Is Toxin-Producing Planktothrix sp. an Emerging Species in Lake Constance?
Abstract
1. Introduction
2. Results
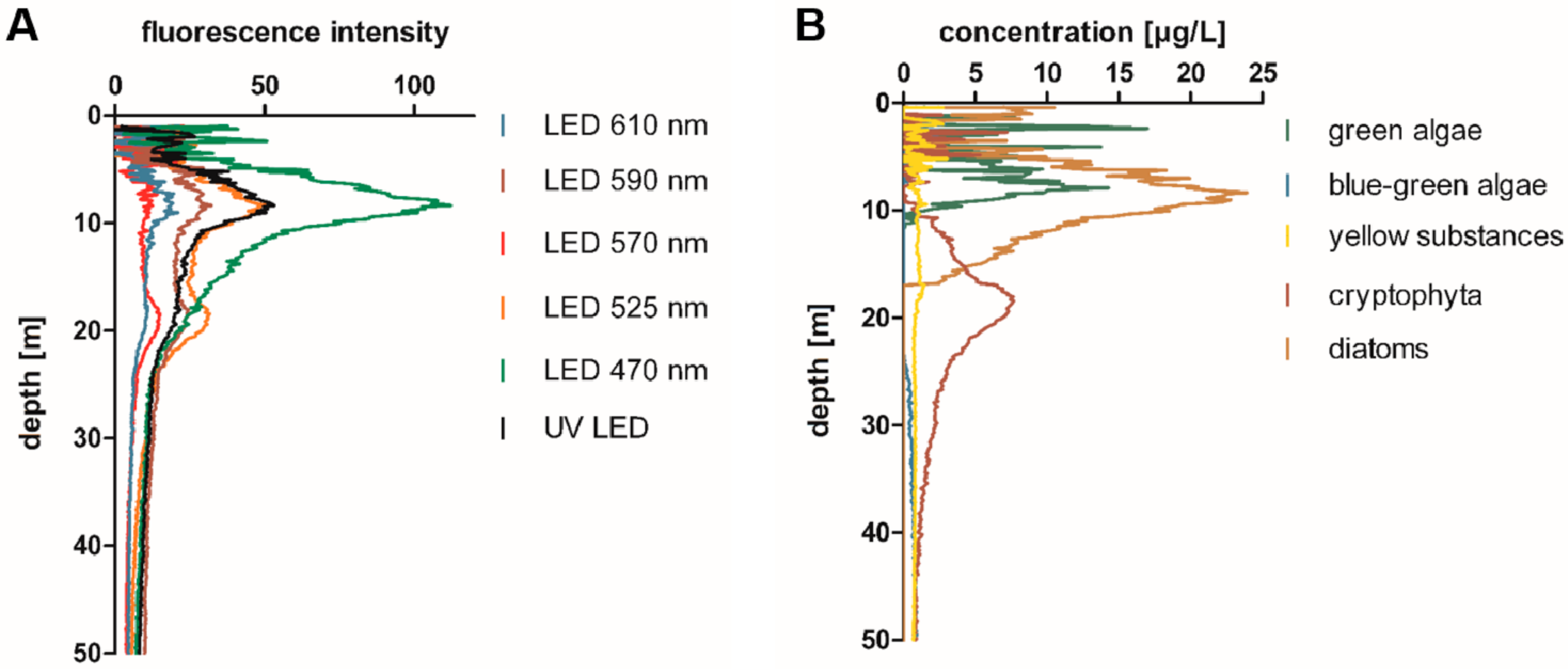
2.1. Blooms of Red-Pigmented Phytoplankton at Water Depths below the Chlorophyll-a Maximum in Lake Constance
2.2. Phycoerythrin-Rich Synechococcus Phylotypes Dominated the DRM Cyanobacterial Community
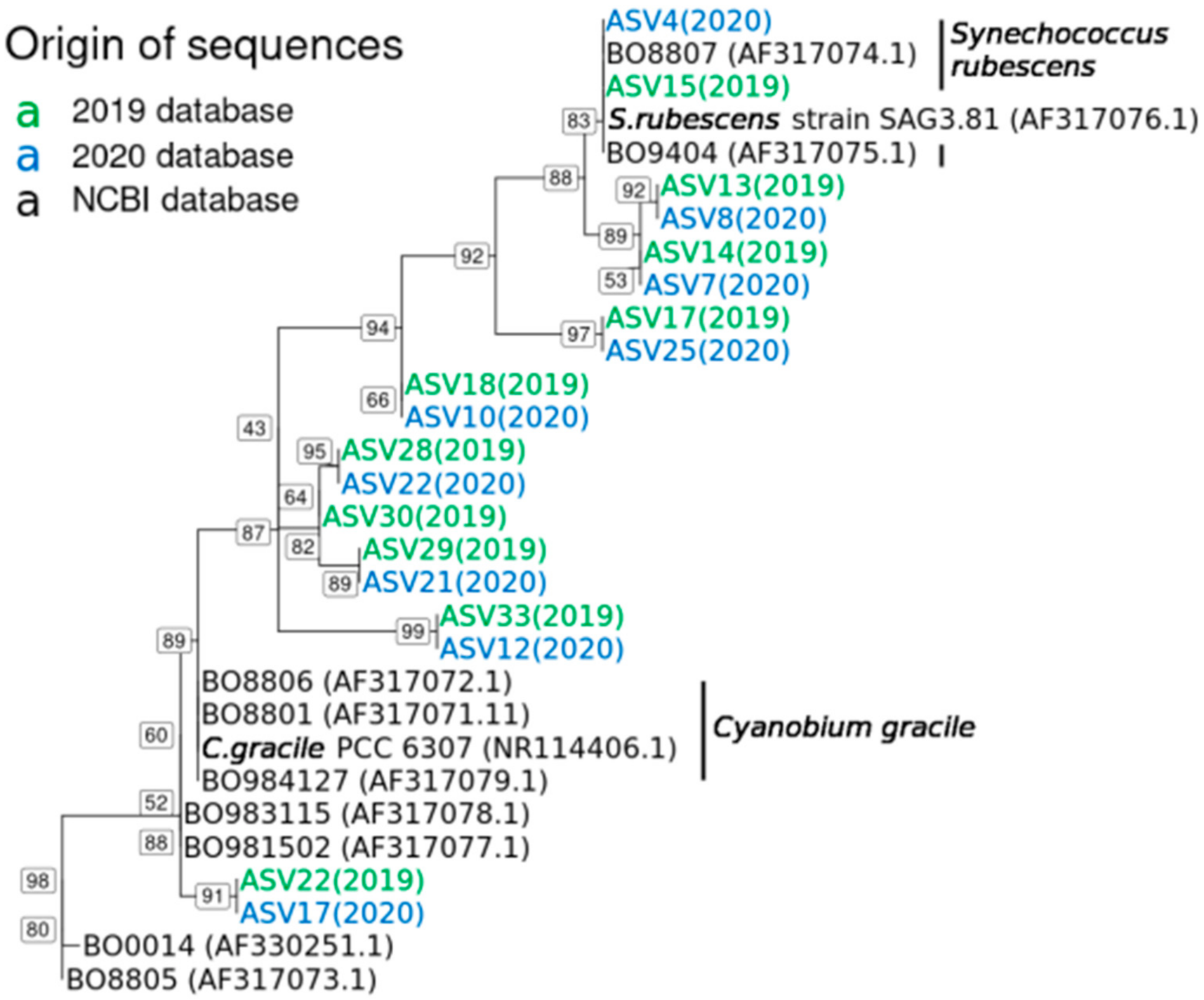
2.3. Synechococcus Rubescens and Cyanobium Gracile Clusters in 2019 and 2020
2.4. Dynamics of Synechococcus ASVs in 2019 and 2020
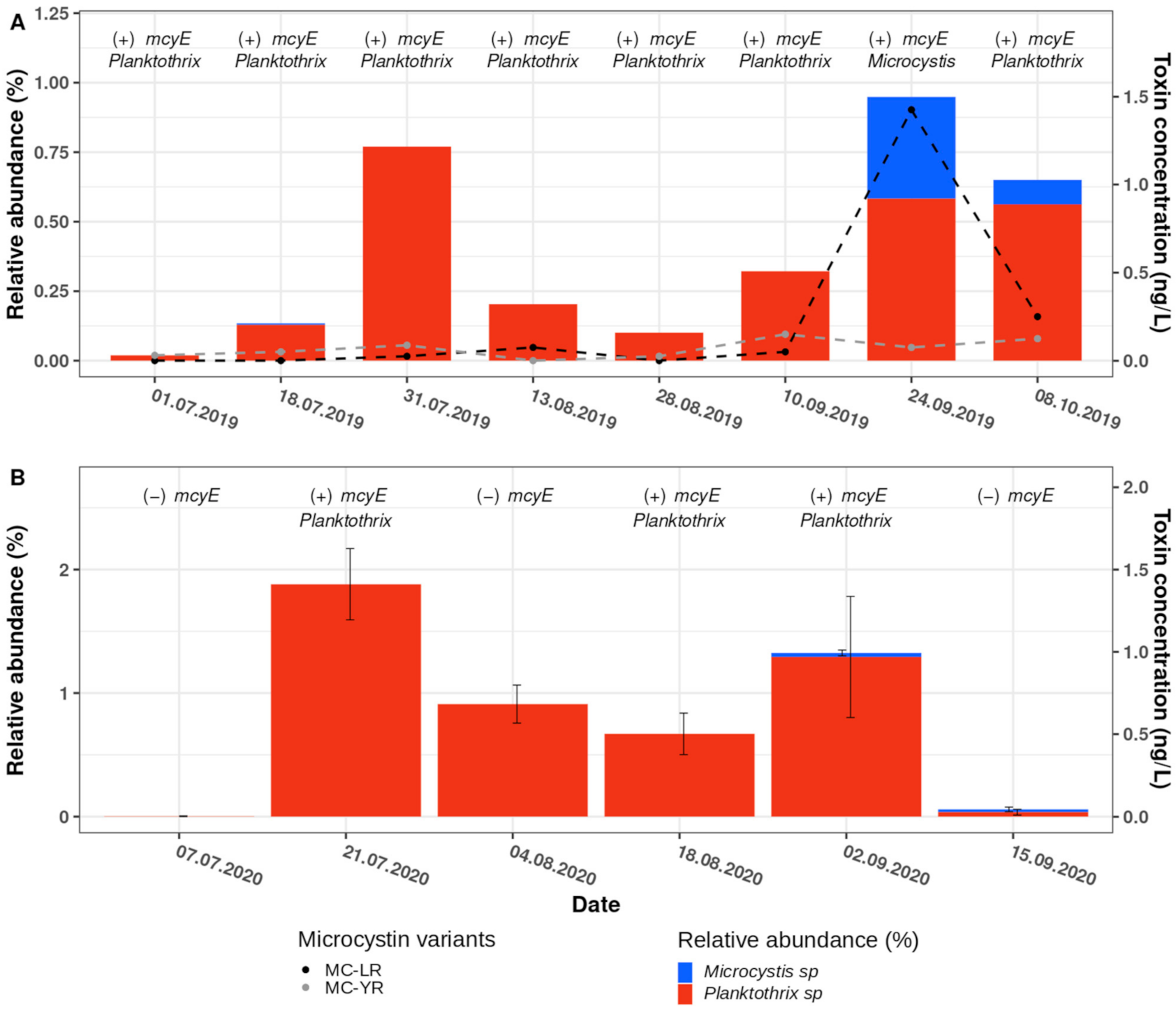
2.5. Dynamics of Planktothrix and Microcystis ASVs, the Abundance of Microcystin Biosynthesis Genes and the Concentration of Microcystins in Samples Taken during 2019 and 2020
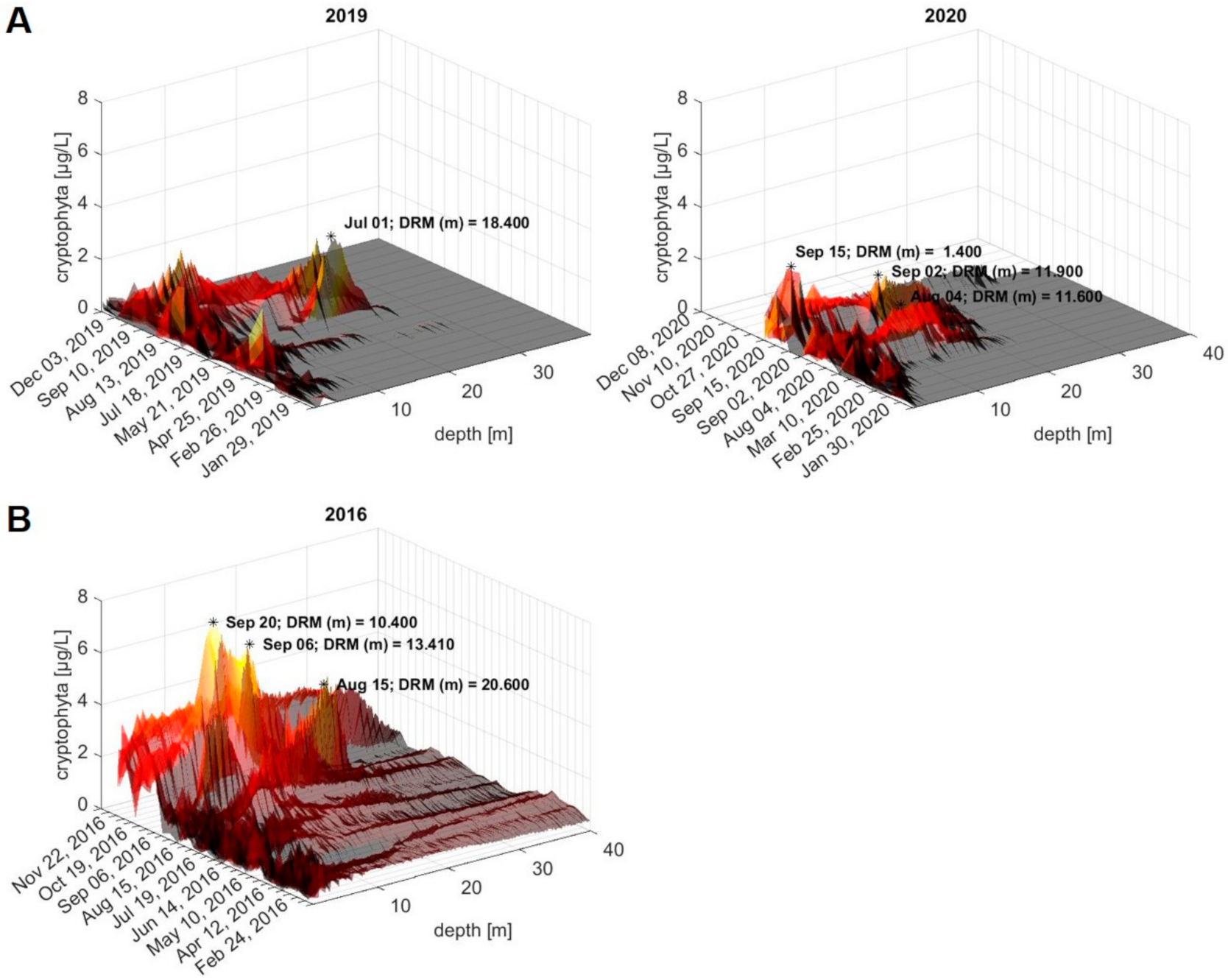
2.6. Retrospective Evaluation of Depth Profiles for the Lake Überlingen Routine Sampling Site
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sample Collection
5.2. DNA Extraction and PCR
5.3. RT-PCR/TNA
5.4. Sanger Sequencing
5.5. Amplification and Illumina Sequencing
5.6. Bioinformatics Pipeline
5.7. Phylogenetic Analysis
5.8. Statistical and Network Analysis
5.9. Toxin Extraction and Analysis
5.10. Evaluation of bbe Moldaenke FluoroProbe Data
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Mantzouki, E.; Campbell, J.; van Loon, E.; Visser, P.; Konstantinou, I.; Antoniou, M.; Giuliani, G.; Machado-Vieira, D.; de Oliveira, A.G.; Maronić, D.Š.; et al. A European multi lake survey dataset of environmental variables, phytoplankton pigments and cyanotoxins. Sci. Data 2018, 5, 1–13. [Google Scholar] [CrossRef]
- Rücker, J.; Wiedner, C.; Zippel, P. Factors controlling the dominance of Planktothrix agardhii and Limnothrix redekei in eutrophic shallow lakes. Hydrobiologia 1997, 342–343, 107–115. [Google Scholar] [CrossRef]
- Van den Wyngaert, S.; Salcher, M.M.; Pernthaler, J.; Zeder, M.; Posch, T. Quantitative dominance of seasonally persistent filamentous cyanobacteria (Planktothrix rubescens) in the microbial assemblages of a temperate lake. Limnol. Oceanogr. 2011, 56, 97–109. [Google Scholar] [CrossRef]
- Walsby, A.E.; Schanz, F. Light-dependent growth rate determines changes in the population of Planktothrix rubescens over the annual cycle in lake Zürich, Switzerland. New Phytol. 2002, 154, 671–687. [Google Scholar] [CrossRef]
