The Natterin Proteins Diversity: A Review on Phylogeny, Structure, and Immune Function
Abstract
:1. Introduction
2. Results and Discussion
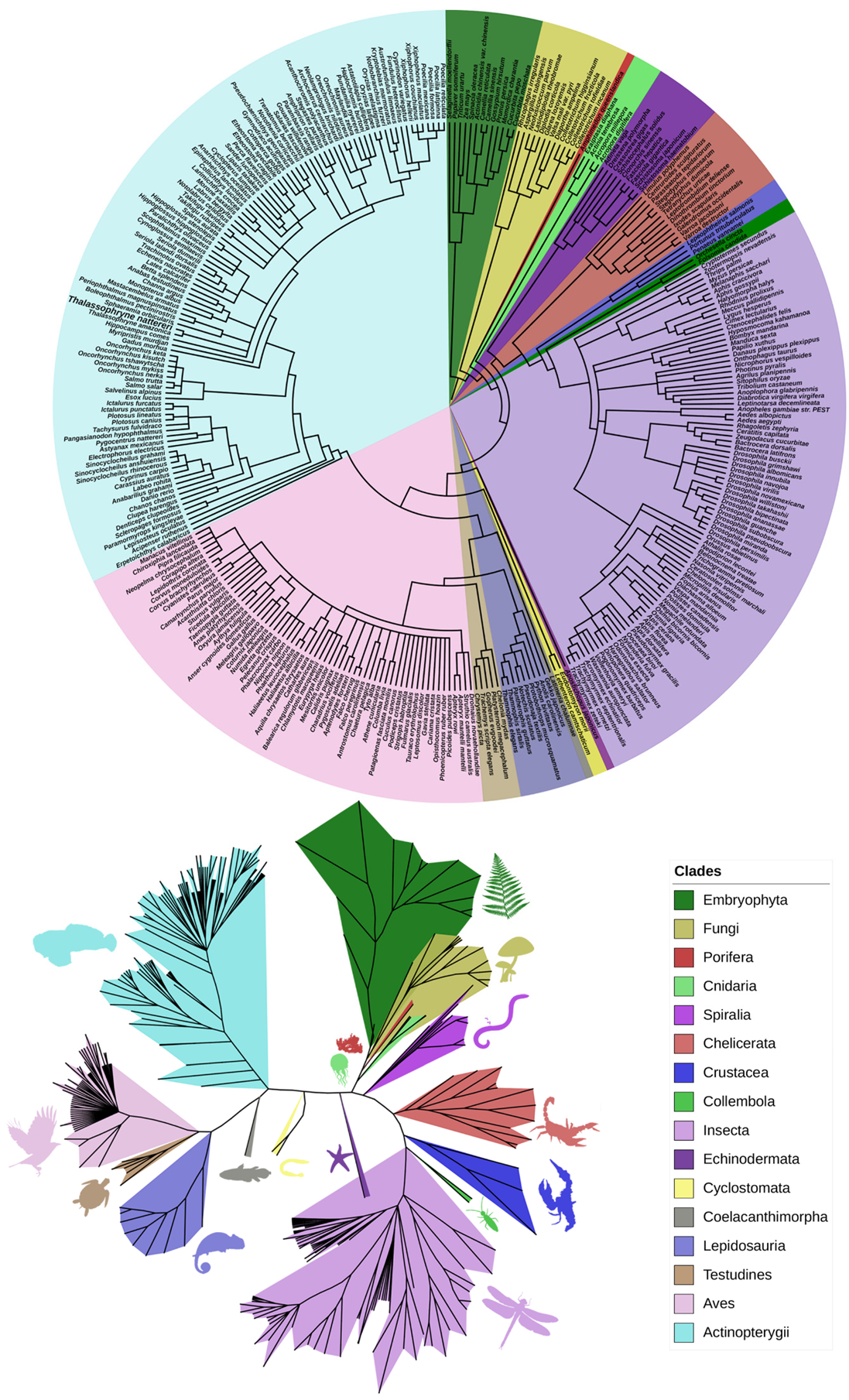
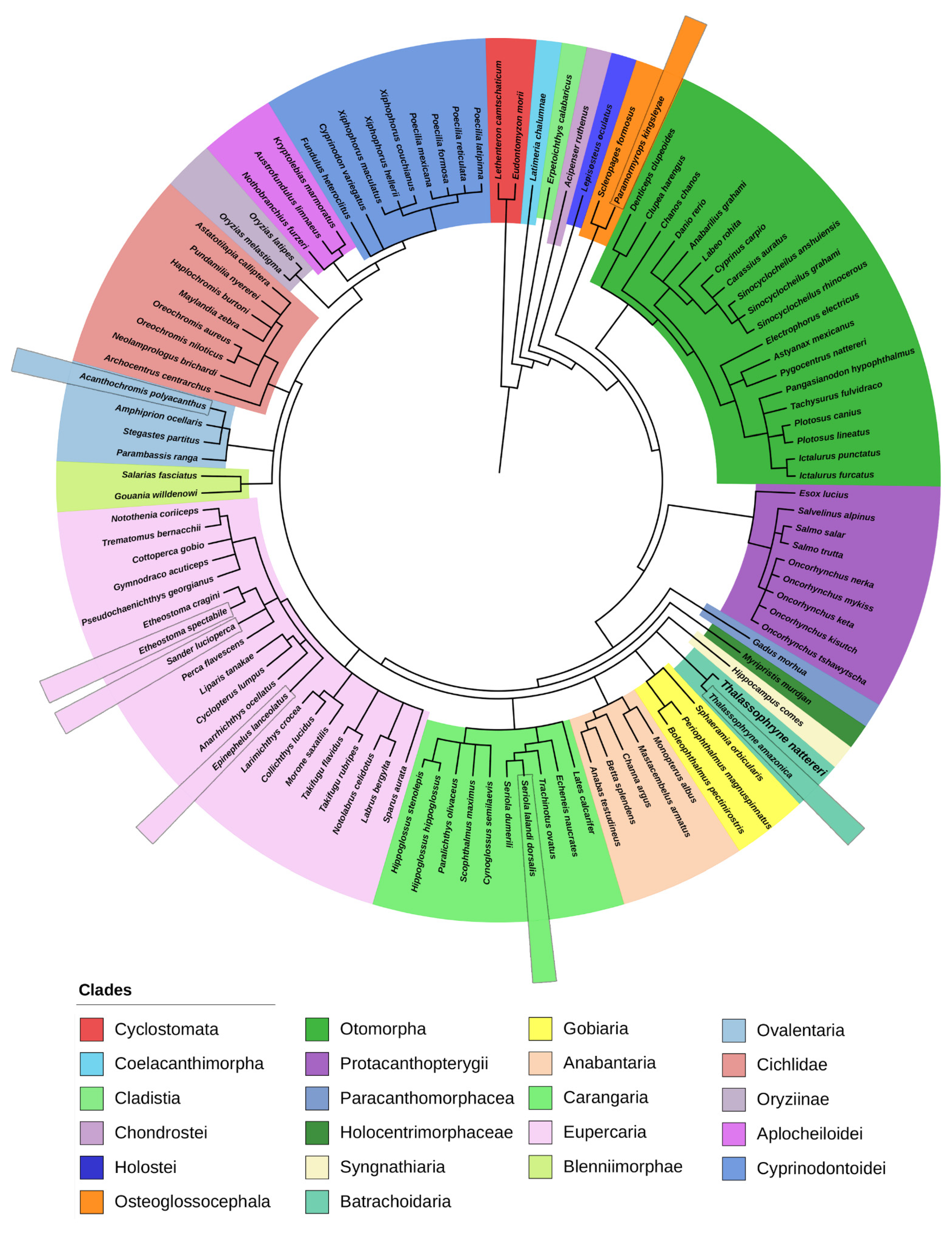
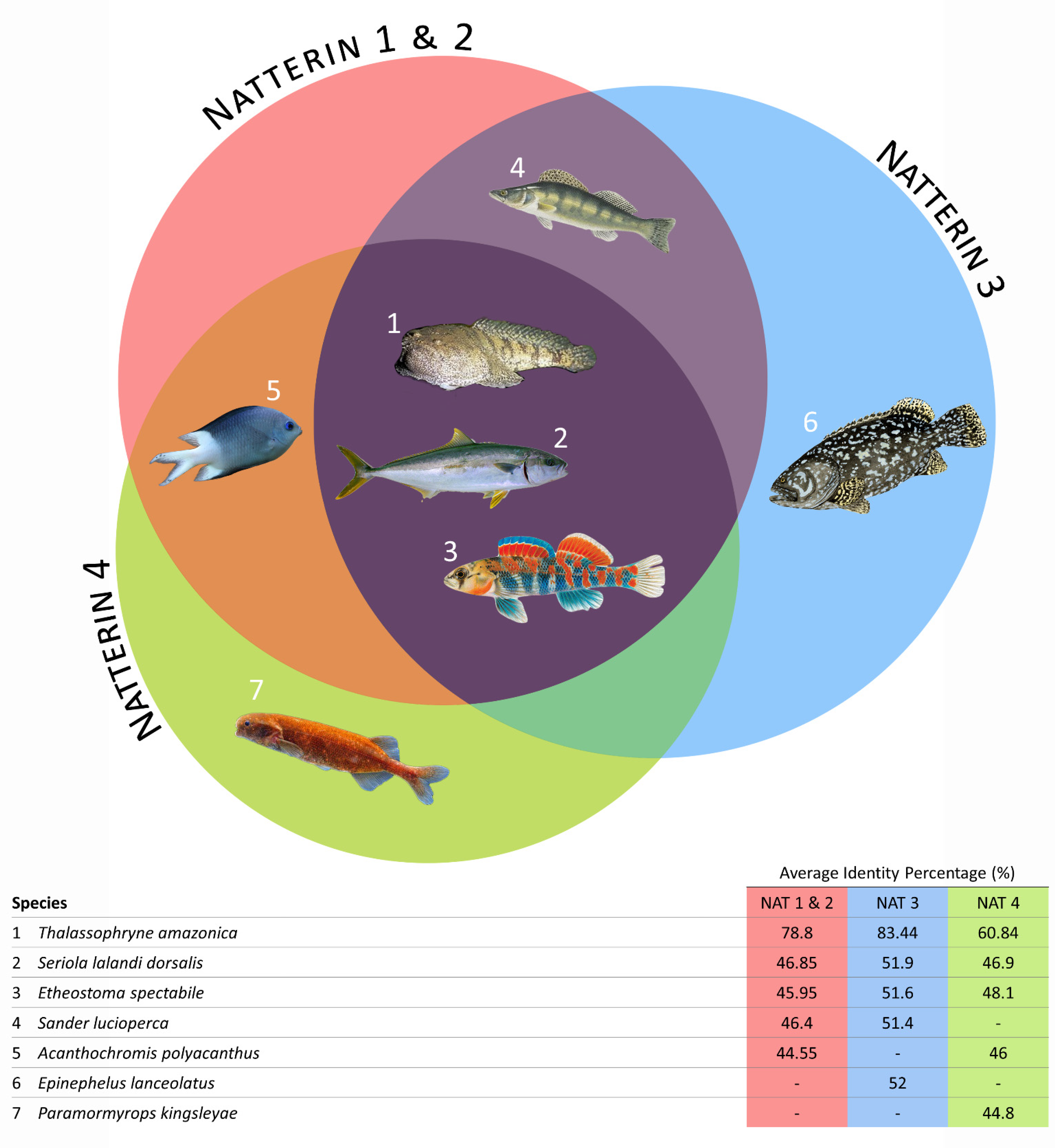
2.1. Phylogenetic Analysis of Natterin-like Proteins
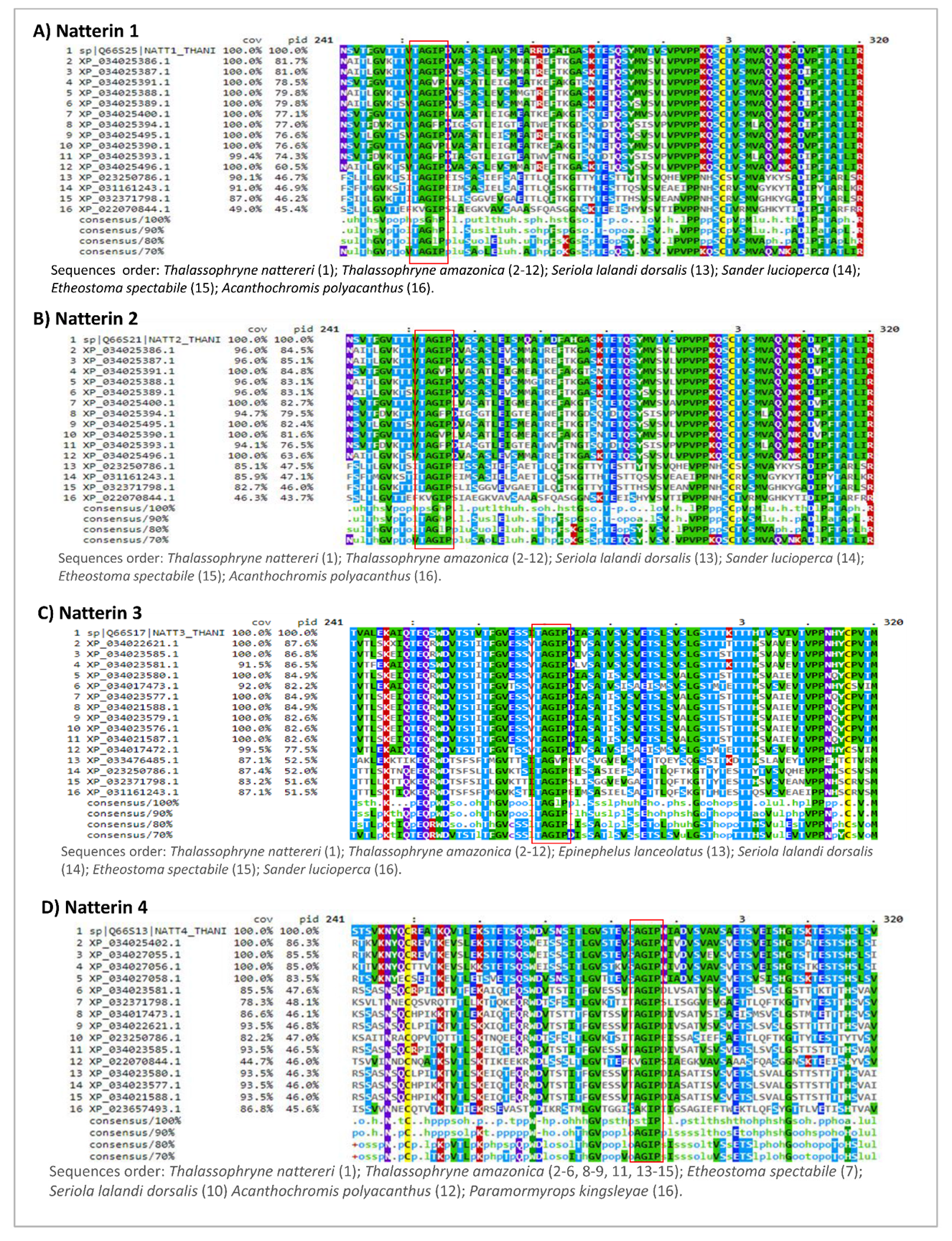
2.2. Multiple Alignments of the 15 Most Similar Members of Natterin-like Proteins
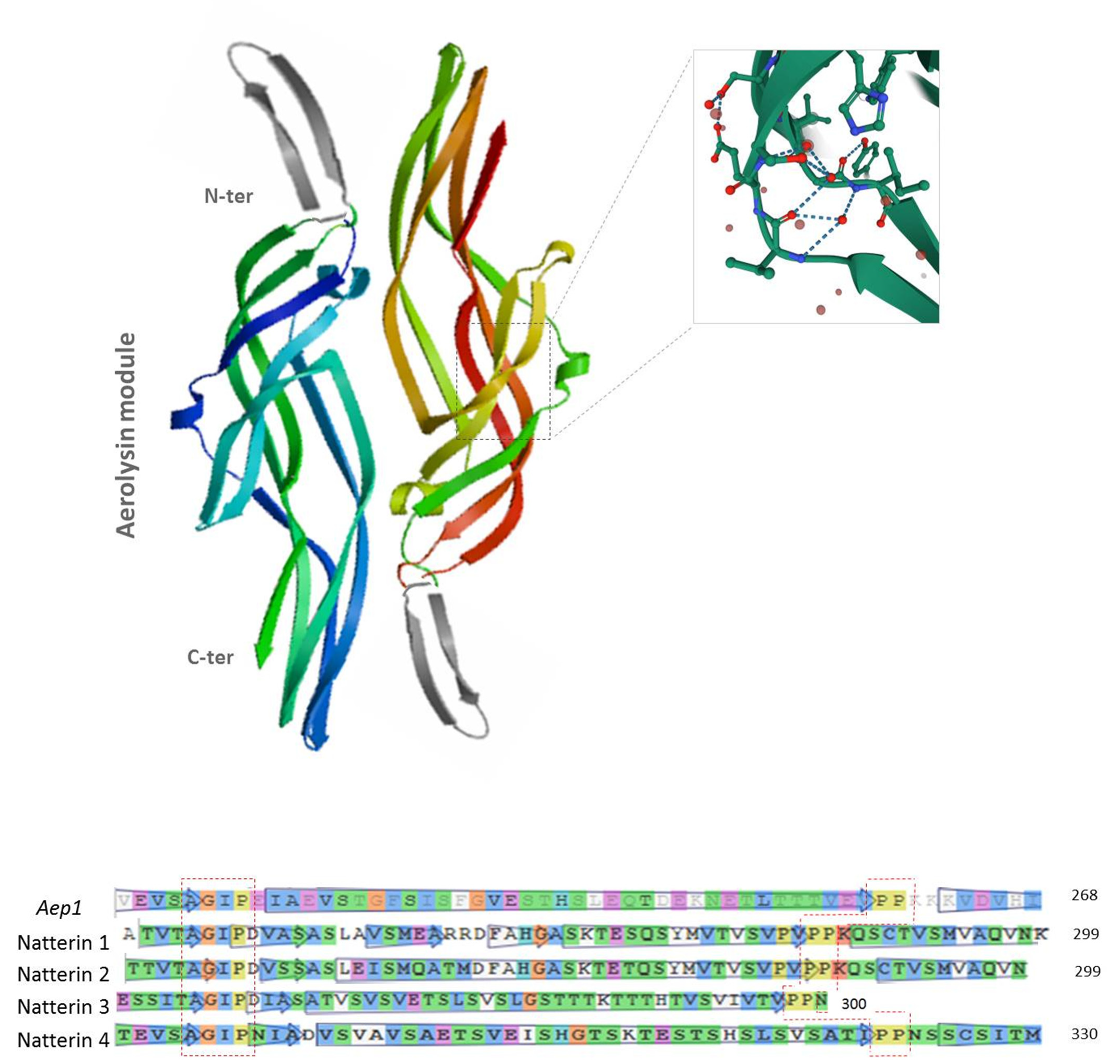
2.3. Conserved Domains of Natterin-like Proteins
2.4. Functional Analysis in the Immune Response
3. Final Considerations
4. Methods
4.1. Protein Diversity and Phylogenetic Analysis
4.2. Multiple Sequence Alignment
4.3. Structural Analysis of Conserved Domains
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magalhães, G.S.; Junqueira-de-Azevedo, I.L.M.; Lopes-Ferreira, M.; Lorenzini, D.M.; Ho, P.L.; Moura-da-Silva, A.M. Transcriptome analysis of expressed sequence tags from the venom glands of the fish Thalassophryne nattereri. Biochimie 2006, 88, 693–699. [Google Scholar] [CrossRef]
- Magalhães, G.S.; Lopes-Ferreira, M.; Junqueira-De-Azevedo, I.L.M.; Spencer, P.J.; Araújo, M.S.; Portaro, F.C.V.; Ma, L.; Valente, R.H.; Juliano, L.; Fox, J.W.; et al. Natterins, a new class of proteins with kininogenase activity characterized from Thalassophryne nattereri fish venom. Biochimie 2005, 87, 687–699. [Google Scholar] [CrossRef]
- Lopes-Ferreira, M.; Emim, J.A.; Oliveira, V.; Puzer, L.; Cezari, M.H.; Araújo, M.S.; Juliano, L.; Lapa, A.J.; Souccar, C.; Moura-da-Silva, A.M. Kininogenase activity of Thalassophryne nattereri fish venom. Biochem. Pharmacol. 2004, 68, 2151–2157. [Google Scholar] [CrossRef] [PubMed]
- Szczesny, P.; Iacovache, I.; Muszewska, A.; Ginalski, K.; van der Goot, F.G.; Grynberg, M. Extending the aerolysin family: From bacteria to vertebrates. PLoS ONE 2011, 6, e20349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Y.; Gou, M.; Yang, K.; Lu, J.; Han, Y.; Teng, H.; Li, C.; Wang, H.; Liu, C.; Zhang, K.; et al. Crystal structure of a cytocidal protein from lamprey and its mechanism of action in the selective killing of cancer cells. Cell Commun. Signal. 2019, 17, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugawara, T.; Yamashita, D.; Kato, K.; Peng, Z.; Ueda, J.; Kaneko, J.; Kamio, Y.; Tanaka, Y.; Yao, M. Structural basis for pore-forming mechanism of staphylococcal α-hemolysin. Toxicon 2015, 15, 226–231. [Google Scholar] [CrossRef] [Green Version]
- Anderluh, G.; Gilbert, R.J. (Eds.) MACPF/CDC Proteins—Agents of Defence, Attack and Invasion; Springer: Dordrecht, The Netherlands, 2014. [Google Scholar]
- Bischofberger, M.; Iacovache, I.; van der Goot, F.G. Pathogenic pore-forming proteins: Function and host response. Cell Host Microbe 2012, 12, 266–275. [Google Scholar] [CrossRef] [Green Version]
- Dal Peraro, M.; van der Goot, F.G. Pore-forming toxins: Ancient, but never really out of fashion. Nat. Rev. Microbiol. 2016, 14, 77–92. [Google Scholar] [CrossRef]
- Jia, N.; Liu, N.; Cheng, W.; Jiang, Y.L.; Sun, H.; Chen, L.L.; Peng, J.; Zhang, Y.; Ding, Y.H.; Zhang, Z.H.; et al. Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein. EMBO Rep. 2016, 17, 235–248. [Google Scholar] [CrossRef] [Green Version]
- Podobnik, M.; Kisovec, M.; Anderluh, G. Molecular mechanism of pore formation by aerolysin-like proteins. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160209. [Google Scholar] [CrossRef]
