Subchronic Oral Cylindrospermopsin Exposure Alters the Host Gut Microbiome and Is Associated with Progressive Hepatic Inflammation, Stellate Cell Activation, and Mild Fibrosis in a Preclinical Study
Abstract
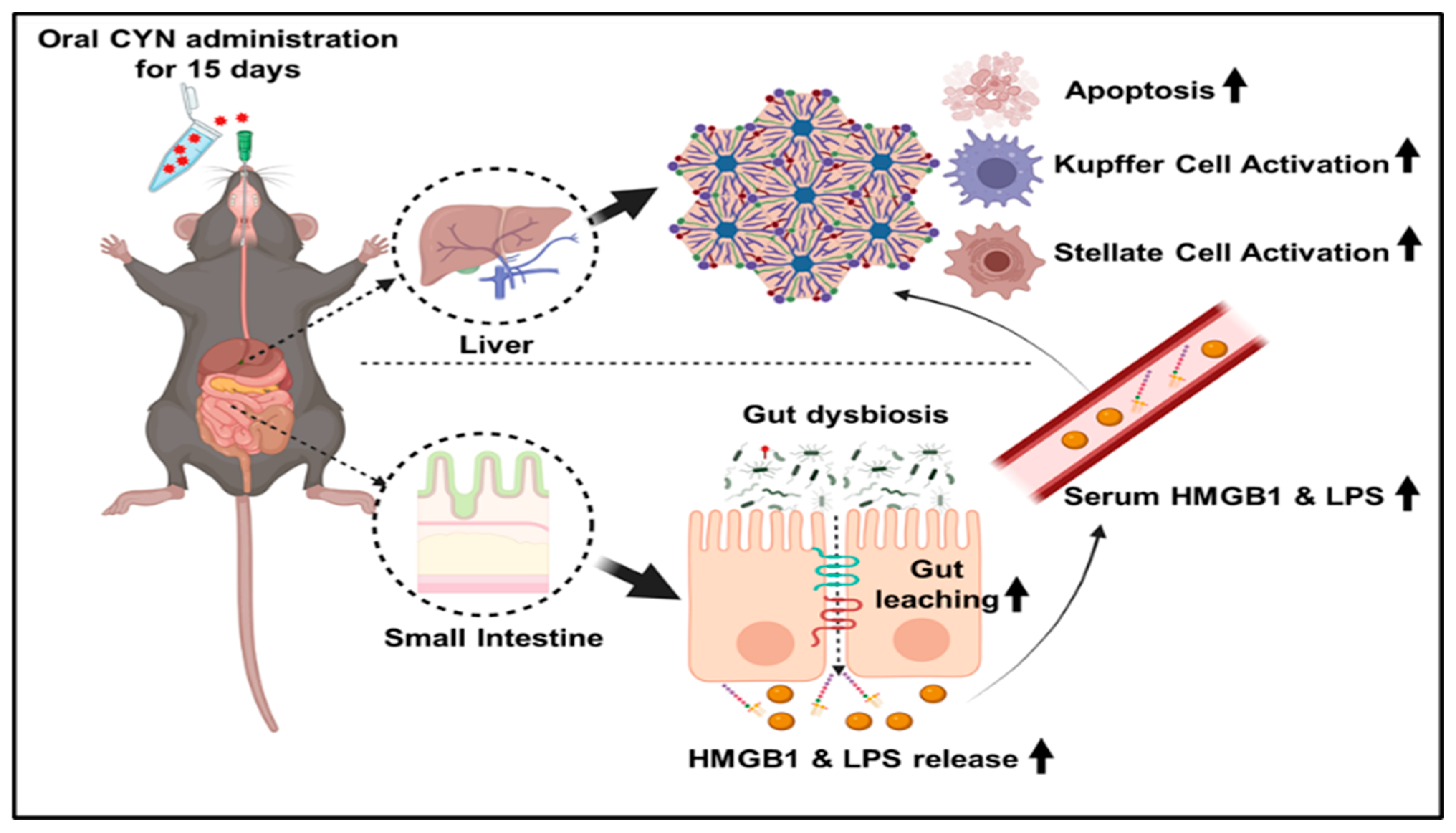
:1. Introduction
2. Results
2.1. CYN Administration in Mice Resulted in an Altered Gut Microbiome Signature
2.2. Exposure to the Environmental Toxin CYN Caused Gut Leaching with a Concomitant Increased Systemic Release of Pathogen-Associated Molecular Pattern (PAMP) and Damage-Associated Molecular Pattern (DAMP) in Mice
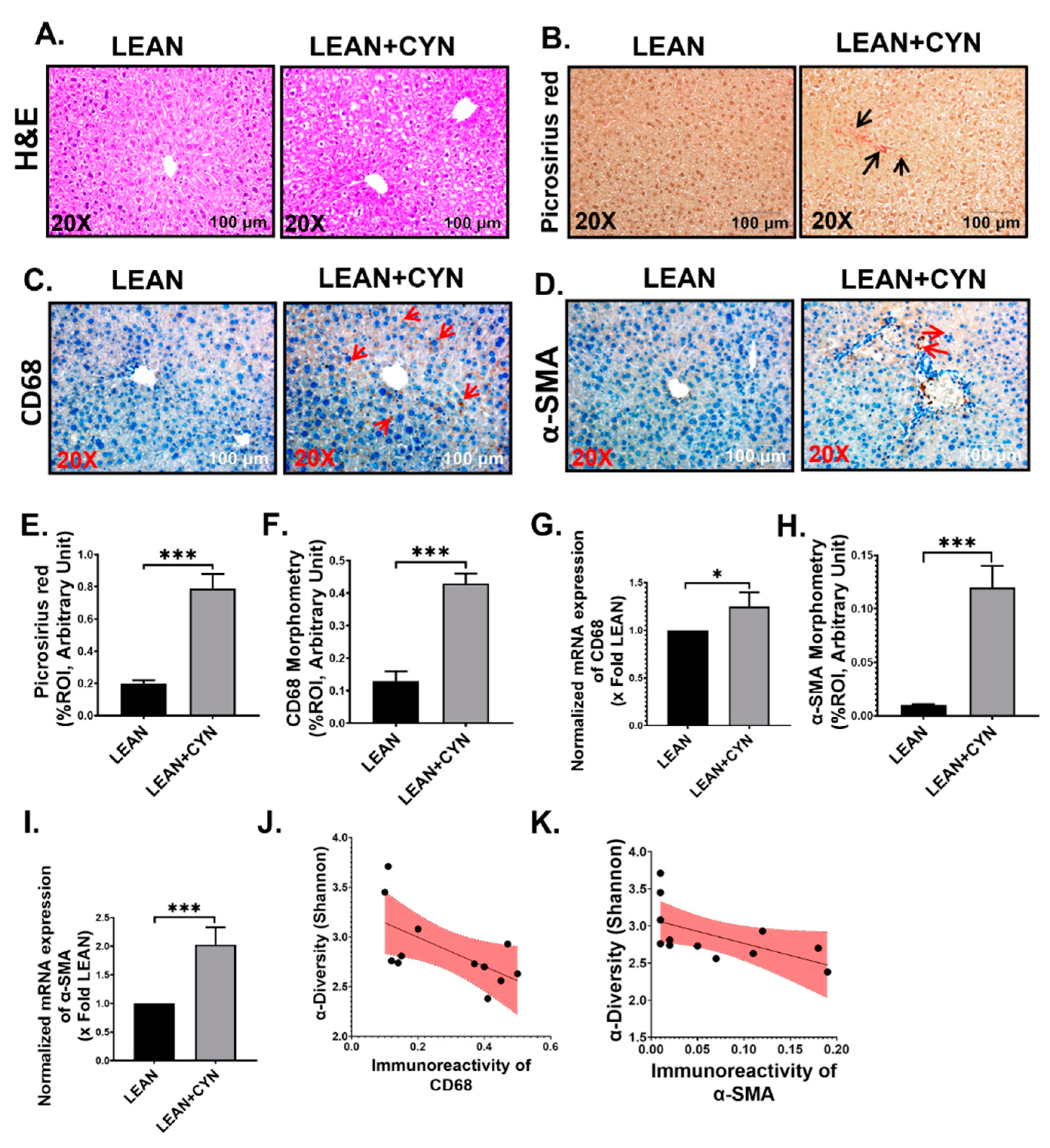
2.3. CYN Administration in Mice Exhibited Markers of Liver Injury, Mild Fibrosis, and Caused Kupffer Cell and Stellate Cell Activation
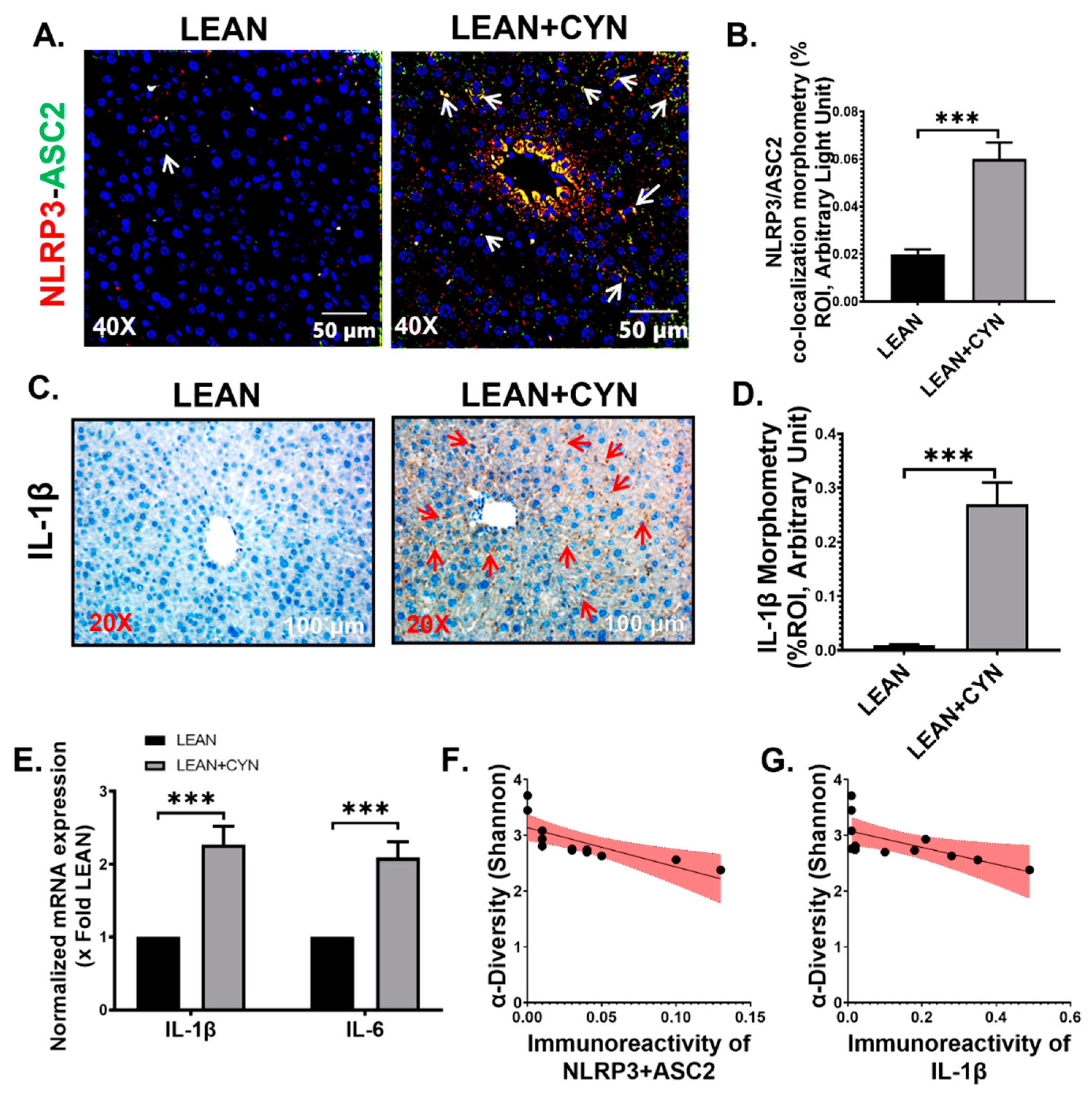
2.4. CYN-Induced Increase of Circulatory PAMP and DAMP Levels Led to Hepatic Inflammasome Activation, Thus Linking Hepatic Inflammation
2.5. CYN Exposure in Mice Increased TGF-β Expression in the Liver, Leading to Smad2/3-Smad4 Mediated Fibrotic Pathway Activation
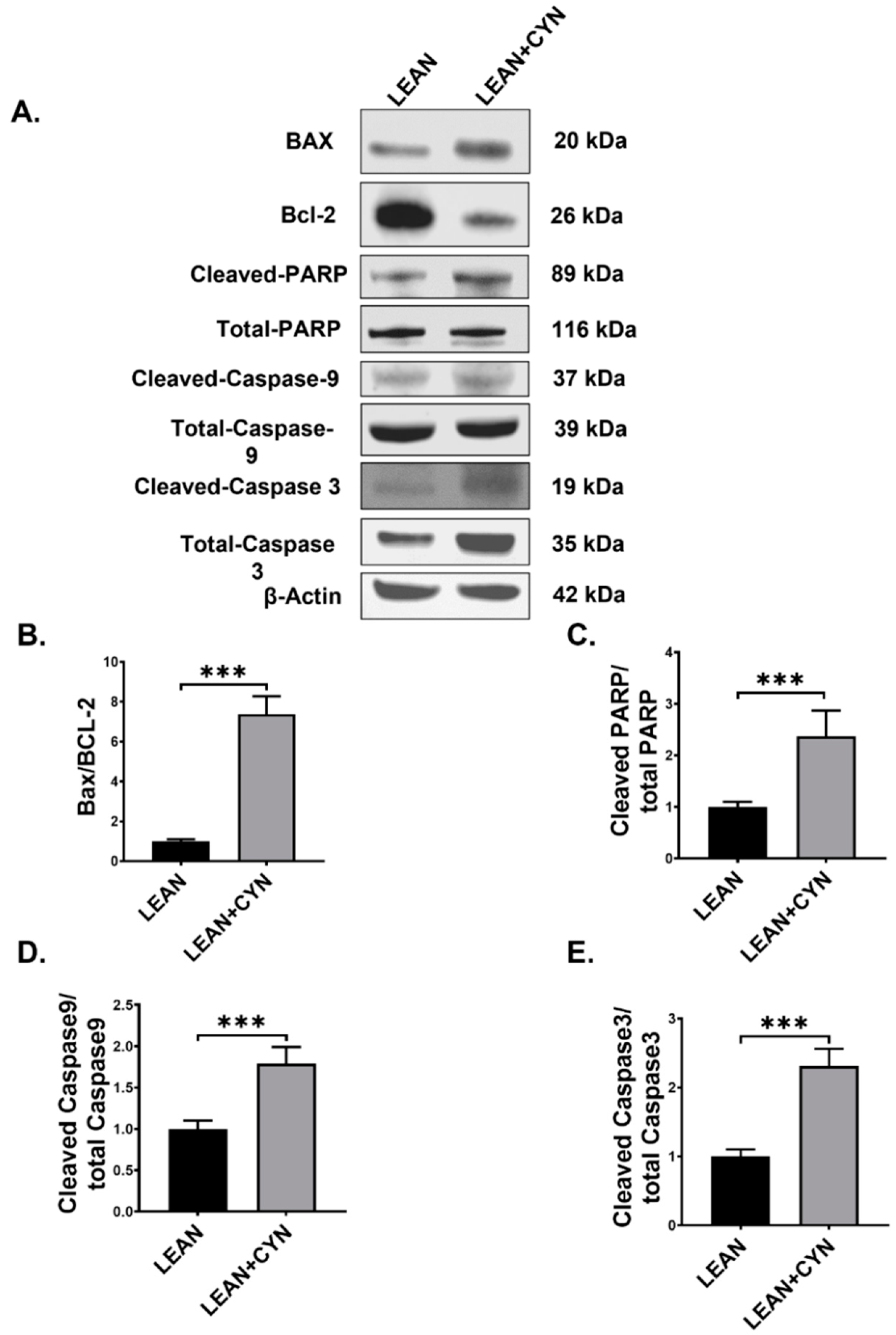
2.6. CYN Treatment in Mice Triggered the Intrinsic Apoptotic Pathway in the Liver
2.7. CYN Administration in Mice Caused Elevated Extracellular HMGB1 Expression and HMGB1-Mediated Receptor Expression in the Liver
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Experimental Murine Model of CYN Exposure