- Jacquet, S.; Briand, J.F.; Leboulanger, C.; Avois-Jacquet, C.; Oberhaus, L.; Tassin, B.; Vinçon-Leite, B.; Paolini, G.; Druart, J.C.; Anneville, O.; et al. The proliferation of the toxic cyanobacterium Planktothrix rubescens following restoration of the largest natural French lake (Lac du Bourget). Harmful Algae 2005, 4, 651–672. [Google Scholar] [CrossRef]
- Salmaso, N. Long-term phytoplankton community changes in a deep subalpine lake: Responses to nutrient availability and climatic fluctuations. Freshw. Biol. 2010, 55, 825–846. [Google Scholar] [CrossRef]
- Kurmayer, R.; Christiansen, G.; Fastner, J.; Börner, T. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 2004, 6, 831–841. [Google Scholar] [CrossRef]
- Posch, T.; Köster, O.; Salcher, M.M.; Pernthaler, J. Harmful filamentous cyanobacteria favoured by reduced water turnover with lake warming. Nat. Clim. Change 2012, 2, 809–813. [Google Scholar] [CrossRef]
- Kurmayer, R.; Jüttner, F. Strategies for the co-existence of zooplankton with the toxic cyanobacterium Planktothrix rubescens in Lake Zurich. J. Plankton Res. 1999, 21, 659–683. [Google Scholar] [CrossRef]
- Kurmayer, R.; Deng, L.; Entfellner, E. Role of toxic and bioactive secondary metabolites in colonization and bloom formation by filamentous cyanobacteria Planktothrix. Harmful Algae 2016, 54, 69–86. [Google Scholar] [CrossRef]
- Salcher, M.M.; Pernthaler, J.; Frater, N.; Posch, T. Vertical and longitudinal distribution patterns of different bacterioplankton populations in a canyon-shaped, deep prealpine lake. Limnol. Oceanogr. 2011, 56, 2027–2039. [Google Scholar] [CrossRef]
- Walsby, A.E.; Ng, G.; Dunn, C.; Davis, P.A. Comparison of the depth where Planktothrix rubescens stratifies and the depth where the daily insolation supports its neutral buoyancy. New Phytol. 2004, 162, 133–145. [Google Scholar] [CrossRef]
- Cuypers, Y.; Vinçon-Leite, B.; Groleau, A.; Tassin, B.; Humbert, J.F. Impact of internal waves on the spatial distribution of Planktothrix rubescens (cyanobacteria) in an alpine lake. ISME J. 2011, 5, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Huisman, J. Climate: Blooms like it hot. Science 2008, 320, 57–58. [Google Scholar] [CrossRef]
- Wejnerowski, Ł.; Rzymski, P.; Kokociński, M.; Meriluoto, J. The structure and toxicity of winter cyanobacterial bloom in a eutrophic lake of the temperate zone. Ecotoxicology 2018, 27, 752–760. [Google Scholar] [CrossRef]
- Legnani, E.; Copetti, D.; Oggioni, A.; Tartari, G.; Palumbo, M.T.; Morabito, G. Planktothrix rubescens’ seasonal dynamics and vertical distribution in Lake Pusiano (North Italy). J. Limnol. 2005, 64, 61–73. [Google Scholar] [CrossRef]