- Gurcel, L.; Abrami, L.; Girardin, S.; Tschopp, J.; van der Goot, F.G. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell 2006, 126, 1135–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greaney, A.J.; Leppla, S.H.; Moayeri, M. Bacterial Exotoxins and the Inflammasome. Front. Immunol. 2015, 10, 570. [Google Scholar] [CrossRef] [Green Version]
- Tamura, S.; Yamakawa, M.; Shiomi, K. Purification, characterization and cDNA cloning of two natterin-like toxins from the skin secretion of oriental catfish Plotosus lineatus. Toxicon 2011, 58, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Liu, X.; Pang, Y.; Yu, T.; Xiao, R.; Jin, M.; Han, Y.; Su, P.; Wang, J.; Lv, L.; et al. Characterization, phylogenetic analysis and cDNA cloning of natterin-like gene from the blood of lamprey, Lampetra japonica. Immunol. Lett. 2012, 148, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Feng, B.; Ren, Y.; Wu, D.; Chen, Y.; Huang, S.; Chen, S.; Xu, A. A pore-forming protein implements VLR-activated complement cytotoxicity in lamprey. Cell Discov. 2017, 3, 17033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gudbrandsson, J.; Ahi, E.P.; Franzdottir, S.R.; Kapralova, K.H.; Kristjansson, B.K.; Steinhaeuser, S.S.; Maier, V.H.; Johannesson, I.M.; Snorrason, S.S.; Jonsson, Z.O.; et al. The developmental transcriptome of contrasting Arctic charr (Salvelinus alpinus) morphs. F1000Research 2015, 4, 136. [Google Scholar] [CrossRef] [Green Version]
- Rajan, B.; Patel, D.M.; Kitani, Y.; Viswanath, K.; Brinchmann, M.F. Novel mannose binding natterin-like protein in the skin mucus of Atlantic cod (Gadus morhua). Fish Shellfish Immunol. 2017, 68, 452–457. [Google Scholar] [CrossRef]
- Lin, K.T.; Wang, W.X.; Ruan, H.T.; Dai, J.G.; Sun, J.J.; Liu, L.; Huang, X.D. Transcriptome analysis of differentially expressed genes in the fore- and hind-intestine of ovate pompano Trachinotus ovatus. Aquaculture 2019, 508, 76–82. [Google Scholar] [CrossRef]
- van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, W.C.; Starcevic, A.; Baranasic, D.; Diminic, J.; Zucko, J.; Gacesa, R.; van Oppen, M.J.; Hranueli, D.; Cullum, J.; Long, P.F. KEGG orthology-based annotation of the predicted proteome of Acropora digitifera: ZoophyteBase—An open access and searchable database of a coral genome. BMC Genom. 2013, 14, 509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leprêtre, M.; Almunia, C.; Armengaud, J.; Salvador, A.; Geffard, A.; Palos-Ladeiro, M. The immune system of the freshwater zebra mussel, Dreissena polymorpha, decrypted by proteogenomics of hemocytes and plasma compartments. J. Proteom. 2019, 202, 103366. [Google Scholar] [CrossRef]
- Unno, H.; Matsuyama, K.; Tsuji, Y.; Goda, S.; Hiemori, K.; Tateno, H.; Hirabayashi, J.; Hatakeyama, T. Identification, characterization, and x-ray crystallographic analysis of a novel type of mannose-specific lectin cgl1 from the pacific oyster Crassostrea gigas. Sci. Rep. 2016, 6, 29135. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Wang, L.; Huang, M.; Jia, Z.; Weinert, T.; Warkentin, E.; Liu, C.; Song, X.; Zhang, H.; Witt, J.; et al. DM9 Domain containing protein functions as a pattern recognition receptor with broad microbial recognition spectrum. Front. Immunol. 2017, 8, 1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.M.; Undheim, E.A.B.; Jauss, R.T.; Jenner, R.A. Venomics of remipede crustaceans reveals novel peptide diversity and illuminates the venom’s biological role. Toxins 2017, 9, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drukewitz, S.H.; Bokelmann, L.; Undheim, E.A.B.; von Reumont, B.M. Toxins from scratch? Diverse, multimodal gene origins in the predatory robber fly Dasypogon diadema indicate a dynamic venom evolution in dipteran insects. GigaScience 2019, 8, giz081. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.A.; Madio, B.; Jin, J.; Undheim, E.A.B.; Fry, B.G.; King, G.F. Melt with this kiss: Paralyzing and liquefying venom of the assassin bug Pristhesancus plagipennis (Hemiptera: Reduviidae). Mol. Cell. Proteom. 2017, 16, 552–566. [Google Scholar] [CrossRef] [Green Version]
