMP P-I/III, PLA2 | [40] |
| C. horridus | 5′-NT, BPP, CNP, CRiSP, Dis, EGF-like, GC, Hya, Kun, LAAO, MYO, neurotrophic factor, NGF, PDE, PLA2, SVMP P-I/III, SVSP, VEGF, vespryn | [29,119,120] |
| C. lepidus | 5′NT, CRiSP, CTL, Dis, LAAO, PDE, PLA2, SVMP-P-I/III, SVSP (TLE, kallikrein) | [49,121,122,123] |
| C. mitchelli | LAAO, SVSP, PLA2 (CRTX/MTX) | [40,124,125] |
| C. molossus | Dis, LAAO, MYO, PLA2, SVMP P-I/III, SVSP (TLE) | [58,98,126,127,128,129,130] |
| C. oreganus | ANP/BNP, BPP, CNP, CRiSP, CTL, Dis, Hya, Kun, LAAO, MYO, NGF, PLA2 (D49), PLA2, SVMP P-II/III, SVSP, VEGF, vespryn | [51,56,57,131,132,133,134,135,136,137] |
| C. polystictus | BIPs, CRiSPs, CTL, Dis, GC, Hya, LAAO, NGF, PDE, PLA2, PLB, SVMP P-I/II/III, SVSP (kallikrein, TLE), vespryn | [48,69] |
| C. ruber | CTL, Dis, LAAO, PDE, PLA2, SVMP P-I/III, SVSP (kallikrein) | [40,47,138,139,140,141,142,143] |
| C. scutulatus | 5′-NT, APase, BPPs, CRiSP, CTL, Dis, Hya, Kun, LAAO, MYO, NGF, PDE, PLA2 (MTX, non-CRTX), SVMP P-I/II/III, SVSP, VEGF, vespryn | [41,52,63,144,145,146,147,148] |
| C. simus | 3FTX, 5′-NT, BIPs, BPPs, CRiSP, CTL, Dis, GC, Hya, Kaz, Kun, LAAO, MYO, NGF, OHA, PDE, PLA2 (CRTX, non-CRTX), PLB, SVMP P-I/III, SVSP, VEGF, WAP | [7,54,68,149,150,151] |
| C. tigris | CRiSP, Dis, PLA2 (MTX), SVMP P-III, SVSP, VEGF | [55,152,153,154] |
| C. vegrandis | 5′-NT, ATPase, BIP, BPP, carboxypeptidase, CNP, CRiSP, CTL, Dis, endonuclease (DNAse, RNAse), exendin4-like protein, glutathione peroxidase, Hya, LAAO, MYO, NGF, PDE, PLA2 (CRTX), PLB, SVMP P-II/III, SVSP | [77,155,156,157,158,159] |
| C. viridis | 5′-NT, APase, BPP, CRiSP, CTL, Dis, GC, LAAO, MYO, OHA, PDE, PLA2 (CRTX, non-CRTX), PLB, SVMP inhibitor, SVMP P-I/II/III, SVSP (TLE, kallikrein) | [42,60,61,62,160,161,162,163] |
| C. willardi | CRiSP, CTL, Dis, LAAO, PDE, PLA2, SVMP P-I/III, SVSP (TLE, kallikrein) | [49,153] |
| C. tortugenesis | N/A | |
| C. stejnegeri | N/A | |
| C. tancitarensis | N/A | |
| C. lannomi | N/A | |
| C. pusillus | N/A | |
| C. transversus | N/A | |
| C. triseriatus | N/A | |
| C. unicolor | N/A | |
| C. intermedius | N/A |
| Rule No. | Protein (Predictor) | Protein (Predicted) | Support | Confidence | Lift |
|---|---|---|---|---|---|
