New Cytoplasmic Virus-Like Elements (VLEs) in the Yeast Debaryomyces hansenii
Abstract
1. Introduction
2. Results and Discussion
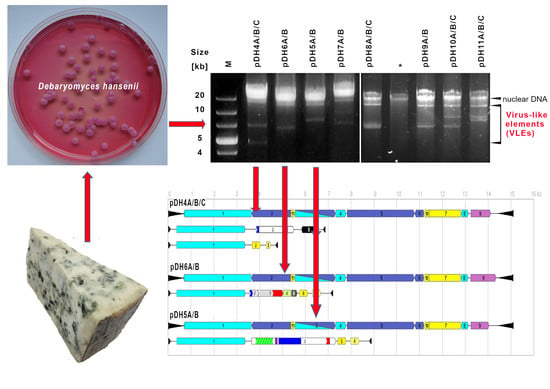
2.1. Frequency of VLEs in D. hansenii Yeast
2.2. The Characteristics of Selected VLEs Systems
| ORF | Protein Length [aa] | Predicted Function/Similarity 1 |
|---|---|---|
| 1 | 998 | B-type DNA polymerase |
| 2 | 565 | Similarity to putative mRNA capping-enzymes from M. acaciae pPac1-1 and S. etchellsii pPE1B |
| 3 | 586 | Helicase-DEXDc and HELICc domain containing protein |
| 4 | 161 | Similarity to putative ssDNA binding protein from M. acaciae pPac1-1 and S. etchellsii pPE1B |
| 5 | 987 | RNA polymerase larger subunit |
| 6 | 130 | Similarity to putative RNA-polymerase subunits from S. etchellsii pPE1B (ORF7) and M. acaciae pPac1-1 (ORF7) |
| 7 | 463 | Unknown |
| 8 | 105 | DNA-binding protein TRF1 (terminal region recognition factor 1) |
| 9 | 286/261/367 | Similarity to immunity determinants to killer toxins encoded by M. acaciae pPac1-2 and D. robertsiae pWR1A, fragment of M. farinosa chromosome F (XP_004202381) |
| 10 | 77 | Unknown, high similarity to product of ORF8 from: L. kluyveri pSKL, M. acaciae pPac1-1, K. lactis pGKL2, S. etchellsii pPE1B |
| 11 | 62 | Unknown, high similarity to products of: S. etchellsii pPE1B ORF3 M. acaciae pPac1-1 ORF11 |
| VLE | ORF | DNA Strand | ORF Length [bp] | Stop Codon | UCS | UCS Position [bp] 1 |
|---|---|---|---|---|---|---|
| pDH4A | 1 | + | 3003 | TGA | ATG TGA | −25 |
| 2 | + | 363 | TAA | ATA TGA | −27 | |
| 3 | + | 222 | TGA | ATG TGA | −26 | |
| pDH4B | 1 | + | 3003 | TGA | ATG TGA | −25 |
| 2 | + | 1647 | TAA | ATA TGA | −254 | |
| 3 | − | 585 | TAG | ATG TGA | −28 | |
| pDH5A | 1 | + | 3000 | TGA | ATG TGA | −25 |
| 2 | + | 3681 | TAA | ATA TGA | −27 | |
| 3 | + | 363 | TAA | ATG TGA | −26 | |
| 4 | + | 369 | TGA | ATG TGA | −26 | |
| 1 | + | 3003 | TGA | ATG TGA | −27 | |
| pDH6A | 2 | + | 396 | TAA | ATT TGA | −34 |
| 3 | + | 1092 | TGA | TTA TGA | −66 | |
| 4 | + | 360 | TGA | ATG TGA | −24 | |
| 5 | + | 264 | TAA | ATA TGA | −40 | |
| 6 | + | 363 | TAA | ATG TGA | −26 | |
| 7 | + | 369 | TAA | ATG TGA | −26 |
| VLE | ORF | Protein Length [aa] | Subcellular Localization Prediction 1 | Predicted Function or Functional Domains/Similarity 2 |
|---|---|---|---|---|
| pDH4A | 1 | 1000 | Cytoplasm | B-type DNA polymerase |
| 2 | 120 | Cytoplasm | Unknown, identical to pDH5A ORF3p and pDH6A ORF6p, high homology to pDH4A ORF3p, pDH5A ORF4p, pDH6A ORF7p and B. inositovora XP_018982205 | |
| 3 | 73 | Nucleoplasm | Unknown, identical to the part of pDH5A ORF4p and pDH6A ORF7p | |
| pDH4B | 1 | 1000 | Cytoplasm | B-type DNA polymerase |
| 2 | 548 | Peroxisome membrane | Similarity to GH18 chitinase-like superfamily proteins, fragment of DNase NucA/NucB | |
| 3 | 194 | Cytoplasm | Unknown, some similarities to ORF1p from autonomous plasmids pSKL and pGKL1 | |
| pDH5A | 1 | 999 | Cytoplasm | B-type DNA polymerase |
| 2 | 1226 3 | Extracellular | Fragment of autolysin domain, full chitin binding 1 domain (ChtBD1), full GH18 chitinase-like domain, fragment of deoxyribonuclease NucA/NucB domain | |
| 3 | 120 | Cytoplasm | as described for pDH4A ORF2p | |
| 4 | 122 | Nucleoplasm | Unknown, identical to pDH6A ORF7p, high homology to pDH4A ORF2p, pDH5A ORF3p and pDH6A ORF6p | |