- Özbek, R.; Wielsch, N.; Vogel, H.; Lochnit, G.; Foerster, F.; Vilcinskas, A.; von Reumont, B.M. Proteo-transcriptomic characterization of the venom from the endoparasitoid wasp Pimpla turionellae with aspects on its biology and evolution. Toxins 2019, 11, 721. [Google Scholar] [CrossRef] [Green Version]
- Fingerhut, L.C.H.W.; Strugnell, J.M.; Faou, P.; Labiaga, A.R.; Zhang, J.; Cooke, I.R. Shotgun proteomics analysis of saliva and salivary gland tissue from the common octopus Octopus vulgaris. J. Proteome Res. 2018, 17, 3866–3876. [Google Scholar] [CrossRef] [Green Version]
- Baum, D. Reading a Phylogenetic Tree: The Meaning of Monophyletic Groups. Nat. Educ. 2008, 1, 190. [Google Scholar]
- Baum, D. Trait evolution on a phylogenetic tree: Relatedness, similarity, and the myth of evolutionary advancement. Nat. Educ. 2008, 1, 191. [Google Scholar]
- Banks, J.A.; Nishiyama, T.; Hasebe, M.; Bowman, J.L.; Gribskov, M.; dePamphilis, C.; Albert, V.A.; Aono, N.; Aoyama, T. The Selaginella Genome Identifies Genetic Changes Associated with the Evolution of Vascular Plants. Science 2011, 332, 960–963. [Google Scholar] [CrossRef] [Green Version]
- Nigel, E.S. How many species of insects and other terrestrial arthropods are there on earth? Annu. Rev. Entomol. 2018, 63, 31–45. [Google Scholar]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Kjer, K.M.; Zhou, X. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- Barrowclough, G.F.; Cracraft, J.; Klicka, J.; Zink, R.M. How many kinds of birds are there and why does it matter? PLoS ONE 2016, 11, e0166307. [Google Scholar] [CrossRef]
- Castro, R.; Tafalla, C. Overview of fish immunity. In Mucosal Health in Aquaculture; Beck, B.H., Peatman, E., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. 3–54. [Google Scholar]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [Green Version]
- Kuraku, S.; Meyer, A.; Kuratani, S. Timing of genome duplications relative to the origin of the vertebrates: Did cyclostomes diverge before or after? Mol. Biol. Evol. 2009, 26, 47–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, J.S. Fishes of the World, 4th ed.; John Wiley & Sons: New York, NY, USA, 2006. [Google Scholar]
- Srivastava, M.; Simakov, O.; Chapman, J.; Fahey, B.; Gauthier, M.E.A.; Mitros, T.; Richards, G.S.; Conaco, C.; Dacre, M.; Hellsten, U.; et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 2010, 466, 720–726. [Google Scholar] [CrossRef]
- Park, E.; Hwang, D.; Lee, J.; Song, J.; Seo, T.; Won, Y. Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol. Phylogenet. Evol. 2012, 62, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Schendel, V.; Rash, L.D.; Jenner, R.A.; Undheim, E.A.B. The diversity of venom: The importance of behavior and venom system morphology in understanding its ecology and evolution. Toxins 2019, 11, 666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, Y.; Fredman, D.; Szczesny, P.; Grynberg, M.; Technau, U. Recurrent Horizontal Transfer of Bacterial Toxin Genes to Eukaryotes. Mol. Biol. Evol. 2012, 29, 2223–2230. [Google Scholar] [CrossRef] [Green Version]