| 1 | CRiSP, LAAO, SVSP | CTL | 0.61 | 1 | 1.5 |
| 2 | Dis, LAAO, SVSP | CTL | 0.67 | 0.93 | 1.4 |
| 3 | LAAO, SVMP P-III, SVSP | CTL | 0.67 | 0.93 | 1.4 |
| 4 | BPP | CRiSP | 0.52 | 1 | 1.4 |
| 5 | CRiSP, LAAO | CTL | 0.61 | 0.92 | 1.39 |
| 6 | CRiSP, SVSP | CTL | 0.61 | 0.92 | 1.39 |
| 7 | CTL | CRiSP | 0.61 | 0.92 | 1.3 |
| 8 | PDE | LAAO | 0.52 | 1 | 1.23 |
| 9 | PDE | Dis | 0.52 | 1 | 1.23 |
| 10 | BPP | LAAO | 0.52 | 1 | 1.23 |
| 11 | BPP | Dis | 0.52 | 1 | 1.23 |
| 12 | CTL | LAAO | 0.67 | 1 | 1.23 |
| 13 | CTL | Dis | 0.66 | 1 | 1.23 |
| 14 | CRiSP | Dis | 0.71 | 1 | 1.23 |
| 15 | LAAO, SVMP P-I | Dis | 0.57 | 1 | 1.23 |
| 16 | Dis, SVMP P-I | LAAO | 0.57 | 1 | 1.23 |
| 17 | LAAO, SVMP P-III | Dis | 0.76 | 1 | 1.23 |
| 18 | LAAO | Dis | 0.76 | 0.94 | 1.16 |
| 19 | Dis | LAAO | 0.76 | 0.94 | 1.16 |
| 20 | CRiSP | LAAO | 0.67 | 0.93 | 1.15 |
| Rules No. | Protein (Predictor) | Protein (Predicted) | Support | Confidence | Lift |
|---|---|---|---|---|---|
| 1 | SVMP_PI | LAAO | 0.6 | 1 | 1.5 |
| 2 | BPP, CRiSP | LAAO | 0.6 | 1 | 1.5 |
| 3 | CRiSP, CTL | LAAO | 0.67 | 1 | 1.5 |
| 5 | CRiSP, CTL | SVMP_PI | 0.6 | 0.9 | 1.5 |
| 6 | CTL | BPP | 0.67 | 0.9 | 1.36 |
| 7 | CTL | LAAO | 0.67 | 0.9 | 1.36 |
| 8 | CRiSP | LAAO | 0.67 | 0.9 | 1.36 |
| 9 | SVMP_PI | CTL | 0.6 | 1 | 1.36 |
| 10 | SVMP_PI | CRiSP | 0.6 | 1 | 1.36 |
| 14 | SVMP_PII, SVMP_PIII | CTL | 0.53 | 1 | 1.36 |
| 15 | CTL, SVMP_PII | SVMP_PIII | 0.53 | 1 | 1.36 |
| 16 | SVMP_PII, SVSP | SVMP_PIII | 0.53 | 1 | 1.36 |
| 17 | SVMP_PII, SVSP | CTL | 0.53 | 1 | 1.36 |
| 18 | BPP | LAAO | 0.6 | 0.9 | 1.35 |
| 20 | PLA2(Other), SVSP | SVMP_PIII | 0.73 | 0.91 | 1.25 |
| 21 | PLA2(Other), SVSP | CTL | 0.73 | 0.91 | 1.25 |
| 22 | PLA2(Other), SVSP | CRiSP | 0.73 | 0.91 | 1.25 |
| 23 | SVMP_PIII | CTL | 0.67 | 0.9 | 1.23 |
| 25 | SVMP_PIII | CRiSP | 0.67 | 0.9 | 1.23 |
| 26 | CRiSP | SVMP_PIII | 0.67 | 0.9 | 1.24 |
| Species | Venom Components | Reference |
|---|---|---|
| S. catenatus | 3FTx, 5′-NT, BPP, CNP, CRiSP, CTL, Dis, GC, LAAO, MYO, NGF, PDE, PLA2 (CRTX, non-CRTX), PLB, Renin-like Aspartic Protease, SVMP P-I/II/III, SVSP, VEGF | [29,70,168,169,170,171,172,173] |
| S. miliarius miliarius | BPP, CRiSP, CTL, Dis, NGF, PLA2, SVMP P-I/III, SVMP-inhibitor, SVSP | [71] |
