Superantigens, a Paradox of the Immune Response
Abstract
1. Introduction
2. Staphylococcal SAgs
2.1. An Overall Description
2.2. Production and Detection of Staphylococcal SAgs
2.3. Superantigens and Human Diseases
2.3.1. Toxic Shock Syndrome
2.3.2. Infection Endocarditis
2.3.3. Pneumonia
2.3.4. Staphylococcal Food Poisoning
2.3.5. Autoimmune Pathologies
Kawasaki Disease
Diabetes Mellitus
Rheumatoid Arthritis
Atopic Dermatitis
2.4. Staphylococcal Classification, Structure and Molecular Targets
2.5. Actions of SAgs on the Immune System Cells
2.6. Superantigens as Therapeutic Tools
3. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- White, J.; Herman, A.; Pullen, A.M.; Kubo, R.; Kappler, J.W.; Marrack, P. The Vβ-specific superantigen staphylococcal enterotoxin B: Stimulation of mature T cells and clonal deletion in neonatal mice. Cell 1989, 56, 27–35. [Google Scholar] [CrossRef]
- Marrack, P.; Kappler, J. The Staphylococcal Enterotoxins and Their Relatives. Science 1990, 248, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Schutzer, S.E.; Fischetti, V.A.; Zabriskie, J.B. Toxic Shock Syndrome and Lysogeny in Staphylococcus aureus. Obstet. Gynecol. Surv. 1983, 220, 316–318. [Google Scholar]
- Quimby, F.; Nguyen, H.T.; Bergdoll, M.S. Animal studies of toxic shock syndrome. Crit. Rev. Microbiol. 1985, 12, 1–44. [Google Scholar] [CrossRef]
- Gaventa, S.; Reingold, A.L.; Hightower, A.W.; Broome, C.V.; Schwartz, B.; Hoppe, C.; Harwell, J.; Lefkowitz, L.K.; Makintubee, S.; Cundiff, D.R.; et al. Active surveillance for toxic shock syndrome in the united states, 1986. Rev. Infect. Dis. 1989, 11, S28–S34. [Google Scholar] [CrossRef]
- Blank, C.; Luz, A.; Bendigs, S.; Erdmann, A.; Wagner, H.; Heeg, K. Superantigen and endotoxin synergize in the induction of lethal shock. Eur. J. Immunol. 1997, 27, 825–833. [Google Scholar] [CrossRef]
- Dalpke, A.H.; Heeg, K. Synergistic and antagonistic interactions between LPS and superantigens. J. Endotoxin Res. 2003, 9, 51–54. [Google Scholar] [CrossRef]
- MacIsaac, C.M.; Curtis, N.; Cade, J.; Visvanathan, K. Superantigens in sepsis. Int. Congr. Ser. 2006, 1289, 121–124. [Google Scholar] [CrossRef]
- Cole, B.C.; Atkin, C.L. The Mycoplasma arthritidis T-cell mitogen MAM: A model superantigen. Immunol. Today 1991, 12, 271–276. [Google Scholar] [CrossRef]
- Igwe, E.I.; Shewmaker, P.L.; Facklam, R.R.; Farley, M.M.; Van Beneden, C.; Beall, B. Identification of superantigen genes speM, ssa, and smeZ in invasive strains of beta-hemolytic group C and G streptococci recovered from humans. FEMS Microbiol. Lett. 2003, 229, 259–264. [Google Scholar] [CrossRef]
- Abe, J.; Takeda, T.; Watanabe, Y.; Nakao, H.; Kobayashi, N.; Leung, D.Y.; Kohsaka, T. Evidence for superantigen production by Yersinia pseudotuberculosis. J. Immunol. 1993, 151, 4183. [Google Scholar] [PubMed]
- Wei, B.; Huang, T.; Dalwadi, H.; Sutton, C.L.; Bruckner, D.; Braun, J. Pseudomonas fluorescens encodes the Crohn’s disease-associated I2 sequence and T-cell superantigen. Infect. Immun. 2002, 70, 6567–6575. [Google Scholar] [CrossRef] [PubMed]
- Huber, B.T.; Hsu, P.N.; Sutkowski, N. Virus-encoded superantigens. Microbiol. Rev. 1996, 60, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Sutkowski, N.; Conrad, B.; Thorley-Lawson, D.A.; Huber, B.T. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 2001, 15, 579–589. [Google Scholar] [CrossRef]
- Azar, G.A.; Thibodeau, J. Human endogenous retrovirus IDDMK1,222 and mouse mammary tumor virus superantigens differ in their ability to stimulate murine T cell hybridomas. Immunol. Lett. 2002, 81, 87–91. [Google Scholar] [CrossRef]
- Tuffs, S.; Haeryfar, S.; McCormick, J. Manipulation of Innate and Adaptive Immunity by Staphylococcal Superantigens. Pathogens 2018, 7, 53. [Google Scholar] [CrossRef]
- Kluytmans, J.; Van Belkum, A.; Verbrugh, H. Nasal Carriage of Staphylococcus aureus: Epidemiology, Underlying Mechanisms, and Associated Risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [CrossRef]
- Laux, C.; Peschel, A.; Krismer, B. Staphylococcus aureus Colonization of the Human Nose and Interaction with Other Microbiome Members. Microbiol. Spectr. 2019, 7, GPP3-0029-2018. [Google Scholar] [CrossRef]
- Verhoeven, P.O.; Gagnaire, J.; Botelho-Nevers, E.; Grattard, F.; Carricajo, A.; Lucht, F.; Pozzetto, B.; Berthelot, P. Detection and clinical relevance of Staphylococcus aureus nasal carriage: An update. Expert Rev. Anti. Infect. Ther. 2014, 12, 75–89. [Google Scholar] [CrossRef]
- Williams, R.E. Healthy carriage of Staphylococcus aureus: Its prevalence and importance. Bacteriol. Rev. 1963, 27, 56–71. [Google Scholar] [CrossRef]
- Armstrong-Esther, C.A.; Smith, J.E. Carriage patterns of staphylococcus aureus in a healthy non-hospital population of adults and children. Ann. Hum. Biol. 1976, 3, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Gagnaire, J.; Verhoeven, P.O.; Grattard, F.; Rigaill, J.; Lucht, F.; Pozzetto, B.; Berthelot, P.; Botelho-Nevers, E. Epidemiology and clinical relevance of Staphylococcus aureus intestinal carriage: A systematic review and meta-analysis. Expert Rev. Anti. Infect. Ther. 2017, 15, 767–785. [Google Scholar] [CrossRef] [PubMed]
- Ridley, M. Perineal carriage of staph. aureus. Br. Med. J. 1959, 1, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.L.; Verveer, J.; Boelens, H.A.M.; Van Belkum, A.; Verbrugh, H.A.; Vos, M.C. Effect of mupirocin treatment on nasal, pharyngeal, and perineal carriage of Staphylococcus aureus in healthy adults. Antimicrob. Agents Chemother. 2005, 49, 1465–1467. [Google Scholar] [CrossRef] [PubMed]
- von Eiff, C.; Becker, K.; Machka, K.; Stammer, H.; Peters, G. Nasal Carriage as a Source of Staphylococcus aureus Bacteremia. N. Engl. J. Med. 2001, 344, 11–16. [Google Scholar] [CrossRef]
- Bergdoll, M.S.; Borja, C.R.; Avena, R.M. Identification of a new enterotoxin as enterotoxin C. J. Bacteriol. 1965, 90, 1481–1485. [Google Scholar] [CrossRef]
- Bergdoll, M.S.; Borja, C.R.; Robbins, R.N.; Weiss, K.F. Identification of enterotoxin E. Infect. Immun. 1971, 4, 593–595. [Google Scholar] [CrossRef]
- Casman, E.P.; Bennett, R.W.; Dorsey, A.E.; Issa, J.A. Identification of a fourth staphylococcal enterotoxin, enterotoxin D. J. Bacteriol. 1967, 94, 1875–1882. [Google Scholar] [CrossRef]
- Casman, E.P.; Bennett, R.W. Culture medium for the production of staphylococcal enterotoxin A. J. Bacteriol. 1963, 86, 18–23. [Google Scholar] [CrossRef]
- Casman, E.P.; Bergdoll, M.S.; Robinson, J. Designation of staphylococcal enterotoxins. J. Bacteriol. 1963, 85, 715–716. [Google Scholar] [CrossRef]
- Jones, C.L.; Khan, S.A. Nucleotide Sequence of the Enterotoxin B Gene from Staphylococcus aureus. J. Bacteriol. 1986, 166, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Peavy, D.L.; Adler, W.H.; Smith, R.T. The mitogenic effects of endotoxin and staphylococcal enterotoxin B on mouse spleen cells and human peripheral lymphocytes. J. Immunol. 1970, 105, 1453–1458. [Google Scholar] [PubMed]
- Spaulding, A.R.; Salgado-Pabón, W.; Kohler, P.L.; Horswill, A.R.; Leung, D.Y.M.; Schlievert, P.M. Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 2013, 26, 422–447. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Bannan, J.D.; Pancholi, V.; Cheung, A.L.; Robbins, J.C.; Fischetti, V.A.; Zabriskie, J.B. Characterization and Biological Properties of a New Staphylococcal Exotoxin. J. Exp. Med. 1994, 180, 1675–1683. [Google Scholar] [CrossRef]
- Zhang, S.; Iandolo, J.J.; Stewart, G.C. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 1998, 168, 227–233. [Google Scholar] [CrossRef]
- Munson, S.H.; Tremaine, M.T.; Betley, M.J.; Welch, R.A. Identification and Characterization of Staphylococcal Enterotoxin Types G and I from Staphylococcus aureus Identification and Characterization of Staphylococcal Enterotoxin Types G and I from Staphylococcus aureus. Infect. Immun. 1998, 66, 3337–3348. [Google Scholar] [CrossRef]
- Jarraud, S.; Peyrat, M.A.; Lim, A.; Tristan, A.; Bes, M.; Mougel, C.; Etienne, J.; Vandenesch, F.; Bonneville, M.; Lina, G. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 2001, 166, 669–677. [Google Scholar] [CrossRef]
- Fitzgerald, J.R.; Monday, S.R.; Foster, T.J.; Bohach, G.A.; Hartigan, P.J.; Meaney, W.J.; Smyth, C.J. Characterization of a Putative Pathogenicity Island from Bovine Staphylococcus aureus Encoding Multiple Superantigens. J. Bacteriol. 2001, 183, 63–70. [Google Scholar] [CrossRef]
- Orwin, P.M.; Leung, D.Y.M.; Donahue, H.L.; Novick, R.P.; Schlievert, P.M. Biochemical and Biological Properties of Staphylococcal Enterotoxin K. Infect. Immun. 2001, 69, 360–366. [Google Scholar] [CrossRef]
- Correction. J. Immunol. 2001, 166, 4260. [CrossRef]
- Letertre, C.; Perelle, S.; Dilasser, F.; Fach, P. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J. Appl. Microbiol. 2003, 95, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.Y.; Jarraud, S.; Lemercier, B.; Cozon, G.; Echasserieau, K.; Etienne, J.; Gougeon, M.L.; Lina, G.; Vandenesch, F. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect. Immun. 2006, 74, 4724–4734. [Google Scholar] [CrossRef] [PubMed]
