Monitoring Cyanobacterial Blooms during the COVID-19 Pandemic in Campania, Italy: The Case of Lake Avernus
Abstract
:1. Introduction
2. Results
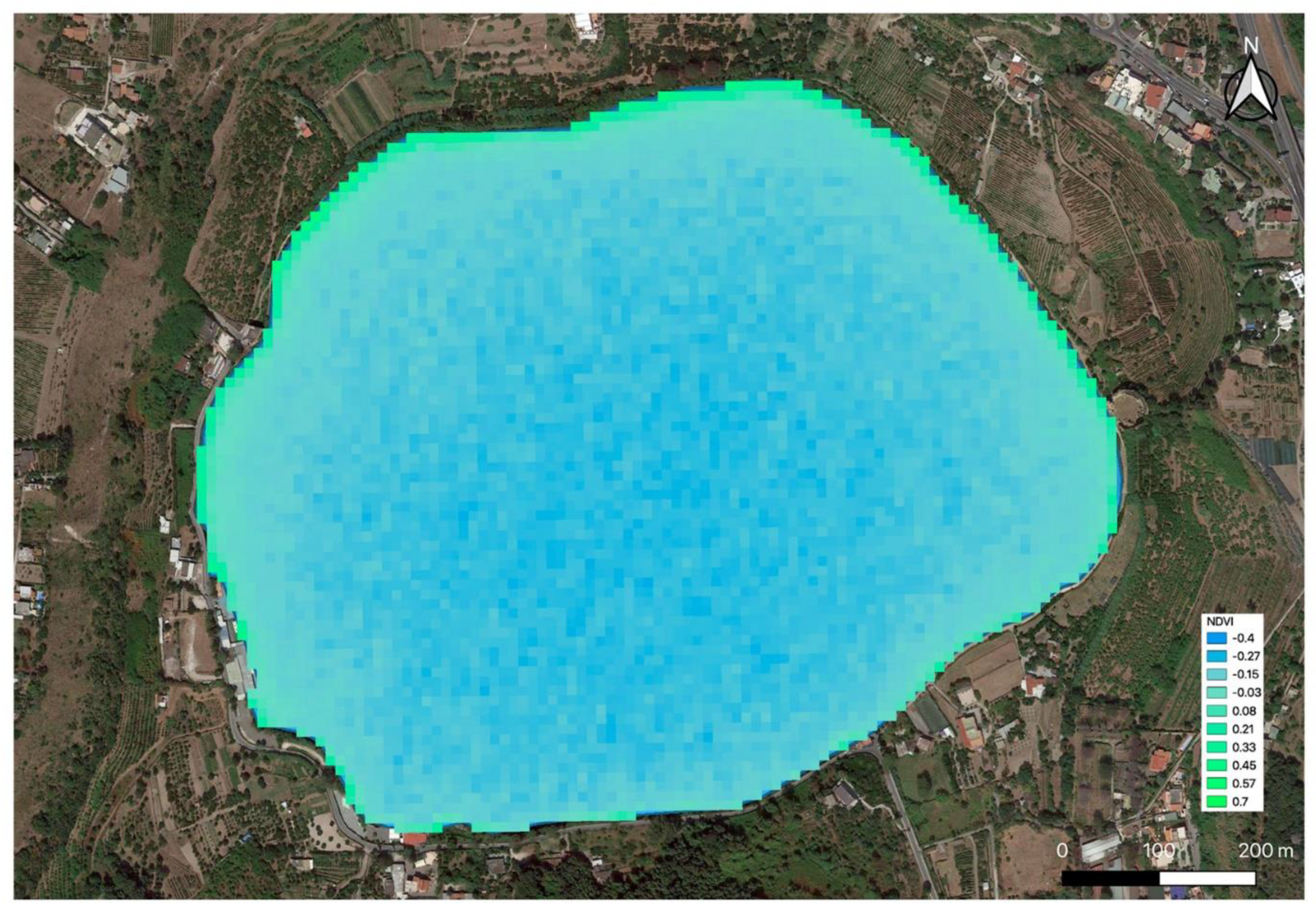
2.1. Remote and Proximal Sensing Analysis
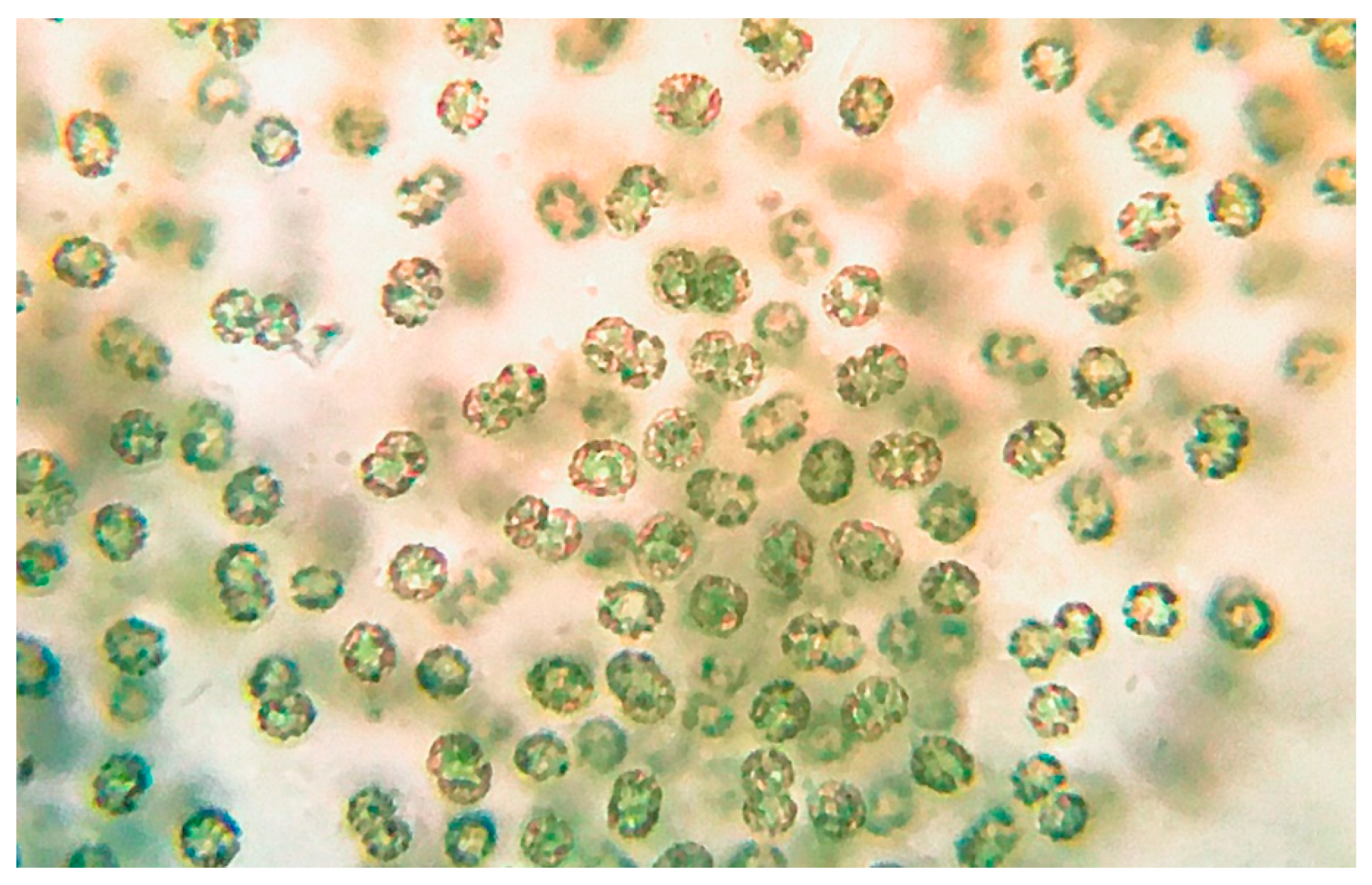
2.2. Sampling and Morphological Identification
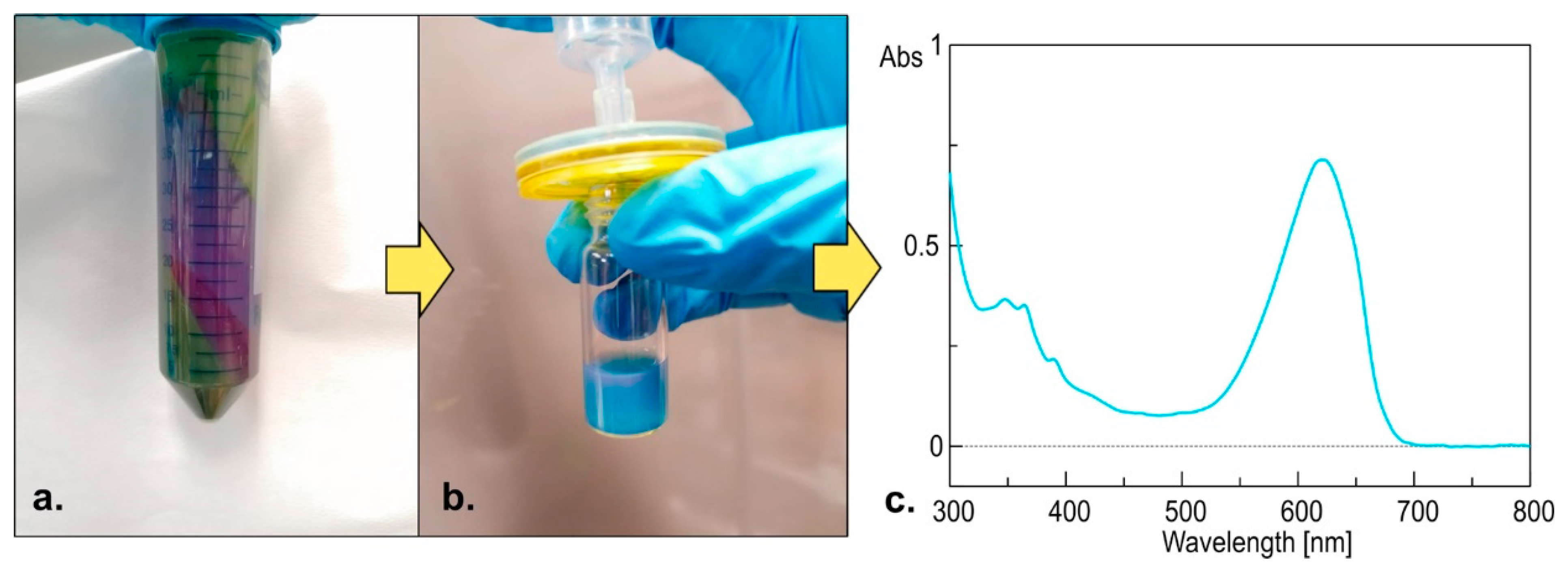
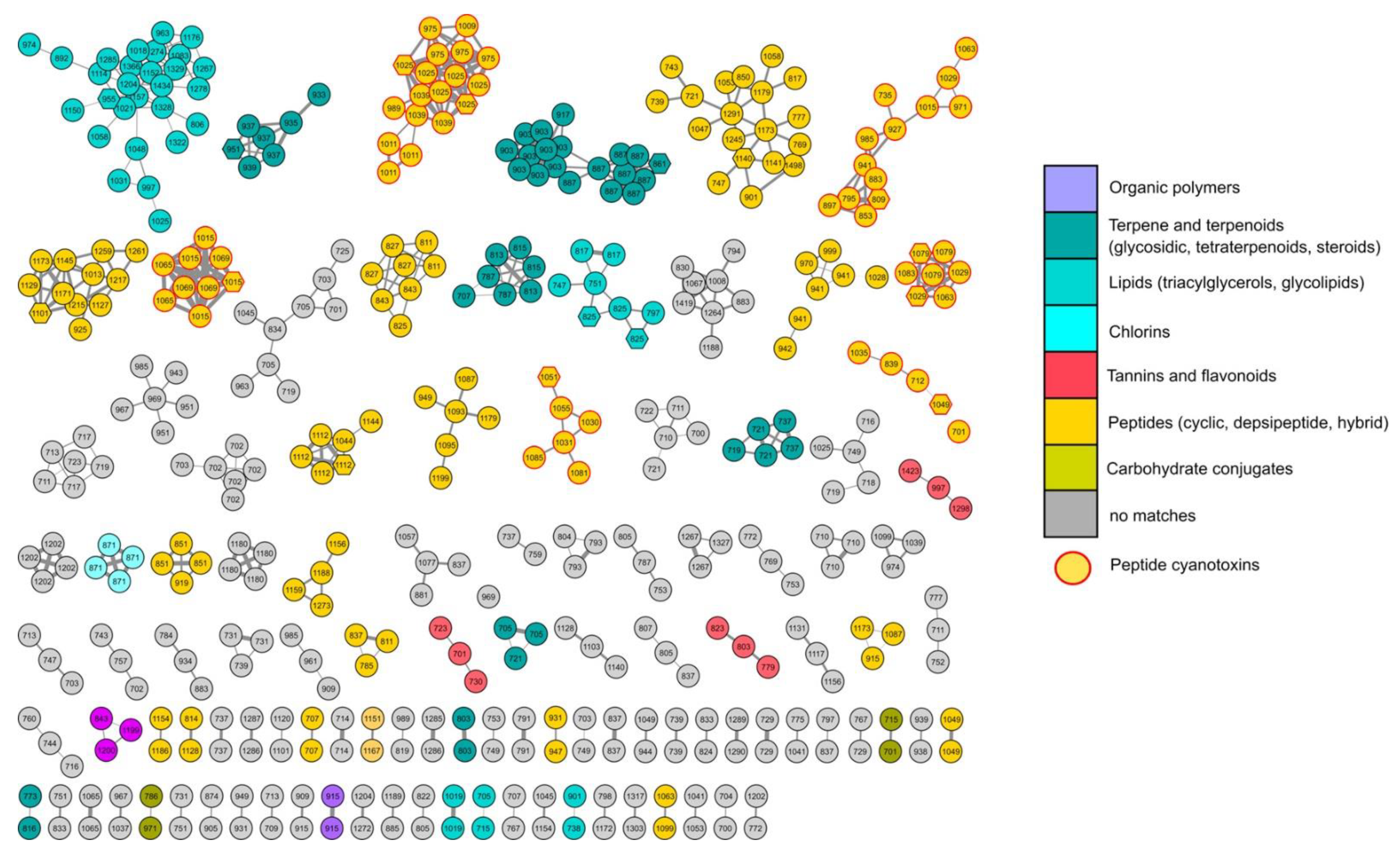
2.3. Chemical Analysis of the Bloom
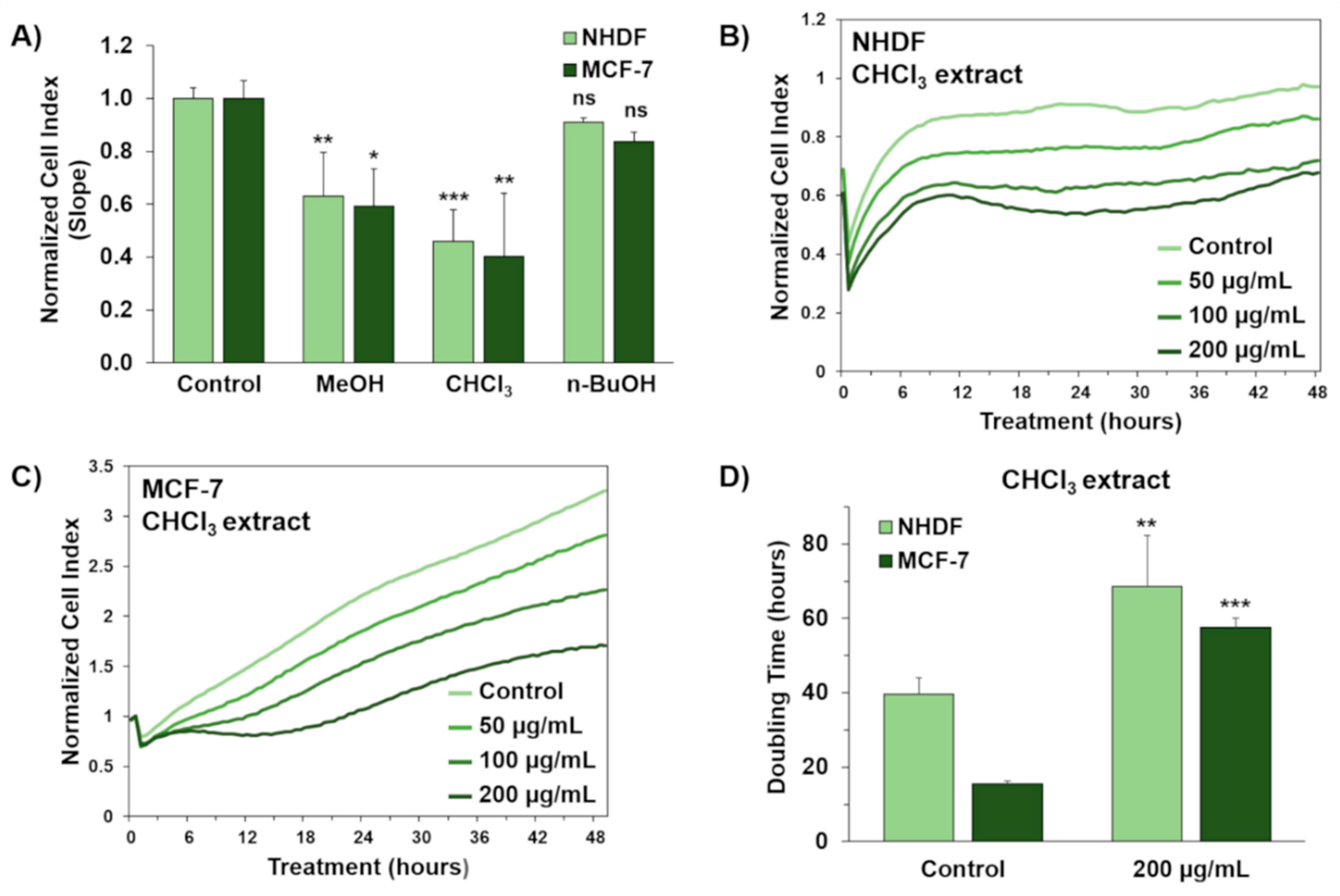
2.4. In Vitro Cytotoxicity of Organic Extracts from the Cyanobacterial Bloom in Lake Avernus
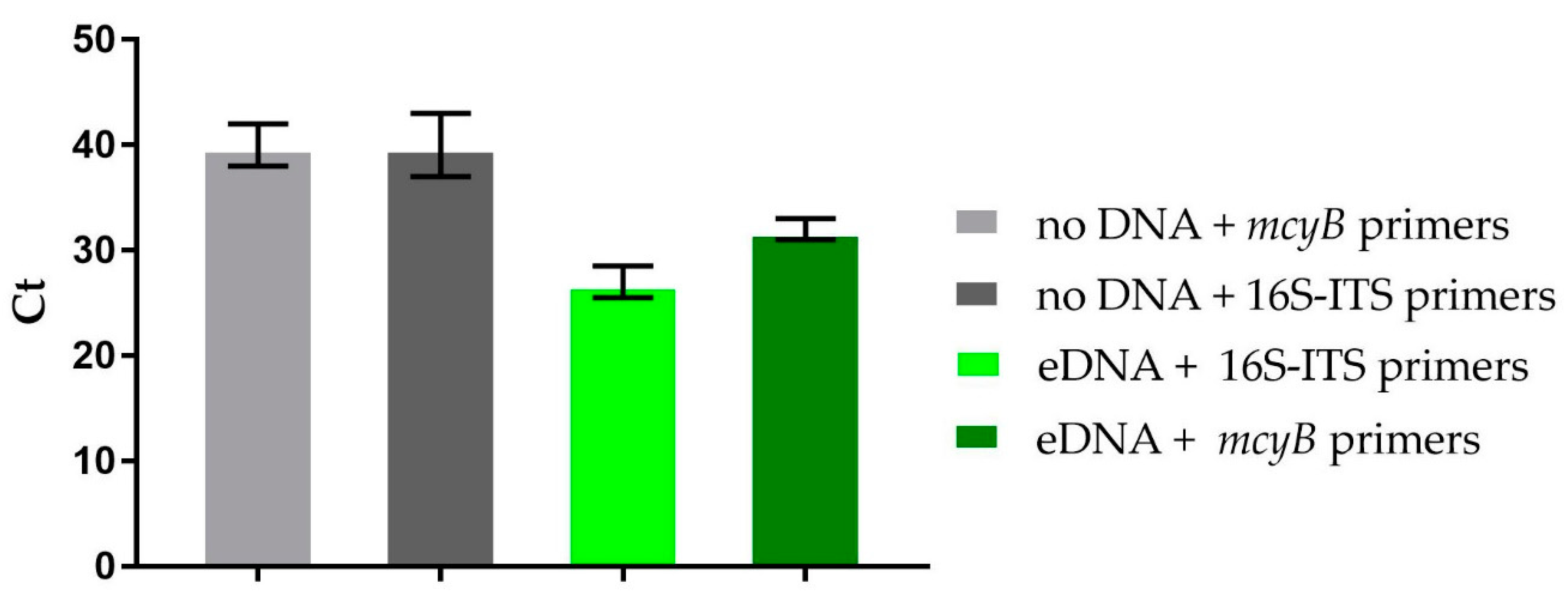
2.5. Metagenomic Detection of the mcy Gene
3. Discussion
4. Materials and Methods
4.1. Remote and Proximal Sensing Analysis
4.2. Sampling, Extraction, LC-MS, and Molecular Networking Analysis
4.3. Cell Viability Assays
4.4. RT-PCR Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- AllTrails. Available online: https://www.alltrails.com/trail/italy/campania/lake-avernus-lago-di-averno (accessed on 25 May 2021).