- Walls, J.T.; Wyatt, K.H.; Doll, J.C.; Rubenstein, E.M.; Rober, A.R. Hot and toxic: Temperature regulates microcystin release from cyanobacteria. Sci. Total Environ. 2018, 610–611, 786–795. [Google Scholar] [CrossRef]
- Kleinteich, J.; Wood, S.A.; Küpper, F.C.; Camacho, A.; Quesada, A.; Frickey, T.; Dietrich, D.R. Temperature-related changes in polar cyanobacterial mat diversity and toxin production. Nat. Clim. Change 2012, 2, 356–360. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Bd. 2/Part 2: Oscillatoriales. In Süßwasserflora von Mitteleuropa, Bd. 19/2: Cyanoprokaryota; Springer: Berlin/Heidelberg, Germany, 2005; p. 759. ISBN 978-3-8274-1914-9. [Google Scholar]
- Buratti, F.M.; Manganelli, M.; Vichi, S.; Stefanelli, M.; Scardala, S.; Testai, E.; Funari, E. Cyanotoxins: Producing organisms, occurrence, toxicity, mechanism of action and human health toxicological risk evaluation. Arch. Toxicol. 2017, 91, 1049–1130. [Google Scholar] [CrossRef]
- Jang, M.H.; Ha, K.; Joo, G.J.; Takamura, N. Toxin production of cyanobacteria is increased by exposure to zooplankton. Freshw. Biol. 2003, 48, 1540–1550. [Google Scholar] [CrossRef]
- Jang, M.H.; Jung, J.M.; Takamura, N. Changes in microcystin production in cyanobacteria exposed to zooplankton at different population densities and infochemical concentrations. Limnol. Oceanogr. 2007, 52, 1454–1466. [Google Scholar] [CrossRef]
- Weisbrod, B.; Riehle, E.; Helmer, M.; Martin-Creuzburg, D.; Dietrich, D.R. Can toxin warfare against fungal parasitism influence short-term Dolichospermum bloom dynamics? A field observation. Harmful Algae 2020, 99, 101915. [Google Scholar] [CrossRef] [PubMed]
- Davis, T.W.; Berry, D.L.; Boyer, G.L.; Gobler, C.J. The effects of temperature and nutrients on the growth and dynamics of toxic and non-toxic strains of Microcystis during cyanobacteria blooms. Harmful Algae 2009, 8, 715–725. [Google Scholar] [CrossRef]
- Dziallas, C.; Grossart, H.P. Increasing oxygen radicals and water temperature select for toxic Microcystis sp. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Berry, M.A.; Davis, T.W.; Cory, R.M.; Duhaime, M.B.; Johengen, T.H.; Kling, G.W.; Marino, J.A.; Den Uyl, P.A.; Gossiaux, D.; Dick, G.J.; et al. Cyanobacterial harmful algal blooms are a biological disturbance to Western Lake Erie bacterial communities. Environ. Microbiol. 2017, 19, 1149–1162. [Google Scholar] [CrossRef]