- Gacesa, R.; Hung, J.Y.; Bourne, D.G.; Long, P.F. Horizontal transfer of a natterin-like toxin encoding gene within the holobiont of the reef building coral Acropora digitifera (Cnidaria: Anthozoa: Scleractinia) and across multiple animal lineages. J. Venom Res. 2020, 10, 7–12. [Google Scholar] [PubMed]
- Undheim, E.; Jenner, R.A. Phylogenetic analyses suggest centipede venom arsenals were repeatedly stocked by horizontal gene transfer. Nat. Commun. 2021, 12, 818. [Google Scholar] [CrossRef] [PubMed]
- Hejnol, A.; Obst, M.; Stamatakis, A.; Ott, M.; Rouse, G.W.; Edgecombe, G.D. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. Biol. Sci. 2009, 276, 4261–4270. [Google Scholar] [CrossRef] [Green Version]
- Shimeld, S.M.; Donoghue, P.C.J. Evolutionary crossroads in developmental biology: Cyclostomes (lamprey and hagfish). Development 2012, 139, 2091–2099. [Google Scholar] [CrossRef] [Green Version]
- Spencer, V.; Venza, Z.N.; Harrison, C.J. What can lycophytes teach us about plant evolution and development? Modern perspectives on an ancient lineage. Evol. Dev. 2020, 9, e12350. [Google Scholar]
- Friedman, M.; Coates, M.I.; Anderson, P. First discovery of a primitive coelacanth fin fills a major gap in the evolution of lobed fins and limbs. Evol. Dev. 2007, 9, 329–337. [Google Scholar] [CrossRef]
- Johanson, Z.; Long, J.A.; Talent, J.A.; Janvier, P.; Warren, J.W. Oldest coelacanth, from the Early Devonian of Australia. Biol. Lett. 2006, 2, 443–446. [Google Scholar] [CrossRef] [Green Version]
- Holder, M.T.; Erdmann, M.V.; Wilcox, T.P.; Caldwell, R.L.; Hillis, D.M. Two living species of coelacanths? Proc. Natl. Acad. Sci. USA 1999, 96, 12616–12620. [Google Scholar] [CrossRef] [Green Version]
- Clarke, J.T.; Lloyd, G.T.; Friedman, M. Little evidence for enhanced phenotypic evolution in early teleosts relative to their living fossil sister group. Proc. Natl. Acad. Sci. USA 2016, 113, 11531–11536. [Google Scholar] [CrossRef] [Green Version]
- Harter, T.S.; Brauner, C.J. The O2 and CO2 transport system in teleosts and the specialized mechanisms that enhance Hb–O2 unloading to tissues. Fish Physiol. 2017, 36, 1–106. [Google Scholar]
- Witten, P.W.; Hall, B.K. Teleost skeletal plasticity: Modulation, adaptation, and remodelling. Copeia 2015, 103, 727–739. [Google Scholar] [CrossRef]
- Near, T.J.; Eytan, R.I.; Dornburg, A.; Kuhn, K.L.; Moore, J.A.; Davis, M.P.; Wainwright, P.C.; Friedman, M.; Smith, W.L. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA 2012, 109, 13698–13703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Nishida, M. Teleost fish with specific genome duplication as unique models of vertebrate evolution. Environ. Biol. Fishes 2010, 88, 169–188. [Google Scholar] [CrossRef]
- Braasch, I.; Bobe, J.; Guiguen, Y.; Postlethwait, J.H. Reply to: ‘Subfunctionalization versus neofunctionalization after whole-genome duplication’. Nat. Genet. 2018, 50, 910–911. [Google Scholar] [CrossRef]
- Cusack, B.P.; Wolfe, K.H. When gene marriages don’t work out: Divorce by subfunctionalization. Trends Genet. 2007, 23, 270–272. [Google Scholar] [CrossRef]
- Rastogi, S.; Liberles, D.A. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 2005, 14, 5–28. [Google Scholar]
- Sandve, S.R.; Rohlfs, R.V.; Hvidsten, T.R. Subfunctionalization versus neofunctionalization after whole-genome duplication. Nat. Genet. 2018, 50, 908–909. [Google Scholar] [CrossRef] [PubMed]
- Weitzman, S.H. Teleost Fish Encyclopedia Britannica. 2018. Available online: https://www.britannica.com/animal/teleost (accessed on 12 April 2020).
- Avise, J.C.; Tatarenkov, A. Population genetics and evolution of the mangrove rivulus Kryptolebias marmoratus, the world’s only self-fertilizing hermaphrodite vertebrate. J. Fish Biol. 2015, 87, 519–538. [Google Scholar] [CrossRef]