- Omoe, K.; Ichi Imanishi, K.; Hu, D.L.; Kato, H.; Fugane, Y.; Abe, Y.; Hamaoka, S.; Watanabe, Y.; Nakane, A.; Uchiyama, T.; et al. Characterization of Novel Staphylococcal Enterotoxin-Like Toxin Type P. Infect. Immun. 2005, 73, 5540–5546. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.K.; Omoe, K.; Imanishi, K.; Iwakabe, Y.; Hu, D.L.; Kato, H.; Saito, N.; Nakane, A.; Uchiyama, T.; Shinagawa, K. Identification and Characterization of Two Novel Staphylococcal Enterotoxins, Types S and T. Infect. Immun. 2008, 76, 4999–5005. [Google Scholar] [CrossRef] [PubMed]
- Orwin, P.M.; Leung, D.Y.M.; Tripp, T.J.; Bohach, G.A.; Earhart, C.A.; Ohlendorf, D.H.; Schlievert, P.M. Characterization of a novel staphylococcal enterotoxin-like superantigen, a member of the group V subfamily of pyrogenic toxins. Biochemistry 2002, 41, 14033–14040. [Google Scholar] [CrossRef] [PubMed]
- Omoe, K.; Ichi Imanishi, K.; Hu, D.L.; Kato, H.; Takahashi-Omoe, H.; Nakane, A.; Uchiyama, T.; Shinagawa, K. Biological Properties of Staphylococcal Enterotoxin-Like Toxin Type R. Infect. Immun. 2004, 72, 3664–3667. [Google Scholar] [CrossRef]
- Okumura, K.; Shimomura, Y.; Yamagata Murayama, S.; Yagi, J.; Ubukata, K.; Kirikae, T.; Miyoshi-Akiyama, T. Evolutionary paths of streptococcal and staphylococcal superantigens. BMC Genom. 2012, 13, 404. [Google Scholar] [CrossRef]
- Wilson, G.J.; Seo, K.S.; Cartwright, R.A.; Connelley, T.; Chuang-Smith, O.N.; Merriman, J.A.; Guinane, C.M.; Park, J.Y.; Bohach, G.A.; Schlievert, P.M.; et al. A Novel Core Genome-Encoded Superantigen Contributes to Lethality of Community-Associated MRSA Necrotizing Pneumonia. PLoS Pathog. 2011, 7, e1002271. [Google Scholar] [CrossRef]
- Ono, H.K.; Sato’o, Y.; Narita, K.; Naito, I.; Hirose, S.; Hisatsune, J.; Asano, K.; Hu, D.L.; Omoe, K.; Sugai, M.; et al. Identification and characterization of a novel staphylococcal emetic toxin. Appl. Environ. Microbiol. 2015, 81, 7034–7040. [Google Scholar] [CrossRef]
- Spoor, L.E.; Richardson, E.; Richards, A.C.; Wilson, G.J.; Mendonca, C.; Gupta, R.K.; Mcadam, P.R.; Nutbeam-Tuffs, S.; Black, N.S.; O’gara, J.P.; et al. Recombination-mediated remodelling of host-pathogen interactions during Staphylococcus aureus niche adaptation. Microb. Genom. 2015, 1, e000036. [Google Scholar] [CrossRef]
- Hisatsune, J.; Hagiya, H.; Shiota, S.; Sugai, M. Complete Genome Sequence of Systemically Disseminated Sequence Type 8 Staphylococcal Cassette Chromosome mec Type IVl Community-Acquired Methicillin-Resistant Staphylococcus aureus. Genome Announc. 2017, 5, e00852-17. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Ono, H.K.; Shimojima, Y.; Kubota, H.; Kato, R.; Kakuda, T.; Hirose, S.; Hu, D.L.; Nakane, A.; Takai, S.; et al. A novel staphylococcal enterotoxin SE02 involved in a staphylococcal food poisoning outbreak that occurred in Tokyo in 2004. Food Microbiol. 2020, 92, 103588. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.F.; Yang, X.Y.; Zhang, J.; Qin, X.; Huang, X.; Cui, Y.; Zhou, M.; Shi, C.; French, N.P.; Shi, X. Identification and characterization of two novel superantigens among Staphylococcus aureus complex. Int. J. Med. Microbiol. 2018, 308, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Bohach, G.A.; Nair, S.P.; Hiramatsu, K.; Jouvin-Marche, E.; Mariuzza, R. Standard Nomenclature for the Superantigens Expressed by Staphylococcus. J. Infect. Dis. 2004, 189, 2334–2336. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Tao, X.X.; Meng, F.L.; Li, X.P.; Ono-Hisaya, O.H.; Wang, D.; Hu, D.L.; Zhang, J.Z.; Wang, G.Q.; Yan, X.M. Cloning and expression of recombinant truncated SElX protein and evaluation on the related emetic activities. Zhonghua Liu Xing Bing Xue Za Zhi 2020, 41, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.L.; Omoe, K.; Shimoda, Y.; Nakane, A.; Shinagawa, K. Induction of Emetic Response to Staphylococcal Enterotoxins in the House Musk Shrew (Suncus murinus). Infect. Immun. 2003, 71, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Matsuki, N.; Saito, H. Suncus murinus as a new experimental model for motion sickness. Life Sci. 1988, 43, 413–420. [Google Scholar] [CrossRef]
- Omoe, K.; Hu, D.L.; Takahashi-Omoe, H.; Nakane, A.; Shinagawa, K. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol. Lett. 2005, 246, 191–198. [Google Scholar] [CrossRef]
- Hu, D.L.; Ono, H.K.; Isayama, S.; Okada, R.; Okamura, M.; Lei, L.C.; Liu, Z.S.; Zhang, X.C.; Liu, M.Y.; Cui, J.C.; et al. Biological characteristics of staphylococcal enterotoxin Q and its potential risk for food poisoning. J. Appl. Microbiol. 2017, 122, 1672–1679. [Google Scholar] [CrossRef]
- Ono, H.K.; Hirose, S.; Narita, K.; Sugiyama, M.; Asano, K.; Hu, D.L.; Nakane, A. Histamine release from intestinal mast cells induced by staphylococcal enterotoxin A (SEA) evokes vomiting reflex in common marmoset. PLoS Pathog. 2019, 15, e1007803. [Google Scholar] [CrossRef]
- Ferry, T.; Thomas, D.; Genestier, A.L.; Bes, M.; Lina, G.; Vandenesch, F.; Etienne, J. Comparative Prevalence of Superantigen Genes in Staphylococcus aureus Isolates Causing Sepsis with and without Septic Shock. Clin. Infect. Dis. 2005, 41, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Noli Truant, S.; De Marzi, M.C.; Sarratea, M.B.; Antonoglou, M.B.; Meo, A.P.; Iannantuono López, L.V.; Fernández Lynch, M.J.; Todone, M.; Malchiodi, E.L.; Fernández, M.M. egc Superantigens Impair Monocytes/Macrophages Inducing Cell Death and Inefficient Activation. Front. Immunol. 2020, 10, 3008. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, X.; Zhang, M.; Kou, M.; Chang, G.; Wan, Y.; Xu, X.; Ruan, F.; Wang, Y.; Wang, X. Prevalence, Antimicrobial Resistance, and Molecular Characteristics of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus from Retail Ice Cream in Shaanxi Province, China. Foodborne Pathog. Dis. 2022, 19, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Aung, M.S.; Urushibara, N.; Kawaguchiya, M.; Ito, M.; Habadera, S.; Kobayashi, N. Prevalence and Genetic Diversity of Staphylococcal Enterotoxin (-Like) Genes sey, selw, selx, selz, sel26 and sel27 in Community-Acquired Methicillin-Resistant Staphylococcus aureus. Toxins 2020, 12, 347. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Friedrich, A.W.; Lubritz, G.; Weilert, M.; Peters, G.; Von Eiff, C. Prevalence of Genes Encoding Pyrogenic Toxin Superantigens and Exfoliative Toxins among Strains of Staphylococcus aureus Isolated from Blood and Nasal Specimens. J. Clin. Microbiol. 2003, 41, 1434–1439. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.J.; Kilgore, S.H.; Singh, S.B.; Allen, P.D.; Hansen, A.R.; Limoli, D.H.; Schlievert, P.M. High Prevalence of Staphylococcus aureus Enterotoxin Gene Cluster Superantigens in Cystic Fibrosis Clinical Isolates. Genes 2019, 10, 1036. [Google Scholar] [CrossRef]
- Park, K.H.; Greenwood-Quaintance, K.E.; Cunningham, S.A.; Chia, N.; Jeraldo, P.R.; Sampathkumar, P.; Nelson, H.; Patel, R. Lack of correlation of virulence gene profiles of Staphylococcus aureus bacteremia isolates with mortality. Microb. Pathog. 2019, 133, 103543. [Google Scholar] [CrossRef]
- Garbacz, K.; Piechowicz, L.; Podkowik, M.; Mroczkowska, A.; Empel, J.; Bania, J. Infection and Drug Resistance Dovepress emergence and spread of worldwide Staphylococcus aureus clones among cystic fibrosis patients. Infect. Drug Resist. 2018, 11, 247. [Google Scholar] [CrossRef]
- Mempel, M.; Lina, G.; Hojka, M.; Schnopp, C.; Seidl, H.P.; Schäfer, T.; Ring, J.; Vandenesch, F.; Abeck, D. High prevalence of superantigens associated with the egc locus in Staphylococcus aureus isolates from patients with atopic eczema. Eur. J. Clin. Microbiol. Infect. Dis. 2003, 22, 306–309. [Google Scholar] [CrossRef]
- Merriman, J.A.; Mueller, E.A.; Cahill, M.P.; Beck, L.A.; Paller, A.S.; Hanifin, J.M.; Ong, P.Y.; Schneider, L.; Babineau, D.C.; David, G.; et al. Temporal and Racial Differences Associated with Atopic Dermatitis Staphylococcus aureus and Encoded Virulence Factors. Msphere 2016, 1, e00295-16. [Google Scholar] [CrossRef]
- Van Belkum, A.; Melles, D.C.; Snijders, S.V.; Van Leeuwen, W.B.; Wertheim, H.F.L.; Nouwen, J.L.; Verbrugh, H.A.; Etienne, J. Clonal Distribution and Differential Occurrence of the Enterotoxin Gene Cluster, egc, in Carriage-versus Bacteremia-Associated Isolates of Staphylococcus aureus. J. Clin. Microbiol. 2006, 44, 1555–1557. [Google Scholar] [CrossRef] [PubMed]
- Bronner, S.; Monteil, H.; Prévost, G. Regulation of virulence determinants in Staphylococcus aureus: Complexity and applications. FEMS Microbiol. Rev. 2006, 28, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.L.; Bayer, A.S.; Zhang, G.; Gresham, H.; Xiong, Y.Q. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2006, 40, 1–9. [Google Scholar] [CrossRef]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 2019, 7, 7-2. [Google Scholar] [CrossRef]
- Bergdoll, M.S.; Czop, J.K.; Gould, S.S. Enterotoxin Synthesis by the Staphylococci. Ann. N. Y. Acad. Sci. 1974, 236, 307–316. [Google Scholar] [CrossRef]
- Bernardo, K.; Pakulat, N.; Fleer, S.; Schnaith, A.; Utermöhlen, O.; Krut, O.; Müller, S.; Krönke, M. Subinhibitory Concentrations of Linezolid Reduce Staphylococcus aureus Virulence Factor Expression. Antimicrob. Agents Chemother. 2004, 48, 546–555. [Google Scholar] [CrossRef]