4.3. Bacteriome Analysis
4.4. Laboratory Methods
4.4.1. Histopathology
4.4.2. Immunohistochemistry
4.4.3. Immunofluorescence Staining and Microscopy
4.4.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.4.5. Western Blot
4.4.6. Enzyme-Linked Immunosorbent Assay (ELISA)
4.4.7. Endotoxemia Detection by Limulus Amebocyte Lysate (LAL) Assay
4.5. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Form
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tomitani, A.; Knoll, A.H.; Cavanaugh, C.M.; Ohno, T. The evolutionary diversification of cyanobacteria: Molecular-phylogenetic and paleontological perspectives. Proc. Natl. Acad. Sci. USA 2006, 103, 5442–5447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paerl, H.W.; Otten, T.G. Environmental science. Blooms bite the hand that feeds them. Science 2013, 342, 433–434. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.W.; Lazorchak, J.M.; Howard, M.D.; Johnson, M.V.; Morton, S.L.; Perkins, D.A.; Reavie, E.D.; Scott, G.I.; Smith, S.A.; Steevens, J.A. Are harmful algal blooms becoming the greatest inland water quality threat to public health and aquatic ecosystems? Environ. Toxicol. Chem. 2016, 35, 6–13. [Google Scholar] [CrossRef]
- Watson, S.B.; Miller, C.; Arhonditsis, G.; Boyer, G.L.; Carmichael, W.; Charlton, M.N.; Confesor, R.; Depew, D.C.; Hook, T.O.; Ludsin, S.A.; et al. The re-eutrophication of Lake Erie: Harmful algal blooms and hypoxia. Harmful Algae 2016, 56, 44–66. [Google Scholar] [CrossRef]