- Teta, R.; Esposito, G.; De Sterlich, C.; Lega, M.; Costantino, V. Early Detection of Cyanobacterial Blooms and Associated Cyanotoxins using Fast Detection Strategy. J. Vis. Exp. 2021, e61889. [Google Scholar] [CrossRef]
- Esposito, G.; Teta, R.; Marrone, R.; De Sterlich, C.; Casazza, M.; Anastasio, A.; Lega, M.; Costantino, V. A Fast Detection Strategy for Cyanobacterial blooms and associated cyanotoxins (FDSCC) reveals the occurrence of lyngbyatoxin A in campania (South Italy). Chemosphere 2019, 225, 342–351. [Google Scholar] [CrossRef]
- Teta, R.; Romano, V.; Della Sala, G.; Picchio, S.; De Sterlich, C.; Mangoni, A.; Di Tullio, G.; Costantino, V.; Lega, M. Cyanobacteria as indicators of water quality in Campania coasts, Italy: A monitoring strategy combining remote/proximal sensing and in situ data. Environ. Res. Lett. 2017, 12, 024001. [Google Scholar] [CrossRef]
- Teta, R.; Della Sala, G.; Glukhov, E.; Gerwick, L.; Gerwick, W.H.; Mangoni, A.; Costantino, V. Combined LC–MS/MS and Molecular Networking Approach Reveals New Cyanotoxins from the 2014 Cyanobacterial Bloom in Green Lake, Seattle. Environ. Sci. Technol. 2015, 49, 14301–14310. [Google Scholar] [CrossRef] [PubMed]