- Lampert, K.P.; Schartl, M. The origin and evolution of a unisexual hybrid: Poecilia formosa. Philos. Trans. R. Soc. B 2008, 363, 2901–2909. [Google Scholar] [CrossRef] [Green Version]
- Froese, R.; Pauly, D. FishBase 2000: Concepts, Design and Data Sources; ICLARM: Los Baños, Philippines, 2000; p. 344. [Google Scholar]
- Wright, J.J. Evolutionary History of Venom Glands in the Siluriformes. Evol. Venom. Anim. Their Toxins 2015, 1–19. [Google Scholar] [CrossRef]
- Fróes, H.P. Sur un poisson toxiphore brésilien: Le “niquim” Thalassophyne maculosa. Rev. Sudam. Med. Cirurugia 1932, 3, 871–878. [Google Scholar]
- Fróes, H.P. Studies on venomous fishes of tropical countries. J. Trop. Med. Hyg. 1933, 36, 134–135. [Google Scholar]
- Lopes-Ferreira, M.; Grund, L.Z.; Lima, C. Thalassophryne nattereri fish venom: From the envenoming to the understanding of the immune system. J. Venom. Anim. Toxins Incl. Trop. Dis. 2014, 20, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, B.; Garai, G. A review on multiple sequence alignment from the perspective of genetic algorithm. Genomics 2017, 109, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Chatzou, M.; Magis, C.; Chang, J.M.; Kemena, C.; Bussotti, G.; Erb, I.; Notredame, C. Multiple sequence alignment modeling: Methods and applications. Brief. Bioinform. 2016, 17, 1009–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postlethwait, J.H.; Woods, I.G.; Ngo-Hazelett, P.; Yan, Y.L.; Kelly, P.D.; Chu, F.; Huang, H.; Hill-Force, A.; Talbot, W.S. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000, 10, 1890–1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [CrossRef] [PubMed]
- MacCarthy, T.; Bergman, A. The limits of subfunctionalization. BMC Evol. Biol. 2007, 7, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapovalov, M.; Vucetic, S.; Dunbrack, R.L., Jr. A new clustering and nomenclature for beta turns derived from high-resolution protein structures. PLoS Comput. Biol. 2019, 15, e1006844. [Google Scholar] [CrossRef] [Green Version]
- Cirauqui, N.; Abriata, L.A.; van der Goot, F.G.; Dal Peraro, M. Structural, physicochemical and dynamic features conserved within the aerolysin pore-forming toxin family. Sci. Rep. 2017, 7, 13932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiba, T.; Abe, Y.; Kitada, S.; Kusaka, Y.; Ito, A.; Ichimatsu, T.; Katayama, H.; Akao, T.; Higuchi, K.; Mizuki, E.; et al. Crystal structure of the parasporin-2 Bacillus thuringiensis toxin that recognizes cancer cells. J. Mol. Biol. 2009, 386, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.R.; Gibert, M.; Popoff, M.; Moss, D.S.; Titball, R.W.; Basak, A.K. Clostridium perfringens epsilon-toxin shows structural similarity to the pore-forming toxin aerolysin. Nat. Struct. Mol. Biol. 2004, 11, 797–798. [Google Scholar] [CrossRef] [PubMed]
- Degiacomi, M.T.; Iacovache, I.; Pernot, L.; Chami, M.; Kudryashev, M.; Stahlberg, H.; van der Goot, F.G.; Dal Peraro, M. Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism. Nat. Chem. Biol 2013, 9, 623–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deane, C.M.; Allen, F.H.; Taylor, R.; Blundell, T.L. Carbonyl-carbonyl interactions stabilize the partially allowed Ramachandran conformations of asparagine and aspartic acid. Protein Eng. 1999, 12, 1025–1028. [Google Scholar] [CrossRef] [PubMed]
- Mayorov, A.; Dal Peraro, M.; Abriata, L.A. Active site-induced evolutionary constraints follow fold polarity principles in soluble globular enzymes. Mol. Biol. Evol. 2019, 36, 1728–1733. [Google Scholar] [CrossRef]