- Czop, J.K.; Bergdoll, M.S. Staphylococcal Enterotoxin Synthesis during the Exponential, Transitional, and Stationary Growth Phases. Infect. Immun. 1974, 9, 229–235. [Google Scholar] [CrossRef]
- Gaskill, M.E.; Khan, S.A. Regulation of the enterotoxin B gene in Staphylococcus aureus. J. Biol. Chem. 1988, 263, 6276–6280. [Google Scholar] [CrossRef]
- Otero, A.; Garcia, M.L.; Garcia, M.C.; Moreno, B.; Bergdoll, M.S. Production of Staphylococcal Enterotoxins C1 and C2 and Thermonuclease throughout the Growth Cycle. Appl. Environ. Microbiol. 1990, 56, 555–559. [Google Scholar] [CrossRef]
- Regassa, L.B.; Couch, J.L.; Betley, M.J. Steady-State Staphylococcal Enterotoxin Type C mRNA Is Affected by a Product of the Accessory Gene Regulator (agr) and by Glucose. Infect. Immun. 1991, 59, 955–962. [Google Scholar] [CrossRef]
- Sihto, H.M.; Tasara, T.; Stephan, R.; Johler, S. Temporal expression of the staphylococcal enterotoxin D gene under NaCl stress conditions. FEMS Microbiol. Lett. 2015, 362, 24. [Google Scholar] [CrossRef] [PubMed]
- Macori, G.; Bellio, A.; Bianchi, D.M.; Chiesa, F.; Gallina, S.; Romano, A.; Zuccon, F.; Cabrera-Rubio, R.; Cauquil, A.; Merda, D.; et al. Genome-wide profiling of enterotoxigenic Staphylococcus aureus strains used for the production of naturally contaminated cheeses. Genes 2020, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Andrey, D.O.; Jousselin, A.; Villanueva, M.; Renzoni, A.; Monod, A.; Barras, C.; Rodriguez, N.; Kelley, W.L. Impact of the regulators SigB, rot, SarA and sarS on the toxic shock tst promoter and TSST-1 expression in Staphylococcus aureus. PLoS ONE 2015, 10, e0135579. [Google Scholar] [CrossRef] [PubMed]
- Baroja, M.L.; Herfst, C.A.; Kasper, K.J.; Xu, S.X.; Gillett, D.A.; Li, J.; Reid, G.; McCormick, J.K. The SaeRS two-component system is a direct and dominant transcriptional activator of toxic shock syndrome toxin 1 in Staphylococcus aureus. J. Bacteriol. 2016, 198, 2732–2742. [Google Scholar] [CrossRef]
- Nagao, M.; Okamoto, A.; Yamada, K.; Hasegawa, T.; Hasegawa, Y.; Ohta, M. Variations in amount of TSST-1 produced by clinical methicillin resistant Staphylococcus aureus (MRSA) isolates and allelic variation in accessory gene regulator (agr) locus. BMC Microbiol. 2009, 9, 52. [Google Scholar] [CrossRef]
- Derzelle, S.; Dilasser, F.; Duquenne, M.; Deperrois, V. Differential temporal expression of the staphylococcal enterotoxins genes during cell growth. Food Microbiol. 2009, 26, 896–904. [Google Scholar] [CrossRef]
- Stach, C.S.; Vu, B.G.; Merriman, J.A.; Herrera, A.; Cahill, M.P.; Schlievert, P.M.; Salgado-Pabón, W. Novel Tissue Level Effects of the Staphylococcus aureus Enterotoxin Gene Cluster Are Essential for Infective Endocarditis. PLoS ONE 2016, 11, e0154762. [Google Scholar] [CrossRef]
- Strandberg, K.L.; Rotschafer, J.H.; Vetter, S.M.; Buonpane, R.A.; Kranz, D.M.; Schlievert, P.M. Staphylococcal Superantigens Cause Lethal Pulmonary Disease in Rabbits. J. Infect. Dis. 2010, 202, 1690–1697. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Peterson, M.L. Glycerol Monolaurate Antibacterial Activity in Broth and Biofilm Cultures. PLoS ONE 2012, 7, e40350. [Google Scholar] [CrossRef]
- Goerke, C.; Campana, S.; Bayer, M.G.; Döring, G.; Botzenhart, K.; Wolz, C. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 2000, 68, 1304–1311. [Google Scholar] [CrossRef]
- Goerke, C.; Wolz, C. Regulatory and genomic plasticity of Staphylococcus aureus during persistent colonization and infection. Int. J. Med. Microbiol. 2004, 294, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Banks, M.C.; Kamel, N.S.; Zabriskie, J.B.; Larone, D.H.; Ursea, D.; Posnett, D.N. Staphylococcus aureus Express Unique Superantigens Depending on the Tissue Source. J. Infect. Dis. 2003, 187, 77–86. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Omoe, K.; Ishikawa, M.; Shimoda, Y.; Hu, D.L.; Ueda, S.; Shinagawa, K. Detection of seg, seh, and sei genes in Staphylococcus aureus Isolates and Determination of the Enterotoxin Productivities of S. aureus Isolates Harboring seg, seh, or sei Genes. J. Clin. Microbiol. 2002, 40, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Burlak, C.; Hammer, C.H.; Robinson, M.A.; Whitney, A.R.; Mcgavin, M.J.; Kreiswirth, B.N.; Deleo, F.R. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell. Microbiol. 2007, 9, 1172–1190. [Google Scholar] [CrossRef]
- Enany, S.; Yoshida, Y.; Magdeldin, S.; Zhang, Y.; Bo, X.; Yamamoto, T. Extensive proteomic profiling of the secretome of European community acquired methicillin resistant Staphylococcus aureus clone. Peptides 2012, 37, 128–137. [Google Scholar] [CrossRef]
- Pocsfalvi, G.; Cacace, G.; Cuccurullo, M.; Serluca, G.; Sorrentino, A.; Schlosser, G.; Blaiotta, G.; Malorni, A. Proteomic analysis of exoproteins expressed by enterotoxigenic Staphylococcus aureus strains. Proteomics 2008, 8, 2462–2476. [Google Scholar] [CrossRef]
- Enany, S.; Yoshida, Y.; Magdeldin, S.; Bo, X.; Zhang, Y.; Enany, M.; Yamamoto, T. Two dimensional electrophoresis of the exo-proteome produced from community acquired methicillin resistant Staphylococcus aureus belonging to clonal complex 80. Microbiol. Res. 2013, 168, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.S.; Siggins, M.K.; Gierula, M.; Pichon, B.; Turner, C.E.; Lynskey, N.N.; Mosavie, M.; Kearns, A.M.; Edwards, R.J.; Sriskandan, S. Identification of commonly expressed exoproteins and proteolytic cleavage events by proteomic mining of clinically relevant UK isolates of Staphylococcus aureus. Microb. Genom. 2016, 2, e000049. [Google Scholar] [CrossRef]
- Ziebandt, A.K.; Kusch, H.; Degner, M.; Jaglitz, S.; Sibbald, M.J.J.B.; Arends, J.P.; Chlebowicz, M.A.; Albrecht, D.; Pantuček, R.; Doškar, J.; et al. Proteomics uncovers extreme heterogeneity in the Staphylococcus aureus exoproteome due to genomic plasticity and variant gene regulation. Proteomics 2010, 10, 1634–1644. [Google Scholar] [CrossRef]
- Brizzio, A.A.; Tedeschi, F.A.; Zalazar, F.E. Multiplex PCR strategy for the simultaneous identification of Staphylococcus aureus and detection of staphylococcal enterotoxins in isolates from food poisoning outbreaks. Biomed. Rev. Inst. Nac. Salud 2013, 33, 122–127. [Google Scholar] [CrossRef]
- Chiang, Y.C.; Liao, W.W.; Fan, C.M.; Pai, W.Y.; Chiou, C.S.; Tsen, H.Y. PCR detection of Staphylococcal enterotoxins (SEs) N, O, P, Q, R, U, and survey of SE types in Staphylococcus aureus isolates from food-poisoning cases in Taiwan. Int. J. Food Microbiol. 2008, 121, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Hait, J.M.; Tallent, S.M.; Bennett, R.W. Screening, Detection, and Serotyping Methods for Toxin Genes and Enterotoxins in Staphylococcus Strains. J. AOAC Int. 2014, 97, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, D.; Blanco-Valle, K.; Hennekinne, J.A.; Simon, S.; Fenaille, F.; Becher, F.; Nia, Y.; Lefebvre, D.; Blanco-Valle, K.; Hennekinne, J.-A.; et al. Multiplex Detection of 24 Staphylococcal Enterotoxins in Culture Supernatant Using Liquid Chromatography Coupled to High-Resolution Mass Spectrometry. Toxins 2022, 14, 249. [Google Scholar] [CrossRef] [PubMed]
- Sarratea, M.B.; Noli, T.S.; Mitarotonda, R.; Antonoglou, M.B.; Chiappini, S.; Fernández Lynch, M.J.; Romasanta, P.; Vescina, C.; Desimone, M.; De Marzi, M.; et al. Optimized surface plasmon resonance immunoassay for staphylococcal enterotoxin G detection using silica nanoparticles. Biochem. Biophys. Res. Commun. 2021, 558, 168–174. [Google Scholar] [CrossRef]
- Gupta, G.; Singh, P.K.; Boopathi, M.; Kamboj, D.V.; Singh, B.; Vijayaraghavan, R. Surface plasmon resonance detection of biological warfare agent Staphylococcal enterotoxin B using high affinity monoclonal antibody. Thin Solid Film 2010, 519, 1171–1177. [Google Scholar] [CrossRef]
- Hait, J.M.; Nguyen, A.T.; Tallent, S.M. Analysis of the VIDAS® Staph Enterotoxin III (SET3) for detection of staphylococcal enterotoxins G, H, and I in foods. J. AOAC Int. 2018, 101, 1482–1489. [Google Scholar] [CrossRef]
- Nagaraj, S.; Ramlal, S.; Kingston, J.; Batra, H.V. Development of IgY based sandwich ELISA for the detection of staphylococcal enterotoxin G (SEG), an egc toxin. Int. J. Food Microbiol. 2016, 237, 136–141. [Google Scholar] [CrossRef]
- Wu, S.; Duan, N.; Gu, H.; Hao, L.; Ye, H.; Gong, W.; Wang, Z. A Review of the Methods for Detection of Staphylococcus aureus Enterotoxins. Toxins 2016, 8, 176. [Google Scholar] [CrossRef]
- Rasooly, L.; Rasooly, A. Real time biosensor analysis of Staphylococcal enterotoxin A in food. Int. J. Food Microbiol. 1999, 49, 119–127. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Blomster, D.A. Production of Staphylococcal Pyrogenic Exotoxin Type C: Influence of Physical and Chemical Factors. J. Infect. Dis. 1983, 147, 236–242. [Google Scholar] [CrossRef]
- Hajjeh, R.A.; Reingold, A.; Weil, A.; Shutt, K.; Schuchat, A.; Perkins, B.A. Hajjeh Toxic Shock Syndrome in the United States: Surveillance Update, 1979–1996. Emerg. Infect. Dis. 1996, 5, 807. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.P. Toxic-Shock Syndrome Epidemiologic Features, Recurrence, Risk Factors, and Prevention. N. Engl. J. Med. 1986, 314, 144–149. [Google Scholar] [CrossRef]