- Bingham, M.; Sinha, S.K.; Lupi, F. Economic Benefits of Reducing Harmful Algal Blooms in Lake Erie; Environmental Consulting & Technology Inc.: Gainesville, FL, USA, 2015. [Google Scholar]
- Spoof, L.; Catherine, A. Appendix 3: Tables of microcystins and nodularins. In Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis; John Wiley & Sons, Ltd.: Chichester, UK, 2017; pp. 526–537. [Google Scholar]
- Bouaïcha, N.; Miles, C.O.; Beach, D.G.; Labidi, Z.; Djabri, A.; Benayache, N.Y.; Nguyen-Quang, T. Structural diversity, characterization and toxicology of microcystins. Toxins 2019, 11, 714. [Google Scholar] [CrossRef]
- Tillett, D.; Dittmann, E.; Erhard, M.; von Döhren, H.; Börner, T.; Neilan, B.A. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: An integrated peptide-polyketide synthetase system. Chem. Biol. 2000, 7, 753–764. [Google Scholar] [CrossRef]
- Eriksson, J.E.; Toivola, D.; Meriluoto, J.; Karaki, H.; Han, Y.G.; Hartshorne, D. Hepatocyte deformation induced by cyanobacterial toxins reflects inhibition of protein phosphatases. Biochem. Biophys. Res. Commun. 1990, 173, 1347–1353. [Google Scholar] [CrossRef]
- MacKintosh, C.; Beattie, K.A.; Klumpp, S.; Cohen, P.; Codd, G.A.; MacKintosh, C.; Codd, G.A.; Klumpp, S.; Beattie, K.A. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett. 2002, 264, 187–192. [Google Scholar] [CrossRef]
- MacKintosh, R.W.; Dalby, K.N.; Campbell, D.G.; Cohen, P.T.; Cohen, P.; MacKintosh, C. The cyanobacterial toxin microcystin binds covalently to cysteine-273 on protein phosphatase 1. FEBS Lett. 1995, 371, 236–240. [Google Scholar] [CrossRef]
- Feurstein, D.J.; Holst, K.; Fischer, A.; Dietrich, D.R. Oatp-associated uptake and toxicity of microcystins in primary murine whole brain cells. Toxicol. Appl. Pharmacol. 2009, 234, 247–255. [Google Scholar] [CrossRef]
- Fischer, A.; Hoeger, S.J.; Stemmer, K.; Feurstein, D.J.; Knobeloch, D.; Nussler, A.; Dietrich, D.R. The role of organic anion transporting polypeptides (OATPs/SLCOs) in the toxicity of different microcystin congeners in vitro: A comparison of primary human hepatocytes and OATP-transfected HEK293 cells. Toxicol. Appl. Pharmacol. 2010, 245, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Catherine, A.; Quiblier, C.; Yéprémian, C.; Got, P.; Groleau, A.; Vinçon-Leite, B.; Bernard, C.; Troussellier, M. Collapse of a Planktothrix agardhii perennial bloom and microcystin dynamics in response to reduced phosphate concentrations in a temperate lake. FEMS Microbiol. Ecol. 2008, 65, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Yéprémian, C.; Gugger, M.F.; Briand, E.; Catherine, A.; Berger, C.; Quiblier, C.; Bernard, C. Microcystin ecotypes in a perennial Planktothrix agardhii bloom. Water Res. 2007, 41, 4446–4456. [Google Scholar] [CrossRef] [PubMed]