- Van Apeldoorn, M.E.; van Egmond, H.P.; Speijers, G.J.; Bakker, G.J. Toxins of cyanobacteria. Mol. Nutr. Food Res. 2007, 51, 7–60. [Google Scholar] [CrossRef]
- Buratti, F.M.; Manganelli, M.; Vichi, S.; Stefanelli, M.; Scardala, S.; Testai, E.; Funari, E. Cyanotoxins: Producing organisms, occurrence, toxicity, mechanism of action and human health toxicological risk evaluation. Arch. Toxicol. 2017, 91, 1049–1130. [Google Scholar] [CrossRef]
- Boopathi, T.; Ki, J.S. Impact of environmental factors on the regulation of cyanotoxin production. Toxins 2014, 6, 1951–1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, K.I.; Ohtani, I.; Iwamoto, K.; Suzuki, M.; Watanabe, M.F.; Watanabe, M.; Terao, K. Isolation of cylindrospermopsin from a cyanobacterium Umezakia natans and its screening method. Toxicon 1994, 32, 73–84. [Google Scholar] [CrossRef]
- Spoof, L.; Berg, K.A.; Rapala, J.; Lahti, K.; Lepisto, L.; Metcalf, J.S.; Codd, G.A.; Meriluoto, J. First observation of cylindrospermopsin in Anabaena lapponica isolated from the boreal environment (Finland). Environ. Toxicol. 2006, 21, 552–560. [Google Scholar] [CrossRef]