- Parker, M.W.; Buckley, J.T.; Postma, J.P.M.; Tucker, A.D.; Leonard, K.; Pattus, F.; Tsernoglou, D. Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states. Nature 1994, 367, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Mancheño, J.M.; Tateno, H.; Goldstein, I.J.; Martínez-Ripoll, M.; Hermoso, J.A. Structural analysis of the Laetiporus sulphureus hemolytic pore-forming lectin in complex with sugars. J. Biol. Chem. 2005, 280, 17251–17259. [Google Scholar] [CrossRef] [Green Version]
- De Colibus, L.; Sonnen, A.F.P.; Morris, K.J.; Siebert, C.A.; Abrusci, P.; Plitzko, J.; Hodnik, V.; Leippe, M.; Volpi, E.; Anderluh, G.; et al. Structures of lysenin reveal a shared evolutionary origin for pore-forming proteins and its mode of sphingomyelin recognition. Structure 2012, 20, 1498–1507. [Google Scholar] [CrossRef] [Green Version]
- Ponting, C.P.; Mott, R.; Bork, P.; Copley, R.R. Novel protein domains and repeats in Drosophila melanogaster: Insights into structure, function, and evolution. Genome Res. 2001, 11, 1996–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Cruz, A.; Canepa, G.E.; Alves, E.; Silva, T.L.; Williams, A.E.; Nagyal, S.; Yenkoidiok-Douti, L.; Nagata, B.M.; Calvo, E.; Andersen, J.; et al. Plasmodium falciparum evades immunity of Anopheline mosquitoes by interacting with a Pfs47 midgut receptor. Proc. Natl. Acad. Sci. USA 2020, 117, 2597–2605. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, P.; Wang, W.; Dong, M.; Wang, M.; Gong, C.; Jia, Z.; Liu, Z.; Zhang, A.; Wang, L.; et al. A DM9-containing protein from oyster Crassostrea gigas (CgDM9CP-2) serves as a multipotent pattern recognition receptor. Dev. Comp. Immunol. 2018, 84, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, W.; Zhao, Q.; Yuan, P.; Li, J.; Song, X.; Liu, Z.; Ding, D.; Wang, L.; Song, L. A DM9-containing protein from oyster Crassostrea gigas (CgDM9CP-3) mediating immune recognition and encapsulation. Dev. Comp. Immunol. 2021, 116, 10393. [Google Scholar] [CrossRef] [PubMed]
- Zelensky, A.N.; Gready, J.E. The c-type lectin-like domain superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef]
- Wang, X.W.; Wang, J.X. Diversity and multiple functions of lectins in shrimp immunity. Dev. Comp. Immunol. 2013, 39, 27–38. [Google Scholar] [CrossRef]
- Alenton, R.R.; Koiwai, K.; Miyaguchi, K.; Kondo, H.; Hirono, I. Pathogen recognition of a novel C-type lectin from Marsupenaeus japonicus reveals the divergent sugar-binding specificity of QAP motif. Sci. Rep. 2017, 7, 45818. [Google Scholar] [CrossRef] [Green Version]
- Bunn-Moreno, M.; Campos-Neto, A. Lectin(s) extracted from seeds of Artocarpus integrifolia (jackfruit): Potent and selective stimulator(s) of distinct human T and B cell functions. J. Immunol. 1981, 127, 427–429. [Google Scholar] [PubMed]
- Tsaneva, M.; Van Damme, E.J.M. 130 years of Plant Lectin Research. Glycoconj. J. 2020, 37, 533–551. [Google Scholar] [CrossRef]
- Mancheño, J.M.; Tateno, H.; Sher, D.; Goldstein, I.J. Laetiporus sulphureus lectin and aerolysin protein family. Adv. Exp. Med. Biol. 2010, 677, 67–80. [Google Scholar]
- Alves, G.G.; Machado de Avila, R.A.; Chavez-Olortegui, C.D.; Lobato, F.C. Clostridium perfringens epsilon toxin: The third most potent bacterial toxin known. Anaerobe 2014, 30, 102–107. [Google Scholar] [CrossRef]
- Fontes, W.; Sousa, M.V.; Aragao, J.B.; Morhy, L. Determination of the amino acid sequence of the plant cytolysin enterolobin. Arch. Biochem. Biophys. 1997, 347, 201–207. [Google Scholar] [CrossRef]