| 1 | 1000 | Cytoplasm | B-type DNA polymerase | |
| pDH6A | 2 | 131 | Mitochondrion matrix | Contains fragment of GH18 chitinase domain |
| 3 | 363 | Extracellular | Contains fragment of DNase NucA/NucB | |
| 4 | 119 | Cytoplasm | Unknown, homology to proteins of unknown function of D. hansenii CBS 767T, pDH1A ORF4p and to the part of pPE1A ORF3p | |
| 5 | 87 | Extracellular | Unknown, homology to the part of pDH1A ORF3p | |
| 6 | 120 | Cytoplasm | As described for pDH4A ORF2p | |
| 7 | 122 | Nucleoplasm | As described for pDH5A ORF4p |
2.3. The Relationship between New VLEs and Killer Activity
2.4. The Relationship between Plasmids and Halotolerance
3. Conclusions
4. Materials and Methods
4.1. Microorganisms
4.2. New Yeast Strain Isolation
4.3. Yeast Isolate Identification
4.4. Plasmid Detection
4.5. Sequencing, Assembly and Annotation
4.6. Phylogenetic Analysis
4.7. VLE Curing
4.8. Antifungal Activity Test
4.9. Halotolerance Test
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fukuhara, H. Linear DNA Plasmids of Yeasts. FEMS Microbiol. Lett. 1995, 131. [Google Scholar] [CrossRef]
- Meinhardt, F.; Klassen, R. Microbial Linear Plasmids; Meinhardt, F., Klassen, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 7, ISBN 978-3-540-72024-9. [Google Scholar]
- Gunge, N.; Sakaguchi, K. Intergeneric Transfer of Deoxyribonucleic Acid Killer Plasmids, pGKl1 and pGKl2, from Kluyveromyces lactis into Saccharomyces cerevisiae by Cell Fusion. J. Bacteriol. 1981, 147, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Kitada, K.; Hishinuma, F. A New Linear DNA Plasmid Isolated from the Yeast Saccharomyces kluyveri. MGG 1987, 206, 377–381. [Google Scholar] [CrossRef]
- Shepherd, H.S.; Ligon, J.M.; Bolen, P.L.; Kurtzman, C.P. Cryptic DNA Plasmids of the Heterothallic Yeast Saccharomycopsis crataegensis. Curr. Genet. 1987, 12, 297–304. [Google Scholar] [CrossRef]
- Ligon, J.M.; Bolen, P.L.; Hill, D.S.; Bothast, R.J.; Kurtzman, C.P. Physical and Biological Characterization of Linear DNA Plasmids of the Yeast Pichia inositovora. Plasmid 1989, 21, 185–194. [Google Scholar] [CrossRef]
- Worsham, P.L.; Bolen, P.L. Killer Toxin Production in Pichia Acaciae Is Associated with Linear DNA Plasmids. Curr. Genet. 1990, 18, 77–80. [Google Scholar] [CrossRef]
- Gunge, N.; Fukuda, K.; Morikawa, S.; Murakami, K.; Takeda, M.; Miwa, A. Osmophilic Linear Plasmids from the Salt-Tolerant Yeast Debaryomyces hansenii. Curr. Genet. 1993, 23, 443–449. [Google Scholar] [CrossRef]
- Cong, Y.-S.; Yarrow, D.; Li, Y.-Y.; Fukuhara, H. Linear DNA Plasmids from Pichia etchellsii, Debaryomyces hansenii and Wingea robertsiae. Microbiology 1994, 140, 1327–1335. [Google Scholar] [CrossRef][Green Version]
- Hagenson, M.J.; Barr, K.A.; Stroman, D.W.; Harpold, M.M.; Klein, R.D.; Gaertner, F.H. Pichia pastoris Linear Plasmids and DNA Fragments Thereof. European Patent Office. European Patent Application: 92115908.3, March 1993. [Google Scholar]
- Banerjee, H.; Kopvak, C.; Curley, D. Identification of Linear DNA Plasmids of the Yeast Pichia pastoris. Plasmid 1998, 40, 58–60. [Google Scholar] [CrossRef]
- Blaisonneau, J.; Nosek, J.; Fukuhara, H. Linear DNA plasmid pPK2 of Pichia kluyveri: Distinction between cytoplasmic and mitochondrial linear plasmids in yeasts. Yeast 1999, 15. [Google Scholar] [CrossRef]