- Preussel, K.; Stuken, A.; Wiedner, C.; Chorus, I.; Fastner, J. First report on cylindrospermopsin producing Aphanizomenon flos-aquae (Cyanobacteria) isolated from two German lakes. Toxicon 2006, 47, 156–162. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, G.; Chen, Y.; Jia, N.; Li, R. Four decades of progress in cylindrospermopsin research: The ins and outs of a potent cyanotoxin. J. Hazard. Mater. 2021, 406, 124653. [Google Scholar] [CrossRef] [PubMed]
- Scarlett, K.R.; Kim, S.; Lovin, L.M.; Chatterjee, S.; Scott, J.T.; Brooks, B.W. Global scanning of cylindrospermopsin: Critical review and analysis of aquatic occurrence, bioaccumulation, toxicity and health hazards. Sci. Total Environ. 2020, 738, 139807. [Google Scholar] [CrossRef] [PubMed]
- Chonudomkul, D.; Yongmanitchai, W.; Theeragool, G.; Kawachi, M.; Kasai, F.; Kaya, K.; Watanabe, M.M. Morphology, genetic diversity, temperature tolerance and toxicity of Cylindrospermopsis raciborskii (Nostocales, Cyanobacteria) strains from Thailand and Japan. FEMS Microbiol. Ecol. 2004, 48, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Kinnear, S. Cylindrospermopsin: A decade of progress on bioaccumulation research. Mar. Drugs 2010, 8, 542–564. [Google Scholar] [CrossRef] [Green Version]
- Hoff-Risseti, C.; Dorr, F.A.; Schaker, P.D.; Pinto, E.; Werner, V.R.; Fiore, M.F. Cylindrospermopsin and saxitoxin synthetase genes in Cylindrospermopsis raciborskii strains from Brazilian freshwater. PLoS ONE 2013, 8, e74238. [Google Scholar] [CrossRef] [Green Version]