- Massey, I.Y.; Yang, F. A Mini Review on Microcystins and Bacterial Degradation. Toxins 2020, 12, 268. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Yi, X.; Liu, W.; Zhang, C.; Massey, I.; Yang, F.; Tian, L. A Review of Nephrotoxicity of Microcystins. Toxins 2020, 12, 693. [Google Scholar] [CrossRef]
- Ferranti, P.; Fabbrocino, S.; Cerulo, M.G.; Bruno, M.; Serpe, L.; Gallo, P. Characterisation of biotoxins produced by a cyanobacteria bloom in Lake Averno using two LC–MS-based techniques. Food Addit. Contam. Part A 2008, 25, 1530–1537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Fiore, V.; Cavuoto, G.; Punzo, M.; Tarallo, D.; Casazza, M.; Guarriello, S.M.; Lega, M. Integrated hierarchical geo-environmental survey strategy applied to the detection and investigation of an illegal landfill: A case study in the Campania Region (Southern Italy). Forensic Sci. Int. 2017, 279, 96–105. [Google Scholar] [CrossRef]
- European Environment Agency. Available online: http://www.eea.europa.eu/ (accessed on 16 December 2011).
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Global Natural Product Social. Available online: https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp (accessed on 20 May 2021).
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.-F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ernst, M.; Bin Kang, K.; Caraballo-Rodríguez, A.M.; Nothias, L.-F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Puddick, J. Spectroscopic Investigations of Oligopeptides from Aquatic Cyanobacteria Characterisation of New Oligopeptides, Development of Microcystin Quantification Tools and Investigations into Mi. Ph.D. Thesis, The University of Waikato, Hamilton, New Zealand, 2013. [Google Scholar]
- Ishida, K.; Matsuda, H.; Murakami, M. Micropeptins 88-A to 88-F, chymotrypsin inhibitors from the cyanobacterium Microcystis aeruginosa (NIES-88). Tetrahedron 1998, 54, 5545–5556. [Google Scholar] [CrossRef]
- Kodani, S.; Ishida, K.; Suzuki, S.; Murakami, M. Five new cyanobacterial peptides from water bloom materials of lake Teganuma (Japan). FEMS Microbiol. Lett. 1999, 178, 343–348. [Google Scholar] [CrossRef]