- Chen, W.-B.; Han, Y.-F.; Jong, S.-C.; Chang, S.-C. Isolation, Purification, and Characterization of a Killer Protein from Schwanniomyces occidentalis. Appl. Environ. Microbiol. 2000, 66, 5348–5352. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Z.; Wang, Q.-M.; Göker, M.; Groenewald, M.; Kachalkin, A.V.; Lumbsch, H.T.; Millanes, A.M.; Wedin, M.; Yurkov, A.M.; Boekhout, T.; et al. Towards an Integrated Phylogenetic Classification of the Tremellomycetes. Stud. Mycol. 2015, 81, 85–147. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Synonymy of the Yeast GeneraWingea AndDebaryomyces. Antonie Leeuwenhoek 1994, 66, 337–342. [Google Scholar] [CrossRef]
- Groenewald, M.; Daniel, H.-M.; Robert, V.; Poot, G.A.; Smith, M.T. Polyphasic Re-Examination of Debaryomyces Hansenii Strains and Reinstatement of D. hansenii, D. fabryi and D. subglobosus. Pers. Mol. Phylogeny Evol. Fungi 2008, 21, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Suzuki, M. Phylogenetic Analysis of Ascomycete Yeasts That Form Coenzyme Q-9 and the Proposal of the New Genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience 2010, 51, 2–14. [Google Scholar] [CrossRef]
- Gunge, N. Linear DNA Killer Plasmids from the YeastKluyveromyces. Yeast 1986, 2, 153–162. [Google Scholar] [CrossRef]
- Stam, J.C.; Kwakman, J.; Meijer, M.; Stuitje, A.R. Efficient Isolation of the Linear DNA Killer Plasmid of Kluyveromyces Lactis: Evidence for Location and Expression in the Cytoplasm and Characterization of Their Terminally Bound Proteins. Nucleic. Acids Res. 1986, 14, 6871–6884. [Google Scholar] [CrossRef]
- Frank, A.C.; Wolfe, K.H. Evolutionary Capture of Viral and Plasmid DNA by Yeast Nuclear Chromosomes. Eukaryot. Cell 2009, 8, 1521–1531. [Google Scholar] [CrossRef]
- Kast, A.; Voges, R.; Schroth, M.; Schaffrath, R.; Klassen, R.; Meinhardt, F. Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef]
- Tommasino, M.; Ricci, S.; Galeotti, C.L. Genome Organization of the Killer Plasmid PGK12 from Kluyveromyces Lactis. Nucleic Acids Res. 1988, 16, 5863–5878. [Google Scholar] [CrossRef]
- Wilson, D.W.; Meacock, P.A. Extranuclear Gene Expression in Yeast: Evidence for a Plasmidencoded RNA Polymerase of Unique Structure. Nucleic Acid Res. 1988, 16, 8097–8112. [Google Scholar]
- Klassen, R.; Jablonowski, D.; Schaffrath, R.; Meinhardt, F. Genome Organization of the Linear Pichia Etchellsii Plasmid PPE1A: Evidence for Expression of an Extracellular Chitin-Binding Protein Homologous to the α-Subunit of the Kluyveromyces Lactis Killer Toxin. Plasmid 2002, 47, 224–233. [Google Scholar] [CrossRef]
- Jeske, S.; Meinhardt, F. Autonomous Cytoplasmic Linear Plasmid pPac1-1 of Pichia acaciae: Molecular Structure and Expression Studies. Yeast 2006, 23, 479–486. [Google Scholar] [CrossRef]
- Schaffrath, R.; Meacock, P.A. An SSB Encoded by and Operating on Linear Killer Plasmids From Kluyveromyces lactis. Yeast 2001, 18, 1239–1247. [Google Scholar] [CrossRef]
- Stark, M.J.R.; Boyd, A.; Pritchard, R.H. The Killer Toxin of Kluyveromyces laclis: Characterization of the Toxin Subunits and Identification of the Genes Which Encode Them. EMBO J. 1986, 5, 1995–2002. [Google Scholar] [CrossRef]
- Klassen, R.; Meinhardt, F. Linear Plasmids pWR1A and pWR1B of the Yeast Wingea robertsiae Are Associated with a Killer Phenotype. Plasmid 2002, 48, 142–148. [Google Scholar] [CrossRef]