- Vico, P.; Bonilla, S.; Cremella, B.; Aubriot, L.; Iriarte, A.; Piccini, C. Biogeography of the cyanobacterium Raphidiopsis (Cylindrospermopsis) raciborskii: Integrating genomics, phylogenetic and toxicity data. Mol. Phylogenet. Evol. 2020, 148, 106824. [Google Scholar] [CrossRef]
- Poniedzialek, B.; Rzymski, P.; Kokocinski, M. Cylindrospermopsin: Water-linked potential threat to human health in Europe. Environ. Toxicol. Pharmacol. 2012, 34, 651–660. [Google Scholar] [CrossRef]
- Loftin, K.A.; Graham, J.L.; Hilborn, E.D.; Lehmann, S.C.; Meyer, M.T.; Dietze, J.E.; Griffith, C.B. Cyanotoxins in inland lakes of the United States: Occurrence and potential recreational health risks in the EPA National Lakes Assessment 2007. Harmful Algae 2016, 56, 77–90. [Google Scholar] [CrossRef] [Green Version]
- Drobac, D.; Tokodi, N.; Simeunovic, J.; Baltic, V.; Stanic, D.; Svircev, Z. Human exposure to cyanotoxins and their effects on health. Arh. Hig. Rada Toksikol. 2013, 64, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Humpage, A.R.; Fenech, M.; Thomas, P.; Falconer, I.R. Micronucleus induction and chromosome loss in transformed human white cells indicate clastogenic and aneugenic action of the cyanobacterial toxin, cylindrospermopsin. Mutat. Res. 2000, 472, 155–161. [Google Scholar] [CrossRef]