- Namikoshi, M.; Rinehart, K. Bioactive compounds produced by cyanobacteria. J. Ind. Microbiol. Biotechnol. 1996, 17, 373–384. [Google Scholar] [CrossRef]
- Harada, K.-I.; Mayumi, T.; Shimada, T.; Suzuki, M.; Kondo, F.; Watanabe, M.F. Occurrence of four depsipeptides, aeruginopeptins, together with microcystins from toxic cyanobacteria. Tetrahedron Lett. 1993, 34, 6091–6094. [Google Scholar] [CrossRef]
- Teta, R.; Della Sala, G.; Mangoni, A.; Lega, M.; Costantino, V. Tracing cyanobacterial blooms to assess the impact of wastewaters discharges on coastal areas and lakes. Int. J. Sustain. Dev. Plan. 2016, 11, 804–811. [Google Scholar] [CrossRef] [Green Version]
- Pineda-Mendoza, R.M.; Zúñiga, G.; Martínez-Jerónimo, F. Microcystin production in Microcystis aeruginosa: Effect of type of strain, environmental factors, nutrient concentrations, and N:P ratio on mcyA gene expression. Aquat. Ecol. 2015, 50, 103–119. [Google Scholar] [CrossRef]
- Gargiulo, F.; Persechino, G.; Lega, M.; Errico, A. IDES Project: A New Effective Tool for Safety and Security in the Environment. In Transactions on Petri Nets and Other Models of Concurrency XV; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2013; Volume 8286, pp. 201–208. [Google Scholar]
- Lillesand, T.; Kiefer, R.W.; Chipman, J. Remote Sensing and Image Interpretation, 6th ed.; John Wiley and Sons: Hoboken, NJ, USA, 2018. [Google Scholar]
- Vermote, E.; Justice, C.; Claverie, M.; Franch, B. Preliminary analysis of the performance of the Landsat 8/OLI land surface reflectance product. Remote Sens. Environ. 2016, 185, 46–56. [Google Scholar] [CrossRef]
- Hu, C.; Lee, Z.; Franz, B. Chlorophyll a algorithms for oligotrophic oceans: A novel approach based on three-band reflectance difference. J. Geophys. Res. Space Phys. 2012, 117, 1011. [Google Scholar] [CrossRef] [Green Version]
- Casazza, M.; Lega, M.; Liu, G.; Ulgiati, S.; Endreny, T.A. Aerosol pollution, including eroded soils, intensifies cloud growth, precipitation, and soil erosion: A review. J. Clean. Prod. 2018, 189, 135–144. [Google Scholar] [CrossRef]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Myers, O.D.; Sumner, S.