- Lee, V.T.P.; Chang, A.H.; Chow, A.W. Detection of Staphylococcal Enterotoxin B among Toxic Shock Syndrome (TSS)-and Non-TSS-Associated Staphylococcus aureus Isolates. J. Infect. Dis. 1992, 166, 911–915. [Google Scholar] [CrossRef] [PubMed]
- De Vries, A.S.; Lesher, L.; Schlievert, P.M.; Rogers, T.; Villaume, L.G.; Danila, R.; Lynfield, R. Staphylococcal toxic shock syndrome 2000–2006: Epidemiology, clinical features, and molecular characteristics. PLoS ONE 2011, 6, e22997. [Google Scholar] [CrossRef]
- Bohach, G.A.; Fast, D.J.; Nelson, R.D.; Schlievert, P.M. Staphylococcal and Streptococcal Pyrogenic Toxins Involved in Toxic Shock Syndrome and Related Illnesses. Crit. Rev. Microbiol. 1990, 17, 251–272. [Google Scholar] [CrossRef]
- Chang, A.; Musser, J.; Chow, A.W. A single clone of S. aureus which produces both TSST-1 and SEA causes the majority of menstrual toxic shock syndrome. In Clinical Research; Slack Inc.: Thorofare, NJ, USA, 1991. [Google Scholar]
- Jarraud, S.; Cozon, G.; Vandenesch, O.I.S.; Etienne, J.; Lina, G. Involvement of Enterotoxins G and I in Staphylococcal Toxic Shock Syndrome and Staphylococcal Scarlet Fever. J. Clin. Microbiol. 1999, 37, 2446–2449. [Google Scholar] [CrossRef]
- Peterson, M.L.; Ault, K.; Kremer, M.J.; Klingelhutz, A.J.; Davis, C.C.; Squier, C.A.; Schlievert, P.M. The innate immune system is activated by stimulation of vaginal epithelial cells with Staphylococcus aureus and toxic shock syndrome toxin 1. Infect. Immun. 2005, 73, 2164–2174. [Google Scholar] [CrossRef]
- Asano, K.; Narita, K.; Hirose, S.; Nakane, A. Contribution of toxic shock syndrome toxin-1 to systemic inflammation investigated by a mouse model of cervicovaginal infection with Staphylococcus aureus. Med. Microbiol. Immunol. 2018, 207, 297–306. [Google Scholar] [CrossRef]
- Asano, K.; Asano, Y.; Ono, H.K.; Nakane, A. Suppression of starvation-induced autophagy by recombinant toxic shock syndrome toxin-1 in epithelial cells. PLoS ONE 2014, 9, e113018. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Davis, C.C. Device-Associated Menstrual Toxic Shock Syndrome. Clin. Microbiol. Rev. 2020, 33, e00032-19. [Google Scholar] [CrossRef]
- Billon, A.; Gustin, M.-P.; Tristan, A.; Bénet, T.; Berthiller, J.; Gustave, C.A.; Vanhems, P.; Lina, G. Association of characteristics of tampon use with menstrual toxic shock syndrome in France. EClinicalMedicine 2020, 21, 100308. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.C.; Baccam, M.; Mantz, M.J.; Osborn, T.W.; Hill, D.R.; Squier, C.A. Use of porcine vaginal tissue ex-vivo to model environmental effects on vaginal mucosa to toxic shock syndrome toxin-1. Toxicol. Appl. Pharmacol. 2014, 274, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Nonfoux, L.; Chiaruzzi, M.; Badiou, C.; Baude, J.; Tristan, A.; Thioulouse, J.; Muller, D.; Prigent-Combaret, C.; Lina, G. Impact of currently marketed tampons and menstrual cups on Staphylococcus aureus growth and toxic shock syndrome toxin 1 production in vitro. Appl. Environ. Microbiol. 2018, 84, e00351-18. [Google Scholar] [CrossRef]
- Schlievert, P.M. Effect of non-absorbent intravaginal menstrual/contraceptive products on Staphylococcus aureus and production of the superantigen TSST-1. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Tierno, P.M.; Hanna, B.A. Propensity of Tampons and Barrier Contraceptives to Amplify Staphylococcus aureus Toxic Shock Syndrome Toxin-1. Infect. Dis. Obstet. Gynecol. 1994, 2, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, T.; Accolla, R.S.; MacDonald, H.R. Different staphylococcal enterotoxins bind preferentially to distinct major histocompatibility complex class ii isotypes. Eur. J. Immunol. 1989, 19, 2171–2174. [Google Scholar] [CrossRef] [PubMed]
- Reingold, A.L.; Hargrett, N.T.; Dan, B.B.; Shands, K.N.; Strickland, B.Y.; Broome, C.V.; Atlanta, G. Nonmenstrual Toxic Shock Syndrome A Review of 130 Cases. Ann. Intern. Med. 1982, 96, 871–874. [Google Scholar] [CrossRef]
- Whiting, J.L.; Rosten, P.M.; Chow, A.W. Determination by Western blot (immunoblot) of seroconversions to toxic shock syndrome (TSS) toxin 1 and enterotoxin A, B, or C during infection with TSS- and Non-TSS-associated Staphylococcus aureus. Infect. Immun. 1989, 57, 231–234. [Google Scholar] [CrossRef]
- Ferro, J.M.; Fonseca, A.C. Infective endocarditis. Handb. Clin. Neurol. 2014, 119, 75–91. [Google Scholar] [CrossRef]
- Fowler, V.G.; Olsen, M.K.; Corey, G.R.; Cheng, A.C.; Dudley, T.; Oddone, E.Z. Clinical Identifiers of Complicated S. aureus bacteremia. Arch. Intern. Med. 2003, 163, 2066–2072. [Google Scholar] [CrossRef]
- Chung, J.W.; Karau, M.J.; Greenwood-Quaintance, K.E.; Ballard, A.D.; Tilahun, A.; Khaleghi, S.R.; David, C.S.; Patel, R.; Rajagopalan, G. Superantigen profiling of Staphylococcus aureus infective endocarditis isolates. Diagn. Microbiol. Infect. Dis. 2014, 79, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Kinney, K.J.; Tran, P.M.; Gibson-Corley, K.N.; Forsythe, A.N.; Kulhankova, K.; Salgado-Pabón, W. Staphylococcal Enterotoxin C promotes Staphylococcus aureus Infective Endocarditis Independent of Superantigen Activity. bioRxiv 2019, 14. [Google Scholar] [CrossRef]
- Salgado-Pabón, W.; Injury, A.K.; Peterson, M.L.; Schlievert, M. Superantigens Are Critical for Staphylococcus aureus Infective Endocarditis, Sepsis, and Acute Kidney Injury. MBio 2013, 4, e00494-13. [Google Scholar] [CrossRef]
- Kulhankova, K. The Superantigen Toxic Shock Syndrome Toxin 1 Alters Human Aortic Endothelial Cell Function. Infect. Immun. 2018, 86, e00848-17. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Kerr, E. Staphylococcus aureus pneumonia: A “superbug” infection in community and hospital settings. Chest 2005, 128, 99–111. [Google Scholar] [CrossRef]
- Torres, A.; Menéndez, R.; Wundernik, R.G. Bacterial pneumonia and lung abscess. Murray Nadel’s Textb. Respir. Med. 2020, 557–582. [Google Scholar] [CrossRef]
- Hyvernat, H.; Pulcini, C.; Carles, D.; Roques, A.; Lucas, P.; Hofman, V.; Hofman, P.; Bernardin, G. Fatal Staphylococcus aureus haemorrhagic pneumonia producing Panton-Valentine leucocidin. Scand. J. Infect. Dis. 2007, 39, 183–185. [Google Scholar] [CrossRef]
- Ippolito, G.; Leone, S.; Lauria, F.N.; Nicastri, E.; Wenzel, R.P. Methicillin-resistant Staphylococcus aureus: The superbug. Int. J. Infect. Dis. 2010, 14, S7–S11. [Google Scholar] [CrossRef]
- Aliberti, S.; Reyes, L.F.; Faverio, P.; Sotgiu, G.; Dore, S.; Rodriguez, A.H.; Soni, N.J.; Restrepo, M.I.; Aruj, P.K.; Attorri, S.; et al. Global initiative for meticillin-resistant Staphylococcus aureus pneumonia (GLIMP): An international, observational cohort study. Lancet Infect. Dis. 2016, 16, 1364–1376. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. JAMA 1999, 282, 1123–1125. [Google Scholar]
- Strandberg, K.L.; Rotschafer, J.H.; Schlievert, P.M. Extreme Pyrexia and Rapid Death Due to Staphylococcus aureus Infection: Analysis of 2 Cases. Clin. Infect. Dis. 2009, 48, 612–614. [Google Scholar] [CrossRef]
- Kulhankova, K.; King, J.; Salgado-Pabón, W. Staphylococcal toxic shock syndrome: Superantigen-mediated enhancement of endotoxin shock and adaptive immune suppression. Immunol. Res. 2014, 59, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Parker, D.; Ryan, C.L.; Alonzo, F.; Torres, V.J.; Planet, P.J.; Prince, A.S. CD4+ T cells promote the pathogenesis of Staphylococcus aureus pneumonia. J. Infect. Dis. 2015, 211, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Chao, G.; Bao, G.; Cao, Y.; Yan, W.; Wang, Y.; Zhang, X.; Zhou, L.; Wu, Y. Prevalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int. J. Food Microbiol. 2015, 211, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Johler, S.; Giannini, P.; Jermini, M.; Hummerjohann, J.; Baumgartner, A.; Stephan, R. Further evidence for staphylococcal food poisoning outbreaks caused by egc-Encoded enterotoxins. Toxins 2015, 7, 997–1004. [Google Scholar] [CrossRef]
- Lv, G.P.; Xu, B.H.; Wei, P.N.; Song, J.; Zhang, H.Y.; Zhao, C.; Qin, L.Y.; Zhao, B.H. Molecular characterization of foodborne-associated Staphylococcus aureus strains isolated in Shijiazhuang, China, from 2010 to 2012. Diagn. Microbiol. Infect. Dis. 2014, 78, 462–468. [Google Scholar] [CrossRef]
- Shen, M.; Li, Y.; Zhang, L.; Dai, S.; Wang, J.; Li, Y.; Zhang, L.; Huang, J. Staphylococcus enterotoxin profile of China isolates and the superantigenicity of some novel enterotoxins. Arch. Microbiol. 2017, 199, 723–736. [Google Scholar] [CrossRef]
- Wattinger, L.; Stephan, R.; Layer, F.; Johler, S. Comparison of staphylococcus aureus isolates associated with food intoxication with isolates from human nasal carriers and human infections. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 455–464. [Google Scholar] [CrossRef]
- Yan, X.; Wang, B.; Tao, X.; Hu, Q.; Cui, Z.; Zhang, J.; Lin, Y.; You, Y.; Shi, X.; Grundmann, H. Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl. Environ. Microbiol. 2012, 78, 6637–6642. [Google Scholar] [CrossRef]