- Zegura, B.; Gajski, G.; Straser, A.; Garaj-Vrhovac, V. Cylindrospermopsin induced DNA damage and alteration in the expression of genes involved in the response to DNA damage, apoptosis and oxidative stress. Toxicon 2011, 58, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa, M.G.; Gutierrez-Praena, D.; Prieto, A.I.; Guzman-Guillen, R.; Jos, A.; Camean, A.M. Neurotoxicity induced by microcystins and cylindrospermopsin: A review. Sci. Total Environ. 2019, 668, 547–565. [Google Scholar] [CrossRef] [PubMed]
- Moosova, Z.; Pekarova, M.; Sindlerova, L.S.; Vasicek, O.; Kubala, L.; Blaha, L.; Adamovsky, O. Immunomodulatory effects of cyanobacterial toxin cylindrospermopsin on innate immune cells. Chemosphere 2019, 226, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Terao, K.; Ohmori, S.; Igarashi, K.; Ohtani, I.; Watanabe, M.F.; Harada, K.I.; Ito, E.; Watanabe, M. Electron microscopic studies on experimental poisoning in mice induced by cylindrospermopsin isolated from blue-green alga Umezakia natans. Toxicon 1994, 32, 833–843. [Google Scholar] [CrossRef]
- Runnegar, M.T.; Kong, S.M.; Zhong, Y.Z.; Lu, S.C. Inhibition of reduced glutathione synthesis by cyanobacterial alkaloid cylindrospermopsin in cultured rat hepatocytes. Biochem. Pharmacol. 1995, 49, 219–225. [Google Scholar] [CrossRef]