- Klassen, R.; Teichert, S.; Meinhardt, F. Novel Yeast Killer Toxins Provoke S-Phase Arrest and DNA Damage Checkpoint Activation. Mol. Microbiol. 2004, 53, 263–273. [Google Scholar] [CrossRef]
- Stark, M.J.R.; Boyd, A.; Mileham, A.J.; Ramonos, M.A. The Plasmid-Encoded Killer System of Kluyveromyces lactis: A Review. Yeast 1990, 6, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Gardini, F.; Suzzi, G.; Lombardi, A.; Galgano, F.; Crudele, M.A.; Andrighetto, C.; Schirone, M.; Tofalo, R. A Survey of Yeasts in Traditional Sausages of Southern Italy. FEMS Yeast Res. 2001, 1. [Google Scholar] [CrossRef] [PubMed]
- Simoncini, N.; Rotelli, D.; Virgili, R.; Quintavalla, S. Dynamics and Characterization of Yeasts during Ripening of Typical Italian Dry-Cured Ham. Food Microbiol. 2007, 24, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Brežná, B.; Ženišová, K.; Chovanová, K.; Chebeňová, V.; Kraková, L.; Kuchta, T.; Pangallo, D. Evaluation of Fungal and Yeast Diversity in Slovakian Wine-Related Microbial Communities. Antonie Leeuwenhoek 2010, 98, 519–529. [Google Scholar] [CrossRef]
- Jacques, N.; Zenouche, A.; Gunde-Cimerman, N.; Casaregola, S. Increased Diversity in the Genus Debaryomyces from Arctic Glacier Samples. Antonie Leeuwenhoek 2015, 107, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Girones, R.; Koutsoumanis, K.; Lindqvist, R.; Nørrung, B.; Robertson, L.; et al. Update of the List of QPS-recommended Biological Agents Intentionally Added to Food or Feed as Notified to EFSA 6: Suitability of Taxonomic Units Notified to EFSA until March 2017. EFSA J. 2017, 15, e04884. [Google Scholar] [CrossRef]
- Wagner, D.; Sander, A.; Bertz, H.; Finke, J.; Kern, W.V. Breakthrough Invasive Infection Due to Debaryomyces hansenii (Teleomorph Candida famata) and Scopulariopsis Brevicaulis in a Stem Cell Transplant Patient Receiving Liposomal Amphotericin B and Caspofungin for Suspected Aspergillosis. Infection 2005, 33, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Chan, G.F.; Sinniah, S.; Tengku Idr, T.I.N.; Ahmad Puad, M.S.; Abd Rahman, A.Z. Multiple Rare Opportunistic and Pathogenic Fungi in Persistent Foot Skin Infection. Pak. J. Biol. Sci. 2013, 16, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Van den Tempel, T.; Jakobsen, M. The Technological Characteristics of Debaryomyces hansenii and Yarrowia lipolytica and Their Potential as Starter Cultures for Production of Danablu. Int. Dairy J. 2000, 10, 263–270. [Google Scholar] [CrossRef]
- Żarowska, B.; Wojtatowicz, M.; Połomska, X.; Juszczyk, P.; Chrzanowska, J. Factors Affecting Killer Activity of Some Yeast Species Occurring in Rokpol Cheese. Folia Microbiol. 2004, 49, 713–717. [Google Scholar] [CrossRef]
- Fröhlich-Wyder, M.-T.; Arias-Roth, E.; Jakob, E. Cheese Yeasts. Yeast 2019, 36, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Marquina, D.; Barroso, J.; Peinado, J.M. (1-6)-Beta-D-Glucan as the Cell Wall Binding Site for Debaryomyces hansenii Killer Toxin. Lett. Appl. Microbiol. 2002, 34, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Żarowska, B. Biosynteza i Charakterystyka Toksyn Killerowych Drożdży Debaryomyces Hansenii; Wydawnictwo Uniwersytetu Przyrodniczego we Wrocławiu: Wrocław, Poland, 2012; pp. 34–49. [Google Scholar]
- Çorbacı, C.; Uçar, F.B. Purification, Characterization and in Vivo Biocontrol Efficiency of Killer Toxins from Debaryomyces hansenii Strains. Int. J. Biol. Macromol. 2018, 119, 1077–1082. [Google Scholar] [CrossRef]