- Blaiotta, G.; Ercolini, D.; Pennacchia, C.; Fusco, V.; Casaburi, A.; Pepe, O.; Villani, F. PCR detection of staphylococcal enterotoxin genes in Staphylococcus spp. strains isolated from meat and dairy products. Evidence for new variants of seG and seI in S. aureus AB-8802. J. Appl. Microbiol. 2004, 97, 719–730. [Google Scholar] [CrossRef]
- McLauchlin, J.; Narayanan, G.L.; Mithani, V.; O’Neill, G. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. 2000, 63, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Viçosa, G.N.; Le Loir, A.; Le Loir, Y.; de Carvalho, A.F.; Nero, L.A. Egc characterization of enterotoxigenic Staphylococcus aureus isolates obtained from raw milk and cheese. Int. J. Food Microbiol. 2013, 165, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Tamate, N.; Yamaguchi, K.; Makino, S.I. Mass outbreak of food poisoning disease caused by small amounts of staphylococcal enterotoxins A and H. Appl. Environ. Microbiol. 2005, 71, 2793–2795. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, H.J.; Mathisen, T.; Løvseth, A.; Omoe, K.; Qvale, K.S.; Loncarevic, S. An outbreak of staphylococcal food poisoning caused by enterotoxin H in mashed potato made with raw milk. FEMS Microbiol. Lett. 2005, 252, 267–272. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, M.F.; Luz, I.D.S.; Silveira-Filho, V.D.M.; Júnior, J.W.P.; Stamford, T.L.; Mota, R.A.; de Sena, M.J.; de Almeida, A.M.P.; de Q. Balbino, V.; Leal-Balbino, T.C. Staphylococcal toxin genes in strains isolated from cows with subclinical mastitis. Pesqui. Veterinária Bras. 2008, 28, 617–621. [Google Scholar] [CrossRef][Green Version]
- Bianchi, D.M.; Gallina, S.; Bellio, A.; Chiesa, F.; Civera, T.; Decastelli, L. Enterotoxin gene profiles of Staphylococcus aureus isolated from milk and dairy products in Italy. Lett. Appl. Microbiol. 2014, 58, 190–196. [Google Scholar] [CrossRef]
- Carfora, V.; Caprioli, A.; Marri, N.; Sagrafoli, D.; Boselli, C.; Giacinti, G.; Giangolini, G.; Sorbara, L.; Dottarelli, S.; Battisti, A.; et al. Enterotoxin genes, enterotoxin production, and methicillin resistance in Staphylococcus aureus isolated from milk and dairy products in Central Italy. Int. Dairy J. 2015, 42, 12–15. [Google Scholar] [CrossRef]
- Rosec, J.P.; Guiraud, J.P.; Dalet, C.; Richard, N. Enterotoxin production by staphylococci isolated from foods in France. Int. J. Food Microbiol. 1997, 35, 213–221. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, A.; Tang, J.; Tang, C.; Chen, J.; Liu, J. Identification and measurement of staphylococcal enterotoxin-like protein I (SEll) secretion from Staphylococcus aureus clinical isolate. J. Appl. Microbiol. 2016, 121, 539–546. [Google Scholar] [CrossRef]
- Cheng, F.H.M.; Andrews, P.L.R.; Moreaux, B.; Ngan, M.P.; Rudd, J.A.; Sam, T.S.W.; Wai, M.K.; Wan, C. Evaluation of the anti-emetic potential of anti-migraine drugs to prevent resiniferatoxin-induced emesis in Suncus murinus (house musk shrew). Eur. J. Pharmacol. 2005, 508, 231–238. [Google Scholar] [CrossRef]
- Hu, D.L.; Zhu, G.; Mori, F.; Omoe, K.; Okada, M.; Wakabayashi, K.; Kaneko, S.; Shinagawa, K.; Nakane, A. Staphylococcal enterotoxin induces emesis through increasing serotonin release in intestine and it is downregulated by cannabinoid receptor 1. Cell. Microbiol. 2007, 9, 2267–2277. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, H.; Hayama, T. Abdominal viscera as site of emetic action for staphylococcal enterotoxin in the monkey. J. Infect. Dis. 1965, 115, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Shupp, J.W.; Jett, M.; Pontzer, C.H. Identification of a transcytosis epitope on staphylococcal enterotoxins. Infect. Immun. 2002, 70, 2178–2186. [Google Scholar] [CrossRef] [PubMed]
- Spiekermann, G.M.; Anderson, C.N. Oral Administration of the Bacterial Superantigen Staphylococcal Enterotoxin B Induces Activation and Cytokine Production by T Cells in Murine Gut-Associated Lymphoid Tissue. J. Immunol. 1998, 161, 5825–5831. [Google Scholar] [CrossRef]
- McKay, D.M.; Benjamin, M.A.; Lu, J. CD4+ T cells mediate superantigen-induced abnormalities in murine jejunal ion transport. Am. J. Physiol.-Gastrointest. Liver Physiol. 1998, 275, 29–38. [Google Scholar] [CrossRef]
- Pinchuk, I.V.; Beswick, E.J.; Reyes, V.E. Staphylococcal enterotoxins. Toxins 2010, 2, 2177–2197. [Google Scholar] [CrossRef]
- Friedman, S.M.; Posnett, D.N.; Tumang, J.R.; Crow, M.K.; Cole, B.C. A potential role for microbial superantigens in the pathogenesis of systemic autoimmune disease. Arthritis Rheum. 1991, 34, 468–480. [Google Scholar] [CrossRef]
- Wucherpfennig, K.W. Mechanisms for the induction of autoimmunity by infectious agents. J. Clin. Investig. 2001, 108, 1097–1104. [Google Scholar] [CrossRef]
- Schiffenbauer, J.; Soos, J.; Johnson, H. The possible role of bacterial superantigens in the pathogenesis of autoimmune disorders. Immunol. Today 1998, 19, 117–120. [Google Scholar] [CrossRef]
- Dietz, S.M.; van Stijn, D.; Burgner, D.; Levin, M.; Kuipers, I.M.; Hutten, B.A.; Kuijpers, T.W. Dissecting Kawasaki disease: A state-of-the-art review. Eur. J. Pediatr. 2017, 176, 995–1009. [Google Scholar] [CrossRef]
- Matsubara, K.; Fukaya, T. The role of superantigens of group A Streptococcus and Staphylococcus aureus in Kawasaki disease. Curr. Opin. Infect. Dis. 2007, 20, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S. Causes of Kawasaki disease—From past to present. Front. Pediatr. 2019, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yeung, R.S.M. The etiology of Kawasaki disease: A superantigen-mediated process. Prog. Pediatr. Cardiol. 2004, 19, 115–122. [Google Scholar] [CrossRef]
- Nagata, S.; Yamashiro, Y.; Ohtsuka, Y.; Shimizu, T.; Sakurai, Y.; Misawa, S.; Ito, T. Heat shock proteins and superantigenic properties of bacteria from the gastrointestinal tract of patients with Kawasaki disease. Immunology 2009, 128, 511–520. [Google Scholar] [CrossRef]
- Hall, M.; Schlievert, P.M. Kawasaki Syndrome-Like Illness Associated with Infection Caused by Enterotoxin B-Secreting Staphylococcus aureus. Clin. Infect. Dis. 1999, 29, 586–589. [Google Scholar] [CrossRef]
- Leung, D.Y.M.; Meissner, H.C.; Fulton, D.; Murray, D.L.; Kotzin, B.; Schlievert, P.M. Toxic shock syndrome toxin-secreting Staphylococcus aureus in Kawasaki syndrome. Lancet 1994, 343, 300. [Google Scholar] [CrossRef]
- Leung, D.Y.M.; Meissner, H.C.; Shulman, S.T.; Mason, W.H.; Gerber, M.A.; Glode, M.P.; Myones, B.L.; Wheeler, J.G.; Ruthazer, R.; Schlievert, P.M. Prevalence of superantigen-secreting bacteria in patients with Kawasaki disease. J. Pediatr. 2002, 140, 742–746. [Google Scholar] [CrossRef]
- Matsubara, K.; Fukaya, T.; Miwa, K.; Shibayama, N.; Nigami, H.; Harigaya, H.; Nozaki, H.; Hirata, T.; Baba, K.; Suzuki, T.; et al. Development of serum IgM antibodies against superantigens of Staphylococcus aureus and Streptococcus pyogenes in Kawasaki disease. Clin. Exp. Immunol. 2006, 143, 427–434. [Google Scholar] [CrossRef]
- Nomura, Y.; Yoshinaga, M.; Masuda, K.; Takei, S.; Miyata, K. Maternal Antibody against Toxic Shock Syndrome Toxin–1 May Protect Infants Younger than 6 Months of Age from Developing Kawasaki Syndrome. J. Infect. Dis. 2002, 185, 1677–1680. [Google Scholar] [CrossRef]
- Terai, M.; Miwa, K.; Williams, T.; Kabat, W.; Fukuyama, M.; Okajima, Y.; Igarashi, H.; Shulman, S.T. The absence of evidence of staphylococcal toxin involvement in the pathogenesis of kawasaki disease. J. Infect. Dis. 1995, 172, 558–561. [Google Scholar] [CrossRef]
- Yim, D.; Ramsay, J.; Kothari, D.; Burgner, D. Coronary artery dilatation in toxic shock-like syndrome: The kawasaki disease shock syndrome. Pediatr. Cardiol. 2010, 31, 1232–1235. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.G.; Stach, C.S.; Kulhankova, K.; Salgado-Pabón, W.; Klingelhutz, A.J.; Schlievert, P.M. Chronic superantigen exposure induces systemic inflammation, elevated bloodstream endotoxin, and abnormal glucose tolerance in rabbits: Possible role in diabetes. MBio 2015, 6, e02554-14. [Google Scholar] [CrossRef] [PubMed]
- Banke, E.; Rödström, K.; Ekelund, M.; Dalla-Riva, J.; Lagerstedt, J.O.; Nilsson, S.; Degerman, E.; Lindkvist-Petersson, K.; Nilson, B. Superantigen activates the gp130 receptor on adipocytes resulting in altered adipocyte metabolism. Metabolism 2014, 63, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.G.; Gourronc, F.A.; Bernlohr, D.A.; Schlievert, P.M.; Klingelhutz, A.J. Staphylococcal superantigens stimulate immortalized human adipocytes to produce chemokines. PLoS ONE 2013, 8, e77988. [Google Scholar] [CrossRef]
- Paliard, X.; West, S.G.; Lafferty, J.A.; Clements, J.R.; Kappler, J.W.; Marrack, P.; Kotzin, B.L. Evidence for the effects of a superantigen in rheumatoid arthritis. Science 1991, 253, 325–329. [Google Scholar] [CrossRef]
- Goodacre, J.A.; Brownlie, C.E.D.; Ross, D.A. Bacterial superantigens in autoimmune arthritis. Rheumatology 1994, 33, 413–419. [Google Scholar] [CrossRef]
- Ataee, R.A.; Ataee, M.H.; Alishiri, G.H.; Esmaeili, D. Staphylococcal enterotoxin C in synovial fluid of patients with rheumatoid arthritis. Iran. Red Crescent Med. J. 2014, 16, e16075. [Google Scholar] [CrossRef][Green Version]
- Ataee, R.A.; Kahani, M.S.; Alishiri, G.H.; Ahamadi, Z. Staphylococcal Enterotoxin A Detection from Rheumatoid Arthritis Patients’ Blood and Synovial Fluid Ramezan. Electron. Physician 2016, 8, 1850. [Google Scholar] [CrossRef][Green Version]