- WHO. Cyanobacterial Toxins: Microcystins—Background Document for Development of WHO Guidelines for Drinking-Water Quality and Guidelines for Safe Recreational Water Environments; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Hoeger, S.J.; Dietrich, D.R.; Hitzfeld, B.C. Effect of ozonation on the removal of cyanobacterial toxins during drinking water treatment. Environ. Health Perspect. 2002, 110, 1127–1132. [Google Scholar] [CrossRef]
- Hoeger, S.J.; Shaw, G.; Hitzfeld, B.C.; Dietrich, D.R. Occurrence and elimination of cyanobacterial toxins in two Australian drinking water treatment plants. Toxicon 2004, 43, 639–649. [Google Scholar] [CrossRef]
- IGKB. Bericht Nr. 42: Limnologischer Zustand des Bodensees; Internationale Gewässerschutzkommission für den Bodensee (IGKB): Baden, Germany, 2018. [Google Scholar]
- Konstanz, S. Burgunderblut-Alge breitet sich im Bodensee aus. Südkurier, 28 March 2017; p. H1. [Google Scholar]
- Nübel, U.; Garcia-Pichel, F.; Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [CrossRef] [PubMed]
- Ernst, A.; Becker, S.; Wollenzien, U.I.A.; Postius, C. Ecosystem-dependent adaptive radiations of picocyanobacteria inferred from 16S rRNA and ITS-1 sequence analysis. Microbiology 2003, 149, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ostermaier, V.; Kurmayer, R. Distribution and abundance of nontoxic mutants of cyanobacteria in lakes of the Alps. Microb. Ecol. 2009, 58, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Jungblut, A.D.; Neilan, B.A. Molecular identification and evolution of the cyclic peptide hepatotoxins, microcystin and nodularin, synthetase genes in three orders of cyanobacteria. Arch. Microbiol. 2006, 185, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, G.; Fastner, J.; Erhard, M.; Börner, T.; Dittmann, E. Microcystin biosynthesis in Planktothrix: Genes, evolution, and manipulation. J. Bacteriol. 2003, 185, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Beversdorf, L.J.; Chaston, S.D.; Miller, T.R.; McMahon, K.D. Microcystin mcyA and mcyE gene abundances are not appropriate indicators of microcystin concentrations in lakes. PLoS ONE 2015, 10, e0125353. [Google Scholar] [CrossRef] [PubMed]