- Dang, L.; Rougé, P.; Van Damme, E.J.M. Amaranthin-Like Proteins with Aerolysin Domains in Plants. Front. Plant Sci. 2017, 8, 1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shogomori, H.; Kobayashi, T. Lysenin: A sphingomyelin specific pore-forming toxin. Biochim. Biophys. Acta 2008, 1780, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Sher, D.; Fishman, Y.; Zhang, M.; Lebendiker, M.; Gaathon, A.; Mancheño, J.M.; Zlotkin, E. Hydralysins, a new category of beta-pore-forming toxins in cnidaria. J. Biol. Chem. 2005, 280, 22847–22855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holm, H.J.; Wadsworth, S.; Bjelland, A.K.; Krasnov, A.; Evensen, Ø.; Skugor, S. Dietary phytochemicals modulate skin gene expression profiles and result in reduced lice counts after experimental infection in Atlantic salmon. Parasit. Vectors 2016, 9, 271. [Google Scholar] [CrossRef] [Green Version]
- Neave, M.J.; Sunarto, A.; McColl, K.A. Transcriptomic analysis of common carp anterior kidney during Cyprinid herpesvirus 3 infection: Immunoglobulin repertoire and homologue functional divergence. Sci. Rep. 2017, 7, 41531. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.L.; Xie, J.; Cao, D.D.; Jia, N.; Li, Y.J.; Sun, H.; Li, W.F.; Hu, B.; Chen, Y.; Zhou, C.Z. The pore-forming protein Aep1 is an innate immune molecule that prevents zebrafish from bacterial infection. Dev. Comp. Immunol. 2018, 82, 49–54. [Google Scholar] [CrossRef]
- Leprêtre, M.; Almunia, C.; Armengaud, J.; Le Guernic, A.; Salvador, A.; Geffard, A.; Palos-Ladeiro, M. Identification of immune-related proteins of Dreissena polymorpha hemocytes and plasma involved in host-microbe interactions by differential proteomics. Sci. Rep. 2020, 10, 6226. [Google Scholar] [CrossRef]
- Patel, D.M.; Brinchmann, M.F. Skin mucus proteins of lumpsucker (Cyclopterus lumpus). Biochem. Biophys. Rep. 2017, 5, 217–225. [Google Scholar] [CrossRef]
- Patel, D.M.; Bhide, K.; Bhide, M.; Iversen, M.H.; Brinchmann, M.F. Proteomic and structural differences in lumpfish skin among the dorsal, caudal and ventral regions. Sci. Rep. 2019, 9, 6990. [Google Scholar] [CrossRef] [Green Version]
- Cokus, S.J.; De La Torre, M.; Medina, E.F.; Rasmussen, J.P.; Ramirez-Gutierrez, J.; Sagasti, A.; Wang, F. Tissue-Specific Transcriptomes Reveal Gene Expression Trajectories in Two Maturing Skin Epithelial Layers in Zebrafish Embryos. G3 Genes Genomes Genet. 2019, 9, 3439–3452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Y.; Li, C.; Wang, S.; Ba, W.; Yu, T.; Pei, G.; Bi, D.; Liang, H.; Pan, X.; Zhu, T.; et al. A novel protein derived from lamprey supraneural body tissue with efficient cytocidal actions against tumor cells. Cell Commun. Signal. 2017, 15, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, X.; Su, P.; Bi, D.; Tai, Z.; Li, Y.; Pang, Y.; Li, Q. Lamprey immune protein-1 (LIP-1) from Lampetra japonica induces cell cycle arrest and cell death in HeLa cells. Fish Shellfish Immunol. 2018, 75, 295–300. [Google Scholar] [CrossRef]
- Komegae, E.M.; Ramos, A.D.; Oliveira, A.K.; Serrano, S.M.T.; Lopes-Ferreira, M.; Lima, C. Insights into the local pathogenesis induced by fish toxins: Role of natterins and nattectin in the disruption of cell–cell and cell–extracellular matrix interactions and modulation of cell migration. Toxicon 2011, 58, 509–517. [Google Scholar] [CrossRef] [Green Version]
- Komegae, E.N.; Grund, L.Z.; Lopes-Ferreira, M.; Lima, C. The longevity of Th2 humoral response induced by proteases natterins requires the participation of long-lasting innate-like B cells and plasma cells in spleen. PLoS ONE 2013, 8, e67135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komegae, E.N.; Grund, L.Z.; Lopes-Ferreira, M.; Lima, C. TLR2, TLR4 and the MyD88 signaling are crucial for the in vivo generation and the longevity of long-lived antibody-secreting cells. PLoS ONE 2013, 8, e71185. [Google Scholar] [CrossRef]