- Ataee, R.A.; Kashefi, R.; Alishiri, G.H.; Esmaieli, D. Staphylococcus aureus enterotoxin D: Absence of bacteria but presence of its toxin. Jundishapur J. Microbiol. 2015, 8, e28395. [Google Scholar] [CrossRef]
- Rashki, M.; Ataee, R.A.; Alishiri, G.H.; Esmaeili, D. Molecular Assay of Staphylococcal Enterotoxin E in Synovial Fluid of Patients with Rheumatoid Arthritis. Int. J. Infect. 2018, 5, 1–4. [Google Scholar] [CrossRef]
- Shokrollahi, M.R.; Noorbakhsh, S.; Aliakbari, M.; Tabatabaei, A. Searching the staphylococcal superantigens: Enterotoxins A, B, C, and TSST1 in synovial fluid of cases with negative culture inflammatory arthritis. Jundishapur J. Microbiol. 2014, 7, e11647. [Google Scholar] [CrossRef] [PubMed]
- Tabarya, D.; Hoffman, W.L. Staphylococcus aureus nasal carriage in rheumatoid arthritis: Antibody response to toxic shock syndrome toxin-1. Ann. Rheum. Dis. 1996, 55, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Origuchi, T.; Eguchi, K.; Kawabe, Y.; Yamashita, I.; Mizokami, A.; Ida, H.; Nagataki, S. Increased levels of serum IgM antibody to staphylococcal enterotoxin B in patients with rheumatoid arthritis. Ann. Rheum. Dis. 1995, 54, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, K.; Tomuschat, C.; Finke, R.; Staege, M.S.; Brütting, C.; Brandt, J.; Jordan, B.; Schwesig, R.; Rosemeier, A.; Delank, K.S.; et al. Experimental Arthritis in the Rat Induced by the Superantigen Staphylococcal Enterotoxin A. Scand. J. Immunol. 2017, 85, 191–196. [Google Scholar] [CrossRef]
- Banaei, M.; Alishiri, G.H.; Ataee, R.A.; Mahalati, A.H. Rheumatoid arthritis mediator CD18 expression by Staphylococcus aureus superantigen C in rats. Iran. J. Microbiol. 2019, 11, 337–344. [Google Scholar] [CrossRef]
- Abdelnour, A.; Bremell, T.; Holmdahl, R.; Tarkowski, A. Clonal expansion of T lymphocytes causes arthritis and mortality in mice infected with toxic shock syndrome toxin-1-producing staphylococci. Eur. J. Immunol. 1994, 24, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Wooley, P.H.; Cingel, B. Staphylococcal enterotoxin B increases the severity of type II collagen induced arthritis in mice. Ann. Rheum. Dis. 1995, 54, 298–304. [Google Scholar] [CrossRef]
- Schwab, J.H.; Brown, R.R.; Anderle, S.K.; Schlievert, P.M. Superantigen can reactivate bacterial cell wall-induced arthritis. J. Immunol. 1993, 150, 4151–4159. [Google Scholar]
- Tsai, C.; Diaz, L.A.; Singer, N.G.; Li, L.L.; Kirsch, A.H.; Mitra, R.; Nickoloff, B.J.; Crofford, L.J.; Fox, D.A. Responsiveness of human T lymphocytes to bacterial superantigens presented by cultured rheumatoid arthritis synoviocytes. Arthritis Rheum. 1996, 39, 125–136. [Google Scholar] [CrossRef]
- Leung, D.Y.M.; Harbeck, R.; Bina, P.; Reiser, R.F.; Yang, E.; Norris, D.A.; Hanifin, J.M.; Sampson, H.A. Presence of IgE antibodies to staphylococcal exotoxins on the skin of patients with atopic dermatitis evidence for a new group of allergens. J. Clin. Investig. 1993, 92, 1374–1380. [Google Scholar] [CrossRef]
- Leyden, J.J.; Marples, R.R.; Kligman, A.M. Staphylococcus aureus in the lesions of atopic dermatitis. Br. J. Dermatol. 1974, 90, 525. [Google Scholar] [CrossRef] [PubMed]
- Skov, L.; Baadsgaard, O. Bacterial superantigens and inflammatory skin diseases. Clin. Exp. Dermatol. 2000, 25, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Jappe, U. Superantigens and their association with dermatological inflammatory diseases: Facts and hypotheses. Acta Derm. Venereol. 2000, 80, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Bunikowski, R.; Mielke, M.; Skarabis, H.; Herz, U.; Bergmann, R.L.; Wahn, U.; Renz, H. Prevalence and role of serum IgE antibodies to the Staphylococcus aureus-derived superantigens SEA and SEB in children with atopic dermatitis. J. Allergy Clin. Immunol. 1999, 103, 119–124. [Google Scholar] [CrossRef]
- Lin, Y.T.; Yang, Y.H.; Hwang, Y.W.; Tsai, M.J.; Tsao, P.N.; Chiang, B.L.; Shau, W.Y.; Wang, L.F. Comparison of serum specific IgE antibodies to staphylococcal enterotoxins between atopic children with and without atopic dermatitis. Allergy Eur. J. Allergy Clin. Immunol. 2000, 55, 641–646. [Google Scholar] [CrossRef]
- Tomczak, H.; Wróbel, J.; Jenerowicz, D.; Sadowska-Przytocka, A.; Wachal, M.; Adamski, Z.; Czarnecka-Operacz, M.M. The role of Staphylococcus aureus in atopic dermatitis: Microbiological and immunological implications. Postep. Dermatol. Alergol. 2019, 36, 485–491. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Case, L.C.; Strandberg, K.L.; Abrams, B.B.; Leung, D.Y.M. Superantigen profile of Staphylococcus aureus isolates from patients with steroid-resistant atopic dermatitis. Clin. Infect. Dis. 2008, 46, 1562–1567. [Google Scholar] [CrossRef]
- Strickland, I.; Hauk, P.J.; Trumble, A.E.; Picker, L.J.; Leung, D.Y.M. Evidence for superantigen involvement in skin homing of T cells in atopic dermatitis. J. Investig. Dermatol. 1999, 112, 249–253. [Google Scholar] [CrossRef]
- McFadden, J.P.; Noble, W.C.; Camp, R.D.R. Superantigenic exotoxin-secreting potential of staphylococci isolated from atopic eczematous skin. Br. J. Dermatol. 1993, 128, 631–632. [Google Scholar] [CrossRef]
- Arad, G.; Levy, R.; Nasie, I.; Hillman, D.; Rotfogel, Z.; Barash, U.; Supper, E.; Shpilka, T.; Minis, A.; Kaempfer, R. Binding of superantigen toxins into the CD28 homodimer interface is essential for induction of cytokine genes that mediate lethal shock. PLoS Biol. 2011, 9, e1001149. [Google Scholar] [CrossRef]
- Ramachandran, G.; Tulapurkar, M.E.; Harris, K.M.; Arad, G.; Shirvan, A.; Shemesh, R.; Detolla, L.J.; Benazzi, C.; Opal, S.M.; Kaempfer, R.; et al. A peptide antagonist of CD28 signaling attenuates toxic shock and necrotizing soft-tissue infection induced by streptococcus pyogenes. J. Infect. Dis. 2013, 207, 1869–1877. [Google Scholar] [CrossRef] [PubMed]
- Levy, R.; Rotfogel, Z.; Hillman, D.; Popugailo, A.; Arad, G.; Supper, E.; Osman, F.; Kaempfer, R. Superantigens hyperinduce inflammatory cytokines by enhancing the B7-2/CD28 costimulatory receptor interaction. Proc. Natl. Acad. Sci. USA 2016, 113, E6437–E6446. [Google Scholar] [CrossRef] [PubMed]
- Ganem, M.B.; De Marzi, M.C.; Fernández-Lynch, M.J.; Jancic, C.; Vermeulen, M.; Geffner, J.; Mariuzza, R.A.; Fernández, M.M.; Malchiodi, E.L. Uptake and Intracellular Trafficking of Superantigens in Dendritic Cells. PLoS ONE 2013, 8, e66244. [Google Scholar] [CrossRef] [PubMed]
- Hovde, C.J.; Marr, J.C.; Hoffmann, M.L.; Hackett, S.P.; Chi, Y.I.; Crum, K.K.; Stevens, D.L.; Stauffacher, C.V.; Bohach, G.A. Investigation of the role of the disulphide bond in the activity and structure of staphylococcal enterotoxin C1. Mol. Microbiol. 1994, 13, 897–909. [Google Scholar] [CrossRef]
- Baker, M.D.; Acharya, K.R. Superantigens: Structure-function relationships. Int. J. Med. Microbiol. 2004, 293, 529–537. [Google Scholar] [CrossRef]
- Petersson, K.; Håkansson, M.; Nilsson, H.; Forsberg, G.; Svensson, L.A.; Liljas, A.; Walse, B. Crystal structure of a superantigen bound to MHC class II displays zinc and peptide dependence. EMBO J. 2001, 20, 3306–3312. [Google Scholar] [CrossRef]
- Jardetzky, T.S.; Brown, J.H.; Gorga, J.C.; Stern, L.J.; Urban, R.G.; Chi, Y.I.; Stauffacher, C.; Strominger, J.L.; Wiley, D.C. Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen. Nature 1994, 368, 711–718. [Google Scholar] [CrossRef]
- Fernández, M.M.; Guan, R.; Swaminathan, C.P.; Malchiodi, E.L.; Mariuzza, R.A. Crystal structure of staphylococcal enterotoxin I (SEI) in complex with a human major histocompatibility complex class II molecule. J. Biol. Chem. 2006, 281, 25356–25364. [Google Scholar] [CrossRef]
- Leder, L.; Llera, A.; Lavoie, P.M.; Lebedeva, M.I.; Li, H.; Sékaly, R.P.; Bohach, G.A.; Gahr, P.J.; Schlievert, P.M.; Karjalainen, K.; et al. A Mutational Analysis of the Binding of Staphylococcal Enterotoxins B and C3 to the T Cell Receptor Beta Chain and Major Histocompatibility Complex Class II. J. Exp. Med. 1998, 187, 823–833. [Google Scholar] [CrossRef]
- Fernández, M.M.; Cho, S.; De Marzi, M.C.; Kerzic, M.C.; Robinson, H.; Mariuzza, R.A.; Malchiodi, E.L. Crystal structure of Staphylococcal Enterotoxin G (SEG) in complex with a mouse T-cell receptor β chain. J. Biol. Chem. 2011, 286, 1189–1195. [Google Scholar] [CrossRef]
- Fernández, M.M.; De Marzi, M.C.; Berguer, P.; Burzyn, D.; Langley, R.J.; Piazzon, I.; Mariuzza, R.A.; Malchiodi, E.L. Binding of natural variants of staphylococcal superantigens SEG and SEI to TCR and MHC class II molecule. Mol. Immunol. 2006, 43, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Saline, M.; Rödström, K.E.J.; Fischer, G.; Orekhov, V.Y.; Karlsson, B.G.; Lindkvist-Petersson, K. The structure of superantigen complexed with TCR and MHC reveals novel insights into superantigenic T cell activation. Nat. Commun. 2010, 1, 119. [Google Scholar] [CrossRef] [PubMed]
- Popugailo, A.; Rotfogel, Z.; Supper, E.; Hillman, D.; Kaempfer, R. Staphylococcal and streptococcal superantigens trigger B7/CD28 costimulatory receptor engagement to hyperinduce inflammatory cytokines. Front. Immunol. 2019, 10, 942. [Google Scholar] [CrossRef]