- Hingsamer, P.; Peeters, F.; Hofmann, H. The consequences of internal waves for phytoplankton focusing on the distribution and production of Planktothrix rubescens. PLoS ONE 2014, 9. [Google Scholar] [CrossRef][Green Version]
- Weisse, T. Dynamics of autotrophic picoplankton in lake Constance. J. Plankton Res. 1988, 10, 1179–1188. [Google Scholar] [CrossRef]
- Weisse, T.; Kenter, U. Ecological Characteristics of Autotrophic Picoplankton in a Prealpine Lake. Int. Rev. Gesamten. Hydrobiol. Hydrogr. 1991, 76, 493–504. [Google Scholar] [CrossRef]
- Six, C.; Thomas, J.C.; Garczarek, L.; Ostrowski, M.; Dufresne, A.; Blot, N.; Scanlan, D.J.; Partensky, F. Diversity and evolution of phycobilisomes in marine Synechococcus spp.: A comparative genomics study. Genome Biol. 2007, 8, R259. [Google Scholar] [CrossRef]
- Salazar, V.W.; Tschoeke, D.A.; Swings, J.; Cosenza, C.A.; Mattoso, M.; Thompson, C.C.; Thompson, F.L. A new genomic taxonomy system for the Synechococcus collective. Environ. Microbiol. 2020, 22, 4557–4570. [Google Scholar] [CrossRef]
- Tan, X.; Gu, H.; Ruan, Y.; Zhong, J.; Parajuli, K.; Hu, J. Effects of nitrogen on interspecific competition between two cell-size cyanobacteria: Microcystis aeruginosa and Synechococcus sp. Harmful Algae 2019, 89, 101661. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Gu, H.; Zhang, X.; Parajuli, K.; Duan, Z. Effects of phosphorus on interspecific competition between two cell-size cyanobacteria: Synechococcus sp. and Microcystis aeruginosa. Bull. Environ. Contam. Toxicol. 2019, 102, 231–238. [Google Scholar] [CrossRef]
- IGKB. Limnologischer Zustand des Bodensees—Jahresbericht 2018/2019. In Jahresbericht Int. Gewässerschutzkommission für den Bodensee—Grüner Bericht; Internationale Gewässerschutzkommission für den Bodensee (IGKB): Baden, Germany, 2020; Volume 43. [Google Scholar]
- Santhakumari, S.; Kannappan, A.; Pandian, S.K.; Thajuddin, N.; Rajendran, R.B.; Ravi, A.V. Inhibitory effect of marine cyanobacterial extract on biofilm formation and virulence factor production of bacterial pathogens causing vibriosis in aquaculture. J. Appl. Phycol. 2016, 28, 313–324. [Google Scholar] [CrossRef]
- Verity, P.G.; Robertson, C.Y.; Tronzo, C.R.; Andrews, M.G.; Nelson, J.R.; Sieracki, M.E. Relationships between cell volume and the carbon and nitrogen content of marine photosynthetic nanoplankton. Limnol. Oceanogr. 1992, 37, 1434–1446. [Google Scholar] [CrossRef]
- Ruber, J.; Geist, J.; Hartmann, M.; Millard, A.; Raeder, U.; Zubkov, M.; Zwirglmaier, K. Spatio-temporal distribution pattern of the picocyanobacterium Synechococcus in lakes of different trophic states: A comparison of flow cytometry and sequencing approaches. Hydrobiologia 2018, 811, 77–92. [Google Scholar] [CrossRef]