- Gonzalez-Juarbe, N.; Bradley, K.M.; Riegler, A.N.; Reyes, L.F.; Brissac, T.; Park, S.S.; Restrepo, M.I.; Orihuela, C.J. Bacterial Pore-Forming Toxins Promote the Activation of Caspases in Parallel to Necroptosis to Enhance Alarmin Release and Inflammation During Pneumonia. Sci. Rep. 2018, 8, 5846. [Google Scholar] [CrossRef]
- McCoy, A.J.; Koizumi, Y.; Higa, N.; Suzuki, T. Differential regulation of caspase-1 activation via NLRP3/NLRC4 inflammasomes mediated by aerolysin and type III secretion system during Aeromonas veronii infection. J. Immunol. 2010, 185, 7077–7084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutterwala, F.S.; Haasken, S.; Cassel, S.L. Mechanism of NLRP3 inflammasome activation. Ann. N. Y. Acad. Sci. 2014, 1319, 82–95. [Google Scholar] [CrossRef]
- Kankkunen, P.; Välimäki, E.; Rintahaka, J.; Palomäki, J.; Nyman, T.; Alenius, H.; Wolff, H.; Matikainen, S. Trichothecene mycotoxins activate NLRP3 inflammasome through a P2X7 receptor and Src tyrosine kinase dependent pathway. Hum. Immunol. 2014, 75, 134–140. [Google Scholar] [CrossRef]
- Ito, M.; Yanagi, Y.; Ichinohe, T. Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog. 2012, 8, e1002857. [Google Scholar] [CrossRef] [Green Version]
- Palm, N.W.; Medzhitov, R. Role of the inflammasome in defense against venoms. Proc. Natl. Acad. Sci. USA 2013, 110, 1809–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Y.; Yan, C.; Guo, X.; Zhou, K.; Li, S.; Gao, Q.; Wang, X.; Zhao, F.; Liu, J.; Lee, W.H.; et al. Host-derived, pore-forming toxin-like protein and trefoil factor complex protects the host against microbial infection. Proc. Natl. Acad. Sci. USA 2014, 111, 6702–6707. [Google Scholar] [CrossRef] [Green Version]
- Lima, C.; Falcao, M.A.P.; Andrade-Barros, A.I.; Seni-Silva, A.C.; Grund, L.Z.; Balogh, E.; Conceiçao, K.; Queniaux, V.F.; Ryffel, B.; Lopes-Ferreira, M. Natterin an aerolysin-like fish toxin drives IL-1β-dependent neutrophilic inflammation mediated by caspase-1 and caspase-11 activated by the inflammasome sensor NLRP6. Int. Immunopharmacol. 2021, 91, 107287. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lima, C.; Disner, G.R.; Falcão, M.A.P.; Seni-Silva, A.C.; Maleski, A.L.A.; Souza, M.M.; Reis Tonello, M.C.; Lopes-Ferreira, M. The Natterin Proteins Diversity: A Review on Phylogeny, Structure, and Immune Function. Toxins 2021, 13, 538. https://doi.org/10.3390/toxins13080538
Lima C, Disner GR, Falcão MAP, Seni-Silva AC, Maleski ALA, Souza MM, Reis Tonello MC, Lopes-Ferreira M. The Natterin Proteins Diversity: A Review on Phylogeny, Structure, and Immune Function. Toxins. 2021; 13(8):538. https://doi.org/10.3390/toxins13080538
Chicago/Turabian StyleLima, Carla, Geonildo Rodrigo Disner, Maria Alice Pimentel Falcão, Ana Carolina Seni-Silva, Adolfo Luis Almeida Maleski, Milena Marcolino Souza, Mayara Cristina Reis Tonello, and Monica Lopes-Ferreira. 2021. "The Natterin Proteins Diversity: A Review on Phylogeny, Structure, and Immune Function" Toxins 13, no. 8: 538. https://doi.org/10.3390/toxins13080538
APA StyleLima, C., Disner, G. R., Falcão, M. A. P., Seni-Silva, A. C., Maleski, A. L. A., Souza, M. M., Reis Tonello, M. C., & Lopes-Ferreira, M. (2021). The Natterin Proteins Diversity: A Review on Phylogeny, Structure, and Immune Function. Toxins, 13(8), 538. https://doi.org/10.3390/toxins13080538