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Teta, R.; Della Sala, G.; Esposito, G.; Via, C.W.; Mazzoccoli, C.; Piccoli, C.; Bertin, M.J.; Costantino, V.; Mangoni, A. A joint molecular networking study of a Smenospongia sponge and a cyanobacterial bloom revealed new antiproliferative chlorinated polyketides. Org. Chem. Front. 2019, 6, 1762–1774. [Google Scholar] [CrossRef]




| tR (min) | m/z | Molecular Formula [M+H]+ | GNPS’ Annotation 1 |
|---|---|---|---|
| 25.3 | 1015.5304 | C51H71O12N10+ | [d-Asp3] MC-FR |
| 25.3 | 1025.5350 | C54H73O12N8+ | MC-LW |
| 25.7 | 1029.5084 | C48H73O13N10S+ | MC-M(O)R |
| 23.8 | 1051.5120 | C55H71O13N8+ | MP-T1 |
| 25.7 | 1079.5288 | C53H75O16N8+ | MP-88B |
| 33.9 | 809.4596 | C41H61O9N8+ | AnaP-C |
| 25.6 | 1049.5165 | C52H73O15N8+ | AerP-228B |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teta, R.; Sala, G.D.; Esposito, G.; Stornaiuolo, M.; Scarpato, S.; Casazza, M.; Anastasio, A.; Lega, M.; Costantino, V. Monitoring Cyanobacterial Blooms during the COVID-19 Pandemic in Campania, Italy: The Case of Lake Avernus. Toxins 2021, 13, 471. https://doi.org/10.3390/toxins13070471
Teta R, Sala GD, Esposito G, Stornaiuolo M, Scarpato S, Casazza M, Anastasio A, Lega M, Costantino V. Monitoring Cyanobacterial Blooms during the COVID-19 Pandemic in Campania, Italy: The Case of Lake Avernus. Toxins. 2021; 13(7):471. https://doi.org/10.3390/toxins13070471
Chicago/Turabian StyleTeta, Roberta, Gerardo Della Sala, Germana Esposito, Mariano Stornaiuolo, Silvia Scarpato, Marco Casazza, Aniello Anastasio, Massimiliano Lega, and Valeria Costantino. 2021. "Monitoring Cyanobacterial Blooms during the COVID-19 Pandemic in Campania, Italy: The Case of Lake Avernus" Toxins 13, no. 7: 471. https://doi.org/10.3390/toxins13070471
APA StyleTeta, R., Sala, G. D., Esposito, G., Stornaiuolo, M., Scarpato, S., Casazza, M., Anastasio, A., Lega, M., & Costantino, V. (2021). Monitoring Cyanobacterial Blooms during the COVID-19 Pandemic in Campania, Italy: The Case of Lake Avernus. Toxins, 13(7), 471. https://doi.org/10.3390/toxins13070471