- Wojtatowicz, M.; Chrzanowska, J.; Juszczyk, P.; Skiba, A.; Gdula, A. Identification and Biochemical Characteristics of Yeast Microflora of Rokpol Cheese. Int. J. Food Microbiol. 2001, 69, 135–140. [Google Scholar] [CrossRef]
- Połomska, X.; Wojtatowicz, M.; Zarowska, B.; Szołtysik, M.; Chrzanowska, J. Freeze-Drying Preservation of Yeast Adjunct Cultures for Cheese Production. Pol. J. Food Nutr. Sci. 2012, 62, 143–150. [Google Scholar] [CrossRef][Green Version]
- Szołtysik, M.; Dâbrowska, A.; Babij, K.; Pokora, M.; Zambrowicz, A.; Połomska, X.; Wojtatowicz, M.; Chrzanowska, J. Biochemical and Microbiological Changes in Cheese Inoculated with Yarrowia lipolytica Yeast. Zywnosc. Nauka. Technol. Jakosc/Food. Sci. Technol. Qual. 2013, 89, 49–64. [Google Scholar] [CrossRef]
- Grzegorczyk, M.; Żarowska, B.; Restuccia, C.; Cirvilleri, G. Postharvest Biocontrol Ability of Killer Yeasts against Monilinia fructigena and Monilinia fructicola on Stone Fruit. Food Microbiol. 2017, 61. [Google Scholar] [CrossRef]
- Czarnecka, M.; Żarowska, B.; Połomska, X.; Restuccia, C.; Cirvilleri, G. Role of Biocontrol Yeasts Debaryomyces hansenii and Wickerhamomyces anomalus in Plants’ Defence Mechanisms against Monilinia fructicola in Apple Fruits. Food Microbiol. 2019, 83, 1–8. [Google Scholar] [CrossRef]
- Żarowska, B.; Połomska, X.; Grzegorczyk, M.; Regiec, P.; Gudarowska, E.; Czaplicka-Pędzich, M.; Sosna, I.; Figiel, A.; Pasławska, M.; Serowik, M.; et al. Method for obtaining a preparation containing killer toxins. Patent of Republic of Poland Pat.229949, September 2018. [Google Scholar]
- Wrent, P.; Rivas, E.-M.; Gil de Prado, E.; Peinado, J.M.; de Silóniz, M.-I. Development of Species-Specific Primers for Rapid Identification of Debaryomyces hansenii. Int. J. Food Microbiol. 2015, 193, 109–113. [Google Scholar] [CrossRef] [PubMed][Green Version]
- O’Donnel, K. Fusarium and Its near Relatives. In The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics; Reynolds, D.R., Taylor, J.W., Eds.; CAB International: Wallingford, UK, 1993; pp. 225–233. [Google Scholar]
- Romero, P.; Patino, B.; Quiros, M.; Gonzalezjaen, M.; Valderrama, M.; Desiloniz, M.; Penaido, J. Differential Detection of Isolated from Intermediate-Moisture Foods by PCR-RFLP of the IGS Region of rDNA. FEMS Yeast Res. 2005, 5, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.-V.; Gaillardin, C.; Neuveglise, C. Differentiation of Debaryomyces hansenii and Candida famata by rRNA Gene Intergenic Spacer Fingerprinting and Reassessment of Phylogenetic Relationships among D. hansenii, C. famata, D. fabryi, C. flareri (D. subglobosus ) and D. prosopidis: Description of D. vietnamensis sp. Nov. Closely Related to D. nepalensis. FEMS Yeast Res. 2009, 9, 641–662. [Google Scholar] [CrossRef][Green Version]
- Fukuda, K.; Jinshan, C.; Kawano, M.; Sudo, K.; Gunge, N. Stress Responses of Linear Plasmids From. FEMS Microbiol. Lett. 2004, 237, 243–248. [Google Scholar] [CrossRef]
- Hishinuma, F.; Hirai, K. Genome Organization of the Linear Plasmid, pSKL, Isolated from Saccharomyces kluyveri. MGG 1991, 226, 97–106. [Google Scholar] [CrossRef]
- Tommasino, M. Killer system of Kluyveromyces lactis: The open reading frame 10 of the pGKL2 plasmid encodes a putative DNA binding protein. Yeast 1991, 7, 245–252. [Google Scholar] [CrossRef]