- Gregory, S.; Zilber, M.T.; Charron, D.; Gelin, C. Human CD1a Molecule Expressed on Monocytes Plays an Accessory Role in the Superantigen-induced Activation of T Lymphocytes. Hum. Immunol. 2000, 61, 193–201. [Google Scholar] [CrossRef]
- Uzunçayır, S.; Vera-Rodriguez, A.; Regenthal, P.; Åbacka, H.; Emanuelsson, C.; Bahl, C.D.; Lindkvist-Petersson, K. Analyses of the complex formation of staphylococcal enterotoxin A and the human gp130 cytokine receptor. FEBS Lett. 2022, 596, 910–923. [Google Scholar] [CrossRef] [PubMed]
- Schlievert, P.M.; Cahill, M.P.; Hostager, B.S.; Brosnahan, A.J.; Klingelhutz, A.J.; Gourronc, F.A.; Bishop, G.A.; Leung, D.Y.M. Staphylococcal superantigens stimulate epithelial cells through CD40 to produce chemokines. MBio 2019, 10, e00214-19. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Gourronc, F.A.; Leung, D.Y.M.; Klingelhutz, A.J. Human Keratinocyte Response to Superantigens. mSphere 2020, 5, e00803-20. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Kilgore, S.H.; Benavides, A.; Klingelhutz, A.J. Pathogen Stimulation of Interleukin-8 from Human Vaginal Epithelial Cells through CD40. Microbiol. Spectr. 2022, 10, e00106-22. [Google Scholar] [CrossRef]
- Garcia, K.C.; Teyton, L.; Wilson, I.A. Structural basis of T cell recognition. Annu. Rev. Immunol. 1999, 17, 369–397. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.C.; Adams, E.J. How the T cell receptor sees antigen—A structural view. Cell 2005, 122, 333–336. [Google Scholar] [CrossRef]
- Smith-Garvin, J.E.; Koretzky, G.A.; Jordan, M.S. T cell activation. Annu. Rev. Immunol. 2009, 27, 592–619. [Google Scholar] [CrossRef] [PubMed]
- Givan, A.L.; Fisher, J.L.; Waugh, M.; Ernstoff, M.S.; Wallace, P.K. A flow cytometric method to estimate the precursor frequencies of cells proliferating in response to specific antigens. J. Immunol. Methods 1999, 230, 99–112. [Google Scholar] [CrossRef]
- Bueno, C.; Criado, G.; McCormick, J.K.; Madrenas, J. T Cell Signalling Induced by Bacterial Superantigens. In Superantigens and Superallergens; KARGER: Basel, Switzerland, 2007; pp. 161–180. [Google Scholar]
- Mccormick, J.K.; Yarwood, J.M.; Schlievert, P.M. Toxic shock syndrome and bacterial superantigens: An update. Annu. Rev. Microbiol. 2001, 55, 77. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, Y.; Ochi, A. Programmed cell death and extrathymic reduction of Vβ8+ CD4+ T cells in mice tolerant to Staphylococcus aureus enterotoxin B. Nature 1991, 349, 245–248. [Google Scholar] [CrossRef]
- MacDonald, H.R.; Baschieri, S.; Lees, R.K. Clonal expansion precedes anergy and death of Vβ8+ peripheral T cells responding to staphylococcal enterotoxin B in vivo. Eur. J. Immunol. 1991, 21, 1963–1966. [Google Scholar] [CrossRef] [PubMed]
- Sundstedt, A.; Höiden, I.; Rosendahl, A.; Kalland, T.; van Rooijen, N.; Dohlsten, M. Immunoregulatory role of IL-10 during superantigen-induced hyporesponsiveness in vivo. J. Immunol. 1997, 158, 180–186. [Google Scholar]
- Taylor, A.L.; Llewelyn, M.J. Superantigen-Induced Proliferation of Human CD4 + CD25− T Cells Is Followed by a Switch to a Functional Regulatory Phenotype. J. Immunol. 2010, 185, 6591–6598. [Google Scholar] [CrossRef]
- Feunou, P.; Poulin, L.; Habran, C.; Le Moine, A.; Goldman, M.; Braun, M.Y. CD4+ CD25+ and CD4+ CD25− T Cells Act Respectively as Inducer and Effector T Suppressor Cells in Superantigen-Induced Tolerance. J. Immunol. 2003, 171, 3475–3484. [Google Scholar] [CrossRef]
- Lee, J.; Park, N.; Park, J.Y.; Kaplan, B.L.F.; Pruett, S.B.; Park, J.W.; Ho Park, Y.; Seo, K.S. Induction of Immunosuppressive CD8+ CD25+ FOXP3+ Regulatory T Cells by Suboptimal Stimulation with Staphylococcal Enterotoxin C1. J. Immunol. 2018, 200, 669–680. [Google Scholar] [CrossRef]
- Taylor, A.L.; Cross, E.L.A.; Llewelyn, M.J. Induction of contact-dependent CD8+ regulatory T cells through stimulation with staphylococcal and streptococcal superantigens*. Immunology 2012, 135, 158–167. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Orlikowsky, T.; Dudhane, A.; Trejo, V.; Dannecker, G.E.; Pernist, B.; Hoffmann, M.K. Staphylococcal enterotoxin B-induced T-cell anergy is mediated by regulatory T cells. Immunology 1998, 94, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Tilahun, A.Y.; Chowdhary, V.R.; David, C.S.; Rajagopalan, G. Systemic Inflammatory Response Elicited by Superantigen Destabilizes T Regulatory Cells Rendering them Ineffective during Toxic Shock Syndrome NIH Public Access. J. Immunol. 2014, 193, 2919–2930. [Google Scholar] [CrossRef] [PubMed]
- Parker, D.; Karauzum, H.; Sähr, A.; Förmer, S.; Hildebrand, D.; Heeg, K. T-cell activation or tolerization: The Yin and Yang of bacterial superantigens. Front. Microbiol. 2015, 6, 1153. [Google Scholar] [CrossRef][Green Version]
- Kato, M.; Nakamura, Y.; Suda, T.; Ozawa, Y.; Inui, N.; Seo, N.; Nagata, T.; Koide, Y.; Kalinski, P.; Nakamura, H.; et al. Enhanced anti-tumor immunity by superantigen-pulsed dendritic cells. Cancer Immunol. Immunother. 2011, 60, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Friedman, S.M.; Cole, B.C.; Nisanian, A.J. Dendritic Cells Are Potent Antigen-presenting Cells for Microbial Superantigens. J. Exp. Med. 1992, 175, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Muraille, E.; De Trez, C.; Pajak, B.; Brait, M.; Urbain, J.; Leo, O. T Cell-Dependent Maturation of Dendritic Cells in Response to Bacterial Superantigens. J. Immunol. 2002, 168, 4352–4360. [Google Scholar] [CrossRef] [PubMed]
- Coutant, K.D.; De Fraissinette, A.D.B.; Cordier, A.; Ulrich, P. Modulation of the activity of human monocyte-derived dendritic cells by chemical haptens, a metal allergen, and a staphylococcal superantigen. Toxicol. Sci. 1999, 52, 189–198. [Google Scholar] [CrossRef]
- Liu, T.; He, S.H.; Zheng, P.Y.; Zhang, T.Y.; Wang, B.Q.; Yang, P.C. Staphylococcal enterotoxin B increases TIM4 expression in human dendritic cells that drives naïve CD4 T cells to differentiate into Th2 cells. Mol. Immunol. 2007, 44, 3580–3587. [Google Scholar] [CrossRef]
- Muralimohan, G.; Vella, A.T. A role for IFNγ in differential superantigen stimulation of conventional versus plasmacytoid DCs. Cell. Immunol. 2006, 23, 9–22. [Google Scholar] [CrossRef]
- Takahashi, M.; Takahashi, M.; Shinohara, F.; Takada, H.; Rikiishi, H. Effects of superantigen and lipopolysaccharide on induction of CD80 through apoptosis of human monocytes. Infect. Immun. 2001, 69, 3652–3657. [Google Scholar] [CrossRef][Green Version]
- Zhang, X.; Shang, W.; Yuan, J.; Hu, Z.; Peng, H.; Zhu, J.; Hu, Q.; Yang, Y.; Liu, H.; Jiang, B.; et al. Positive Feedback Cycle of TNFα Promotes Staphylococcal Enterotoxin B-Induced THP-1 Cell Apoptosis. Front. Cell. Infect. Microbiol. 2016, 6, 109. [Google Scholar] [CrossRef]
- Ulett, G.C.; Adderson, E.E. Regulation of Apoptosis by Gram-Positive Bacteria: Mechanistic Diversity and Consequences for Immunity. Curr. Immunol. Rev. 2006, 2, 119–141. [Google Scholar] [CrossRef]
- Krakauer, T. Staphylococcal Superantigens: Pyrogenic Toxins Induce Toxic Shock. Toxins 2019, 11, 178. [Google Scholar] [CrossRef] [PubMed]
- Grumann, D.; Scharf, S.S.; Holtfreter, S.; Kohler, C.; Steil, L.; Engelmann, S.; Hecker, M.; Völker, U.; Bröker, B.M.; Volker, U.; et al. Immune Cell Activation by Enterotoxin Gene Cluster (egc)-Encoded and Non-egc Superantigens from Staphylococcus aureus. J. Immunol. 2008, 181, 5054–5061. [Google Scholar] [CrossRef] [PubMed]
- Islander, U.; Andersson, A.; Lindberg, E.; Adlerberth, I.; Wold, A.E.; Rudin, A. Superantigenic Staphylococcus aureus stimulates production of interleukin-17 from memory but not naive T cells. Infect. Immun. 2010, 78, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Jupin, C.; Anderson, S.; Damais, C.; Alouf, J.E.; Parant, M. Toxic shock syndrome toxin 1 as an inducer of human tumor necrosis factors and λ interferon. J. Exp. Med. 1988, 167, 752–761. [Google Scholar] [CrossRef]
- Kum, W.W.S.; Cameron, S.B.; Hung, R.W.Y.; Kalyan, S.; Chow, A.W. Temporal Sequence and Kinetics of Proinflammatory and Anti-Inflammatory Cytokine Secretion Induced by Toxic Shock Syndrome Toxin 1 in Human Peripheral Blood Mononuclear Cells. Infect. Immun. 2001, 69, 7544–7549. [Google Scholar] [CrossRef]
- Rust, C.J.; Koning, F. Gamma delta T cell reactivity towards bacterial superantigens. Semin. Immunol. 1993, 5, 41–46. [Google Scholar] [CrossRef]
- Fisher, E.L.; Otto, M.; Cheung, G.Y.C. Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Brandes, M.; Willimann, K.; Moser, B. Immunology: Professional antigen-presentation function by human γδ cells. Science 2005, 309, 264–268. [Google Scholar] [CrossRef]
- Maeurer, M.; Zitvogel, L.; Elder, E.; Storkus, W.J.; Lotze, M.T. Human intestinal V delta 1+ T cells obtained from patients with colon cancer respond exclusively to SEB but not to SEA. Nat. Immun. 1995, 14, 188–197. [Google Scholar] [PubMed]
- Morita, C.T.; Li, H.; Lamphear, J.G.; Rich, R.R.; Fraser, J.D.; Mariuzza, R.A.; Lee, H.K. Superantigen recognition by gamma delta T cells: SEA recognition site for human Vgamma2 T cell receptors. Immunity 2001, 14, 331–344. [Google Scholar] [CrossRef]