- Bodensee Wasserversorgung. Bodensee Wasserversorgung—Startseite. Available online: https://www.bodensee-wasserversorgung.de/startseite.html (accessed on 7 May 2021).
- Christiansen, G.; Kurmayer, R.; Liu, Q.; Börner, T. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl. Environ. Microbiol. 2006, 72, 117–123. [Google Scholar] [CrossRef]
- Altaner, S.; Puddick, J.; Fessard, V.; Feurstein, D.; Zemskov, I.; Wittmann, V.; Dietrich, D.R. Simultaneous Detection of 14 Microcystin Congeners from Tissue Samples Using UPLC-ESI-MS/MS and Two Different Deuterated Synthetic Microcystins as Internal Standards. Toxins 2019, 11, 388. [Google Scholar] [CrossRef] [PubMed]
- Bubak, I.; Śliwińska-Wilczewska, S.; Głowacka, P.; Szczerba, A.; Możdżeń, K. The importance of allelopathic picocyanobacterium Synechococcus sp. on the abundance, biomass formation, and structure of phytoplankton assemblages in three freshwater lakes. Toxins 2020, 12, 259. [Google Scholar] [CrossRef]
- Kovács, A.A.W.; Tóth, V.R.; Pálffy, K. The effects of interspecific interactions between bloom forming cyanobacteria and Scenedesmus quadricauda (Chlorophyta) on their photophysiology. Acta Biol. Hung. 2018, 69, 210–223. [Google Scholar] [CrossRef]
- Costa, M.S.; Costa, M.; Ramos, V.; Leão, P.N.; Barreiro, A.; Vasconcelos, V.; Martins, R. Picocyanobacteria from a Clade of Marine Cyanobium Revealed Bioactive Potential Against Microalgae, Bacteria, and Marine Invertebrates. J. Toxicol. Environ. Health 2015, 78, 432–442. [Google Scholar] [CrossRef]
- Weisbrod, B. Dynamics of Toxic Cyanobacteria in Lakes and Artificial Water Reservoirs. Ph.D. Thesis, University of Konstanz, Konstanz, Germany, 2021. [Google Scholar]
- Fernández Castro, B.; Sepúlveda Steiner, O.; Knapp, D.; Posch, T.; Bouffard, D.; Wüest, A. Inhibited vertical mixing and seasonal persistence of a thin cyanobacterial layer in a stratified lake. Aquat. Sci. 2021, 83, 38. [Google Scholar] [CrossRef]
- Rusch, D.B.; Halpern, A.L.; Sutton, G.; Heidelberg, K.B.; Williamson, S.; Yooseph, S.; Wu, D.; Eisen, J.A.; Hoffman, J.M.; Remington, K.; et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007, 5, e77. [Google Scholar] [CrossRef]
- William, S.; Helene, F.; Copeland, A. Bacterial DNA Isolation CTAB Protocol Bacterial Genomic DNA Isolation Using CTAB; JGI: Walnut Creek, CA, USA, 2012; p. 4.