- Klassen, R.; Meinhardt, F. Structural and Functional Analysis of the Killer Element PPin1-3 from Pichia inositovora. MGG 2003, 270, 190–199. [Google Scholar] [CrossRef]
- Satwika, D.; Klassen, R.; Meinhardt, F. Repeated Capture of a Cytoplasmic Linear Plasmid by the Host Nucleus in Debaryomyces hansenii. Yeast 2012, 29, 145–154. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; de Montigny, J.; Marck, C.; Neuvéglise, C.; Talla, E.; et al. Genome Evolution in Yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef]
- Neuveglise, C.; Feldmann, H.; Bon, E.; Gaillardin, C.; Casaregola, S. Genomic Evolution of the Long Terminal Repeat Retrotransposons in Hemiascomycetous Yeasts. Genome Res. 2002, 12, 930–943. [Google Scholar] [CrossRef]
- Larsen, M.; Meinhardt, F. Kluyveromyces lactis Killer System: Identification of a New Gene Encoded by pGKL2. Curr. Genet. 2000, 38, 271–275. [Google Scholar] [CrossRef]
- Kempken, F.; Hermanns, J.; Osiewacz, H.D. Evolution of Linear Plasmids. J. Mol. Evol. 1992, 35, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, F.; Klassen, R. Linear protein-primed replicating plasmids in eukaryotic microbes. In Microbial Linear Plasmids; Meinhardt, F., Klassen, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 7, pp. 188–227. ISBN 978-3-540-72024-9. [Google Scholar]
- Sýkora, M.; Pospíšek, M.; Novák, J.; Mrvová, S.; Krásný, L.; Vopálenský, V. Transcription Apparatus of the Yeast Virus-like Elements: Architecture, Function, and Evolutionary Origin. PLoS Pathog. 2018, 14, e1007377. [Google Scholar] [CrossRef] [PubMed]
- Vopálenský, V.; Sýkora, M.; Mašek, T.; Pospíšek, M. Messenger RNAs of Yeast Virus-Like Elements Contain Non-Templated 5′ Poly(A) Leaders, and Their Expression Is Independent of EIF4E and Pab1. Front Microbiol. 2019, 10, 2366. [Google Scholar] [CrossRef]
- Gish, W.; States, D.J. Identification of Protein Coding Regions by Database Similarity Search. Nat. Genet. 1993, 3, 266–272. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s Conserved Domain Database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam Protein Families Database: Towards a More Sustainable Future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Schaffrath, R.; Meacock, P.A.; Meinhardt, F. Yeast Killer Plasmid pGKL2: Molecular Analysis of UCS5, a Cytoplasmic Promoter Element Essential for ORF5 Gene Function. MGG 1996, 250, 286–294. [Google Scholar] [CrossRef]
- Meinhardt, F.; Wodara, C.; Larsen, M.; Schickel, J. A Novel Approach to Express a Heterologous Gene on Kluyveromyces Lactis Linear Killer Plasmids: Expression of the Bacterial Aph Gene from a Cytoplasmic Promoter Fragment without In-Phase Fusion to the Plasmid Open Reading Frame. Plasmid 1994, 32, 318–327. [Google Scholar] [CrossRef]
- Schründer, J.; Meinhardt, F. Extrachromosomal Inheritance: Yeast Linear Killer Plasmids as a Tool in Genetic Engineering. In Progress in Botany; Springer: Berlin/Heidelberg, Germany, 1995. [Google Scholar]
- Schründer, J.; Gunge, N.; Meinhardt, F. Extranuclear Expression of the Bacterial Xylose Isomerase (Xyl A) and the UDP-Glucose Dehydrogenase ( Has B) Genes in Yeast with Kluyveromyces lactis Linear Killer Plasmids as Vectors. Curr. Microbiol. 1996, 33, 323–330. [Google Scholar] [CrossRef]