- Hayworth, J.L.; Mazzuca, D.M.; Vareki, S.M.; Welch, I.; McCormick, J.K.; Haeryfar, S.M. CD1d-independent activation of mouse and human i NKT cells by bacterial superantigens. Immunol. Cell Biol. 2012, 90, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Rieder, S.A.; Nagarkatti, P.; Nagarkatti, M. CD1d-independent activation of invariant natural killer T cells by Staphylococcal enterotoxin B through major histocompatibility complex class II/T cell receptor interaction results in acute lung injury. Infect. Immun. 2011, 79, 3141–3148. [Google Scholar] [CrossRef] [PubMed]
- Szabo, P.A.; Rudak, P.T.; Choi, J.; Xu, S.X.; Schaub, R.; Singh, B.; McCormick, J.K.; Haeryfar, S.M.M. Invariant Natural Killer T Cells Are Pathogenic in the HLA-DR4-Transgenic Humanized Mouse Model of Toxic Shock Syndrome and Can Be Targeted to Reduce Morbidity. J. Infect. Dis. 2017, 215, 824–829. [Google Scholar] [CrossRef][Green Version]
- Stohl, W.; Elliott, J.E.; Linsley, P.S. Human T cell-dependent B cell differentiation induced by staphylococcal superantigens. J. Immunol. 1994, 153, 117–127. [Google Scholar]
- Domiati-Saad, R.; Attrep, J.F.; Brezinschek, H.P.; Cherrie, A.H.; Karp, D.R.; Lipsky, P.E. Staphylococcal enterotoxin D functions as a human B cell superantigen by rescuing VH4-expressing B cells from apoptosis. J. Immunol. 1996, 156, 3608–3620. [Google Scholar]
- Domiati-Saad, R.; Lipsky, P.E. Staphylococcal Enterotoxin A Induces Survival of V H 3 Expressing Human B Cells by Binding to the V H Region with Low Affinity. J. Immunol. 1998, 161, 1257–1266. [Google Scholar]
- Gould, H.J.; Takhar, P.; Harries, H.E.; Chevretton, E.; Sutton, B.J. The allergic March from Staphylococcus aureus superantigens to immunoglobulin E. Chem. Immunol. Allergy 2007, 93, 106–136. [Google Scholar] [CrossRef]
- Shin, S.Y.; Choi, G.S.M.; Lee, K.H.; Kim, S.W.; Cho, J.S.; Park, H.S. IgE Response to Staphylococcal Enterotoxins in Adenoid Tissues from Atopic Children. Laryngoscope 2009, 119, 171–175. [Google Scholar] [CrossRef]
- Lotfi-Emran, S.; Ward, B.R.; Le, Q.T.; Pozez, A.L.; Manjili, M.H.; Woodfolk, J.A.; Schwartz, L.B. Human mast cells present antigen to autologous CD4+ T cells. J. Allergy Clin. Immunol. 2018, 141, 311–321.e10. [Google Scholar] [CrossRef]
- Ono, H.K.; Nishizawa, M.; Yamamoto, Y.; Hu, D.L.; Nakane, A.; Shinagawa, K.; Omoe, K. Submucosal mast cells in the gastrointestinal tract are a target of staphylococcal enterotoxin type A. FEMS Immunol. Med. Microbiol. 2012, 64, 392–402. [Google Scholar] [CrossRef]
- Scheuber, P.H.; Denzlinger, C.; Wilker, D.; Beck, G.; Keppler, D.; Hammer, D.K. Staphylococcal enterotoxin B as a nonimmunological mast cell stimulus in primates: The role of endogenous cysteinyl leukotrienes. Int. Arch. Allergy Immunol. 1987, 82, 289–291. [Google Scholar] [CrossRef]
- Shaler, C.R.; Choi, J.; Rudak, P.T.; Memarnejadian, A.; Szabo, P.A.; Tun-Abraham, M.E.; Rossjohn, J.; Corbett, A.J.; McCluskey, J.; McCormick, J.K.; et al. MAIT cells launch a rapid, robust and distinct hyperinflammatory response to bacterial superantigens and quickly acquire an anergic phenotype that impedes their cognate antimicrobial function: Defining a novel mechanism of superantigen-induced immunopathology and immunosuppression. PLoS Biol. 2017, 15, e2001930. [Google Scholar] [CrossRef]
- Godfrey, D.I.; Uldrich, A.P.; Mccluskey, J.; Rossjohn, J.; Moody, D.B. The burgeoning family of unconventional T cells. Nat. Publ. Gr. 2015, 16, 1114–1123. [Google Scholar] [CrossRef]
- D’Souza, C.; Chen, Z.; Corbett, A.J. Revealing the protective and pathogenic potential of MAIT cells. Mol. Immunol. 2018, 103, 46–54. [Google Scholar] [CrossRef]
- Godfrey, D.I.; Koay, H.F.; McCluskey, J.; Gherardin, N.A. The biology and functional importance of MAIT cells. Nat. Immunol. 2019, 20, 1110–1128. [Google Scholar] [CrossRef]
- Langley, R.; Patel, D.; Jackson, N.; Clow, F.; Fraser, J.D. Staphylococcal superantigen super-domains in immune evasion. Crit. Rev. Immunol. 2010, 30, 149–165. [Google Scholar] [CrossRef]
- Langley, R.J.; Ting, Y.T.; Clow, F.; Young, P.G.; Radcliff, F.J.; Choi, J.M.; Sequeira, R.P.; Holtfreter, S.; Baker, H.; Fraser, J.D. Staphylococcal enterotoxin-like X (SElX) is a unique superantigen with functional features of two major families of staphylococcal virulence factors. PLoS Pathog. 2017, 13, e1006549. [Google Scholar] [CrossRef]
- Tuffs, S.W.; James, D.B.A.; Bestebroer, J.; Richards, A.C.; Goncheva, M.I.; O’Shea, M.; Wee, B.A.; Seo, K.S.; Schlievert, P.M.; Lengeling, A.; et al. The Staphylococcus aureus superantigen SElX is a bifunctional toxin that inhibits neutrophil function. PLoS Pathog. 2017, 13, e1006461. [Google Scholar] [CrossRef]
- Spaan, A.N.; Surewaard, B.G.J.; Nijland, R.; van Strijp, J.A.G. Neutrophils versus Staphylococcus aureus: A Biological Tug of War. Annu. Rev. Microbiol. 2013, 67, 629–650. [Google Scholar] [CrossRef]
- Chen, J.Y. Superantigens, superantigen-like proteins and superantigen derivatives for cancer treatment. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1622–1630. [Google Scholar]
- Liu, X.; Zeng, L.; Zhao, Z.; He, J.; Xie, Y.; Xiao, L.; Wang, S.; Zhang, J.; Zou, Z.; He, Y.; et al. PBMC activation via the ERK and STAT signaling pathways enhances the anti-tumor activity of Staphylococcal enterotoxin A. Mol. Cell. Biochem. 2017, 434, 75–87. [Google Scholar] [CrossRef]
- Perabo, F.G.E.; Willert, P.L.; Wirger, A.; Schmidt, D.H.; Von Ruecker, A.; Mueller, S.C. Superantigen-activated Mononuclear Cells Induce Apoptosis in Transitional Cell Carcinoma. Anticancer Res. 2005, 25, 3565–3573. [Google Scholar]
- Kodama, H.; Suzuki, M.; Katayose, Y.; Shinoda, M.; Sakurai, N.; Takemura, S.I.; Yoshida, H.; Saeki, H.; Ichiyama, M.; Tsumoto, K.; et al. Mutated SEA-D227A-conjugated antibodies greatly enhance antitumor activity against MUC1-expressing bile duct carcinoma. Cancer Immunol. Immunother. 2001, 50, 539–548. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, Y.M.; Xu, M.K.; Song, Z.H.; Li, C.Y.; Wang, H.R.; Dai, H.H.; Zhang, Z.P.; Liu, C.X. Anti-tumor activity and immunogenicity of a mutated staphylococcal enterotoxin C2. Pharmazie 2013, 68, 359–364. [Google Scholar] [CrossRef]
- Forsberg, G.; Skartved, N.J.; Wallén-Öhman, M.; Nyhlén, H.C.; Behm, K.; Hedlund, G.; Nederman, T. Naptumomab estafenatox, an engineered antibody-superantigen fusion protein with low toxicity and reduced antigenicity. J. Immunother. 2010, 33, 492–499. [Google Scholar] [CrossRef]
- Golob-Urbanc, A.; Rajčević, U.; Strmšek, Ž.; Jerala, R. Design of split superantigen fusion proteins for cancer immunotherapy. J. Biol. Chem. 2019, 294, 6294–6305. [Google Scholar] [CrossRef]
- Knopick, P.; Terman, D.; Riha, N.; Alvine, T.; Larson, R.; Badiou, C.; Lina, G.; Ballantyne, J.; Bradley, D. Endogenous HLA-DQ8αβ programs superantigens (SEG/SEI) to silence toxicity and unleash a tumoricidal network with long-term melanoma survival. J. Immunother. Cancer 2020, 8, e001493. [Google Scholar] [CrossRef]
- Antonoglou, M.B.; Sánchez Alberti, A.; Redolfi, D.M.; Bivona, A.E.; Fernández Lynch, M.J.; Noli Truant, S.; Sarratea, M.B.; López, L.V.I.; Malchiodi, E.L.; Fernández, M.M. Heterologous Chimeric Construct Comprising a Modified Bacterial Superantigen and a Cruzipain Domain Confers Protection Against Trypanosoma cruzi Infection. Front. Immunol. 2020, 1, 1279. [Google Scholar] [CrossRef]
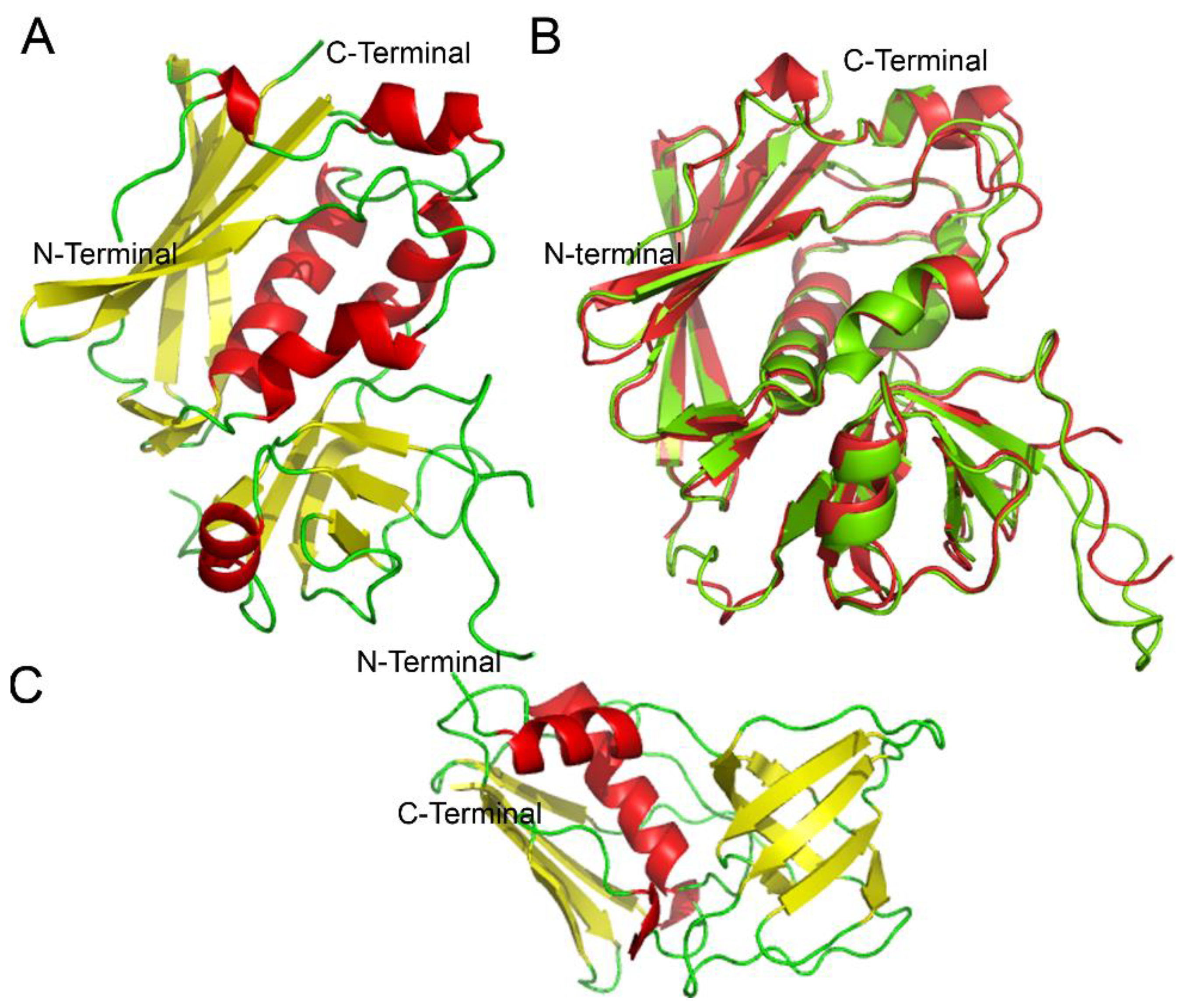

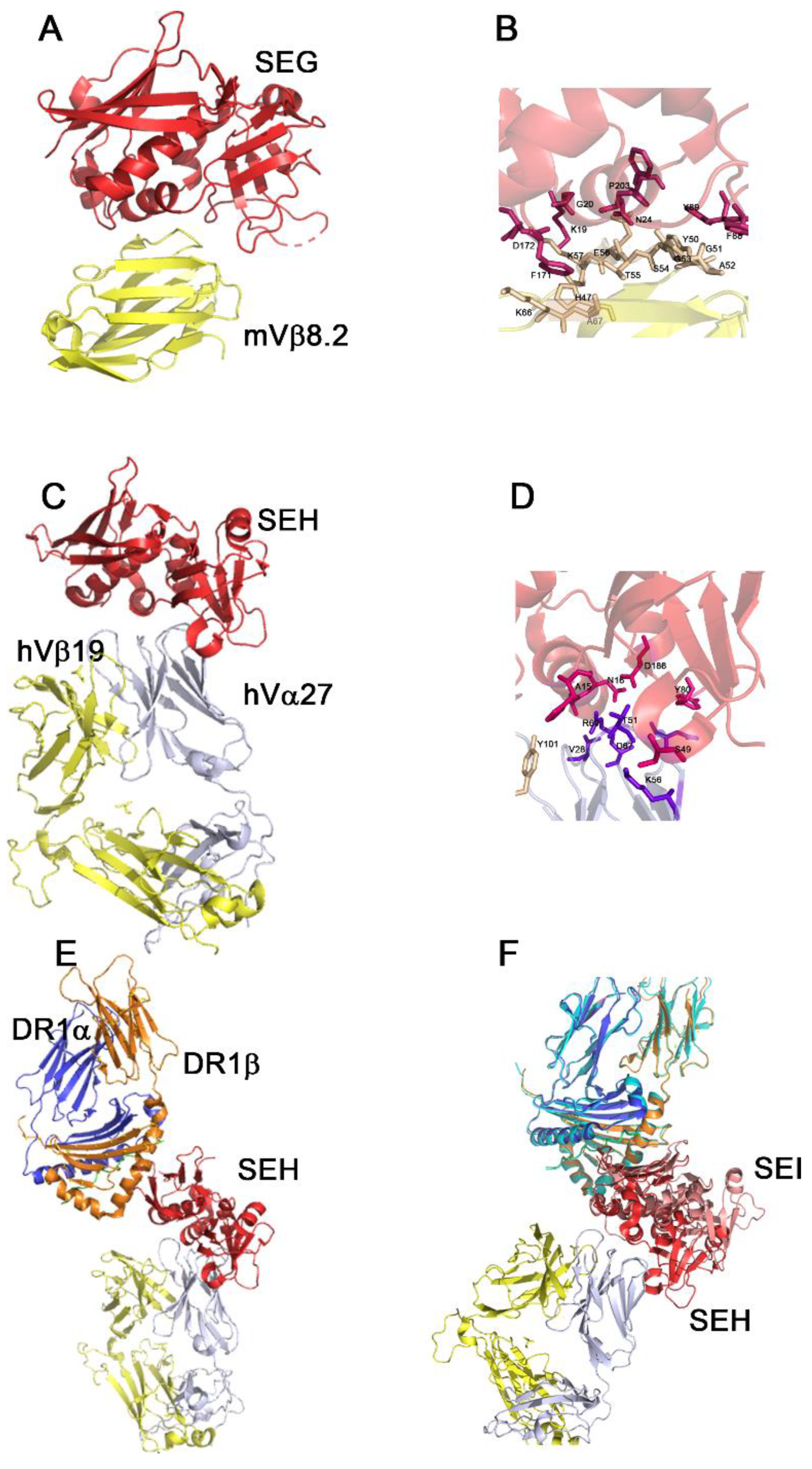
| SAg | Type | Year of Discovery | Phylogenetic Group |
|---|---|---|---|
| SEA | Classical | 1962 | III |
| SEB | Classical | 1962 | II |
| SEC | Classical | 1965 | II |
| SED | Classical | 1967 | III |
| SEE | Classical | 1971 | III |
| TSST-1 (SEF) | Classical | 1981 | II |
| SEG | New | 1998 | I |
| SEH | New | 1994 | III |
| SEI | New | 1998 | V |
| SElJ | New | 1998 | III |
| SEK | New | 2001 | V |
| SEL | New | 2001 | V |
| SEM | New | 2001 | V |
| SEN | New | 2001 | III |
| SEO | New | 2001 | III |
| SEP | New | 2005 | III |
| SEQ | New | 2002 | V |
| SER | New | 2004 | II |
| SES | New | 2008 | III |
| SET | New | 2008 | I |
| SElU | New | 2003 | II |
| SElV | New | 2006 | V |
| SElW | New | 2012 | III |
| SElX | New | 2011 | I |
| SElY | New | 2015 | I |
| SElZ | New | 2015 | I |
| SEl26 | New | 2018 | V |
| SEl27 | New | 2018 | II |
| SE01 | New | 2017 | III |
| SE02 | New | 2020 | II |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noli Truant, S.; Redolfi, D.M.; Sarratea, M.B.; Malchiodi, E.L.; Fernández, M.M. Superantigens, a Paradox of the Immune Response. Toxins 2022, 14, 800. https://doi.org/10.3390/toxins14110800
Noli Truant S, Redolfi DM, Sarratea MB, Malchiodi EL, Fernández MM. Superantigens, a Paradox of the Immune Response. Toxins. 2022; 14(11):800. https://doi.org/10.3390/toxins14110800
Chicago/Turabian StyleNoli Truant, Sofia, Daniela María Redolfi, María Belén Sarratea, Emilio Luis Malchiodi, and Marisa Mariel Fernández. 2022. "Superantigens, a Paradox of the Immune Response" Toxins 14, no. 11: 800. https://doi.org/10.3390/toxins14110800
APA StyleNoli Truant, S., Redolfi, D. M., Sarratea, M. B., Malchiodi, E. L., & Fernández, M. M. (2022). Superantigens, a Paradox of the Immune Response. Toxins, 14(11), 800. https://doi.org/10.3390/toxins14110800