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Schuurman, T.; de Boer, R.F.; Kooistra-Smid, A.M.D.; van Zwet, A.A. Prospective Study of Use of PCR Amplification and Sequencing of 16S Ribosomal DNA from Cerebrospinal Fluid for Diagnosis of Bacterial Meningitis in a Clinical Setting. J. Clin. Microbiol. 2004, 42, 734–740. [Google Scholar] [CrossRef]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 2016, 1. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Simon, A. Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 May 2020).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.; McMurdie, P.; Rosen, M.; Han, A.; Johnson, A.; Holmes, S. DADA2: High resolution sample inference from amplicon data. bioRxiv 2016, 24034. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef] [PubMed]
- R Foundation for Statistical Computing. R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package; R Package Version 2.5-6; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [PubMed]
- Zemskov, I.; Altaner, S.; Dietrich, D.R.; Wittmann, V. Total Synthesis of Microcystin-LF and Derivatives Thereof. J. Org. Chem. 2017, 82, 3680–3691. [Google Scholar] [CrossRef] [PubMed]
- MathWorks. MATLAB; Version R2020a; MathWorks: Natick, MA, USA, 2020. [Google Scholar]


| Name of Forward and Reverse Primers | Sequence (5′-3′) | Target | Reference |
|---|---|---|---|
| HEPF | TTTGGGGTTAACTTTTTTGGGCATAGTC | mcyE | Jungblut and Neilan, 2006 |
| HEPR | AATTCTTGAGGCTGTAAATCGGGTTT | ||
| 16S rDNA PTX fw | ATCCAAGTCTGCTGTTAAAGA | 16S rDNA (only Planktothrix spp.) | Ostermaier and Kurmayer, 2009 |
| 16S rDNA PTX rv | CTCTGCCCCTACTACACTCTAG | ||
| 16S rDNA PTX TaqMan 1 | AAAGGCAGTGGAAACTGGAAG | ||
| mcyBA1 PTX fw | ATTGCCGTTATCTCAAGCGAG | mcyBA1 (only Planktothrix spp.) | Ostermaier and Kurmayer, 2009 |
| mcyBA1 PTX rv | TGCTGAAAAAACTGCTGCATTAA | ||
| mcyBA1 PTX TaqMan 1 | TTTTTGTGGAGGTGAAGCTCTTTCCTCTGA | ||
| CYA359F | GGGGAATYTTCCGCAATGGG | 16S rRNA (Cyanobacteria) | Nübel et al., 1997 |
| CYA784R | ACTACWGGGGTATCTAATCCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fournier, C.; Riehle, E.; Dietrich, D.R.; Schleheck, D. Is Toxin-Producing Planktothrix sp. an Emerging Species in Lake Constance? Toxins 2021, 13, 666. https://doi.org/10.3390/toxins13090666
Fournier C, Riehle E, Dietrich DR, Schleheck D. Is Toxin-Producing Planktothrix sp. an Emerging Species in Lake Constance? Toxins. 2021; 13(9):666. https://doi.org/10.3390/toxins13090666
Chicago/Turabian StyleFournier, Corentin, Eva Riehle, Daniel R. Dietrich, and David Schleheck. 2021. "Is Toxin-Producing Planktothrix sp. an Emerging Species in Lake Constance?" Toxins 13, no. 9: 666. https://doi.org/10.3390/toxins13090666
APA StyleFournier, C., Riehle, E., Dietrich, D. R., & Schleheck, D. (2021). Is Toxin-Producing Planktothrix sp. an Emerging Species in Lake Constance? Toxins, 13(9), 666. https://doi.org/10.3390/toxins13090666