- Schickel, J.; Helmig, C.; Meinhardt, F. Kluyveromyces Lactis Killer System: Analysis of Cytoplasmic Promoters of the Linear Plasmids. Nucleic Acids Res. 1996, 24, 1879–1886. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gunge, N.; Tokunaga, M. Linear DNA Plasmids and Killer System of Kluyveromyces lactis. In Genetics and Biotechnology; Springer: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Mesnage, S.; Dellarole, M.; Baxter, N.J.; Rouget, J.-B.; Dimitrov, J.D.; Wang, N.; Fujimoto, Y.; Hounslow, A.M.; Lacroix-Desmazes, S.; Fukase, K.; et al. Molecular Basis for Bacterial Peptidoglycan Recognition by LysM Domains. Nat. Commun. 2014, 5, 4269. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of Protein Subcellular Localization Using Deep Learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Klassen, R.; Wemhoff, S.; Krause, J.; Meinhardt, F. DNA Repair Defects Sensitize Cells to Anticodon Nuclease Yeast Killer Toxins. MGG 2011, 285, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Wemhoff, S.; Klassen, R.; Meinhardt, F. DNA Damage Induced by the Anticodon Nuclease from a Pichia acaciae Killer Strain Is Linked to Ribonucleotide Reductase Depletion. Cell. Microbiol. 2016, 18, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Wemhoff, S.; Klassen, R.; Beetz, A.; Meinhardt, F. DNA Damage Responses Are Induced by tRNA Anticodon Nucleases and Hygromycin, B. PLoS ONE 2016, 11, e0157611. [Google Scholar] [CrossRef]
- Schaffrath, R.; Meinhardt, F.; Klassen, R. Yeast Killer Toxins: Fundamentals and Applications. In The Mycota. Physiology and Genetics: Selected Basic and Applied Aspects; Anke, T., Schüffler, A., Eds.; Springer International Publishing: New York, NY, USA, 2018; pp. 87–118. [Google Scholar]
- Kall, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of Combined Transmembrane Topology and Signal Peptide Prediction--the Phobius Web Server. Nucleic. Acids Res 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Vega, R.; Dominguez, A. Cell Wall Composition of the Yeast and Mycelial Forms of Yarrowia lipolytica. Arch. Microbiol. 1986, 144, 124–130. [Google Scholar] [CrossRef]
- Andriyanova, D.A.; Smirnova, G.P.; Shashkov, A.S.; Chizhov, A.O.; Galanina, L.A.; Feofilova, E.P.; Usov, A.I. Polysaccharide Composition of Mycelium and Cell Walls of the Fungus Penicillium roqueforti. Russ. J. Bioorganic. Chem. 2011, 37, 356–363. [Google Scholar] [CrossRef]
- McCracken, D.A.; Martin, V.J.; Stark, M.J.R.; Bolen, P.L. The Linear-Plasmid-Encoded Toxin Produced the Yeast Pichia acaciae: Characterization an Comparison with the Toxin of Kluyveromyces lactis. Microbiology 1994, 140, 425–431. [Google Scholar] [CrossRef]
- Çorbaci, C.; Uçar, F.B. Production and Optimization of Killer Toxin in Debaryomyces hansenii Strains. Braz. Arch. Biol. Technol. 2017, 60. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an Enhanced Web Interface to Primer3. Nucleic. Acids Res 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Querol, A.; Barrio, E.; Ramón, D. A Comparative Study of Different Methods of Yeast Strain Characterization. Syst. Appl. Microbiol. 1992, 15, 439–446. [Google Scholar] [CrossRef]
- Woods, D.R.; Bevan, E.A. Studies on the Nature of the Killer Factor Produced by Saccharomyces cerevisiae. J. Gen. Microbiol. 1968, 51, 115–126. [Google Scholar] [CrossRef]




| Culture Collection | Strain Designation | Source of Isolation | ||
|---|---|---|---|---|