| S. miliarius streckeri | BPP, CRiSP, CTL, Dis, NGF, PLA2, SVMP P-I/III, SVMP-inhibitor, SVSP | [71] |
| S. miliarius barbouri | BPP, CNP, CRiSP, Dis, PLA2, SVMP P-I/III, SVSP | [29,30,70,71,173,174,175,176] |
| Rules No. | Protein (Predictor) | Protein (Predicted) | Support | Confidence | Lift |
|---|---|---|---|---|---|
| 1 | SVSP | SVMP inhibitor | 0.5 | 1 | 2 |
| 2 | SVMP inhibitor | SVSP | 0.5 | 1 | 2 |
| 3 | SVSP | CTL | 0.5 | 1 | 1.33 |
| 4 | SVSP | NGF | 0.5 | 1 | 1.33 |
| 5 | SVMP inhibitor | CTL | 0.5 | 1 | 1.33 |
| 6 | SVMP inhibitor | NGF | 0.5 | 1 | 1.33 |
| 7 | CTL | NGF | 0.75 | 1 | 1.33 |
| 8 | NGF | CTL | 0.75 | 1 | 1.33 |
| Rule No. | Protein Predictor | Protein (Predicted) | Support | Confidence | Lift |
|---|---|---|---|---|---|
| 1 | SVMP_PIII | SVMP_PI | 0.5 | 1 | 2 |
| 3 | SVMP_PIII | Vasoactive peptide | 0.5 | 1 | 2 |
| 5 | SVMP_PI | Vasoactive peptide | 0.5 | 1 | 2 |
| 7 | PLA2, CRTX | LAAO | 0.5 | 1 | 1.5 |
| 8 | SVMP_PIII | BPP | 0.5 | 1 | 1.5 |
| 9 | SVMP_PI | BPP | 0.5 | 1 | 1.5 |
| 10 | Vasoactive peptide | BPP | 0.5 | 1 | 1.5 |
| 11 | CNP | BPP | 0.5 | 1 | 1.5 |
| 12 | PLA2, CRTX | CTL | 0.5 | 1 | 1.2 |
| 13 | PLA2, CRTX | NGF | 0.5 | 1 | 1.2 |
| 14 | LAAO | CTL | 0.67 | 1 | 1.2 |
| 15 | LAAO | NGF | 0.67 | 1 | 1.2 |
| 16 | CTL | NGF | 0.83 | 1 | 1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deshwal, A.; Phan, P.; Datta, J.; Kannan, R.; Thallapuranam, S.K. A Meta-Analysis of the Protein Components in Rattlesnake Venom. Toxins 2021, 13, 372. https://doi.org/10.3390/toxins13060372
Deshwal A, Phan P, Datta J, Kannan R, Thallapuranam SK. A Meta-Analysis of the Protein Components in Rattlesnake Venom. Toxins. 2021; 13(6):372. https://doi.org/10.3390/toxins13060372
Chicago/Turabian StyleDeshwal, Anant, Phuc Phan, Jyotishka Datta, Ragupathy Kannan, and Suresh Kumar Thallapuranam. 2021. "A Meta-Analysis of the Protein Components in Rattlesnake Venom" Toxins 13, no. 6: 372. https://doi.org/10.3390/toxins13060372
APA StyleDeshwal, A., Phan, P., Datta, J., Kannan, R., & Thallapuranam, S. K. (2021). A Meta-Analysis of the Protein Components in Rattlesnake Venom. Toxins, 13(6), 372. https://doi.org/10.3390/toxins13060372