| DBFM 1 | AI1a, AI4a, AI4b, AI4c, AI6c, AII2c, AII3b, AII4a, | Dairy products | Rokpol cheese | |
| AII4b, AII4b-1, AII4c, BI6b, EI2b, EI3a, EI4a, EII2a, | ||||
| EII3c, MI1a, MI1a-1, MI1a-2, MI2a, MI4a, MI5b, | ||||
| MI6b, MI7a, OI1a, OI1b, OI5c, OII1c, OII3a, PI1a, | ||||
| PI4a, PI5b, PII3a | ||||
| DRIP1b, DRIP2c, DRIIP1b, | Dorblu cheese | |||
| DRIIIP3c, DRIW4c, DRIIW3b, | ||||
| DRIIW4c, DRIIIW6a, KI2a | ||||
| DNIP4b, DNIIP2b | Danablu cheese | |||
| CIRM-Levures 2 | CLIB 236 | Dairy products and environment | Roncal cheese | |
| CLIB 380, 594 | Goat’s cheese | |||
| CLIB 607 | Camembert | |||
| CLIB 608 | Air | |||
| CLIB 609, 684 | Milk | |||
| CLIB 611 | Dairy brine | |||
| CLIB 613 | Forage | |||
| CLIB 622 | Saint Nectaire | |||
| CLIB 920 | Cheese curd | |||
| CLIB 539, 1302, 1465, 1277, 1298, 1301 | Ice, glacial, and seawater | |||
| CLIB 543, 944, 1086, 1389 | Beverages | |||
| CLIB 545, 907 | Human | |||
| CLIB 542, 1295 | Meat products | |||
| CLIB 1142, 1143 | Insects | |||
| CLIB 195, 1144 | Unknown | |||
| CLIB 1296 | Salted pickles | |||
| Strain | Cheese Type | Country |
|---|---|---|
| 1 (a-d) | Fourme d’Ambert | France |
| 2 (a-d) | Bleu d’Auvergne | France |
| 3 (a-d) | Rokpol Lazur | Poland |
| 4 (a-l) | Edelpilz | Germany |
| 5 (a-f) | Rokpol KG | Poland |
| 6 (a-j) | Dorblu | Germany |
| 7 (a-j) | Bloose | Denmark |
| 8 (a-j) | Turek niebieski | Poland |
| System | Strains | VLE Size |
|---|---|---|
| pDH4A/B/C | 4a, 4c, 4d, 4e, 4f, 4i | 4.8; 6.9; 15.1 |
| pDH5A/B | 5c | 8.9; 15.1 |
| pDH6A/B | 7f, 7g, 7i | 7.2; 15.1 |
| pDH7A/B | 7j | 8.4; 15.1 |
| pDH8A/B/C | 8e | 7.4; 8.0; 15.1 |
| pDH9A/B | 8g | 7.6; 15.1 |
| pDH10A/B/C | 8h | 7.6; 10.0; 15.1 |
| pDH11A/B/C | 8i | 9.2; 10.0; 15.1 |
| ORF | DNA Strand | ORF Length [bp] | ORF Identity [%] | Stop Codon | UCS | UCS Position 1 [bp] |
|---|---|---|---|---|---|---|
| 1 | − | 2997 | 100 | TGA | ATATGA | −34 |
| 2 | − | 1698 | 100 | TAA | ATATGA | −52 |
| 3 | + | 1761 | 99 2 | TAA | ATCTGA | −25 |
| 4 | + | 486 | 100 | TGA | ATGTGA | −108 |
| 5 | + | 2964 | 99 3 | TAA | ATGTGA | −50 |
| 6 | − | 393 | 100 | TAA | ATGTGA | −26 |
| 7 | + | 1392 | 100 | TAA | ATTTGA | −43 |
| 8 | + | 318 | 100 | TAA | ATTTGA | −8 |
| 9 | + | 861/786/1104 | 100 4 | TAA | ATATGA | −38 |
| 10 | − | 234 | 100 | TAA | ATTTGA | −34 |
| 11 | + | 186 | 100 | TAA | ATTTGA | −95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Połomska, X.; Neuvéglise, C.; Zyzak, J.; Żarowska, B.; Casaregola, S.; Lazar, Z. New Cytoplasmic Virus-Like Elements (VLEs) in the Yeast Debaryomyces hansenii. Toxins 2021, 13, 615. https://doi.org/10.3390/toxins13090615
Połomska X, Neuvéglise C, Zyzak J, Żarowska B, Casaregola S, Lazar Z. New Cytoplasmic Virus-Like Elements (VLEs) in the Yeast Debaryomyces hansenii. Toxins. 2021; 13(9):615. https://doi.org/10.3390/toxins13090615
Chicago/Turabian StylePołomska, Xymena, Cécile Neuvéglise, Joanna Zyzak, Barbara Żarowska, Serge Casaregola, and Zbigniew Lazar. 2021. "New Cytoplasmic Virus-Like Elements (VLEs) in the Yeast Debaryomyces hansenii" Toxins 13, no. 9: 615. https://doi.org/10.3390/toxins13090615
APA StylePołomska, X., Neuvéglise, C., Zyzak, J., Żarowska, B., Casaregola, S., & Lazar, Z. (2021). New Cytoplasmic Virus-Like Elements (VLEs) in the Yeast Debaryomyces hansenii. Toxins, 13(9), 615. https://doi.org/10.3390/toxins13090615