- Humpage, A.R.; Fontaine, F.; Froscio, S.; Burcham, P.; Falconer, I.R. Cylindrospermopsin genotoxicity and cytotoxicity: Role of cytochrome P-450 and oxidative stress. J. Toxicol. Environ. Health A 2005, 68, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Praena, D.; Pichardo, S.; Jos, A.; Maria Camean, A. Toxicity and glutathione implication in the effects observed by exposure of the liver fish cell line PLHC-1 to pure cylindrospermopsin. Ecotoxicol. Environ. Saf. 2011, 74, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Praena, D.; Pichardo, S.; Jos, A.; Moreno, F.J.; Camean, A.M. Alterations observed in the endothelial HUVEC cell line exposed to pure Cylindrospermopsin. Chemosphere 2012, 89, 1151–1160. [Google Scholar] [CrossRef]
- Gutierrez-Praena, D.; Pichardo, S.; Jos, A.; Moreno, F.J.; Camean, A.M. Biochemical and pathological toxic effects induced by the cyanotoxin Cylindrospermopsin on the human cell line Caco-2. Water Res. 2012, 46, 1566–1575. [Google Scholar] [CrossRef]
- Bazin, E.; Mourot, A.; Humpage, A.R.; Fessard, V. Genotoxicity of a freshwater cyanotoxin, cylindrospermopsin, in two human cell lines: Caco-2 and HepaRG. Environ. Mol. Mutagen. 2010, 51, 251–259. [Google Scholar] [CrossRef]
- Takser, L.; Benachour, N.; Husk, B.; Cabana, H.; Gris, D. Cyanotoxins at low doses induce apoptosis and inflammatory effects in murine brain cells: Potential implications for neurodegenerative diseases. Toxicol. Rep. 2016, 3, 180–189. [Google Scholar] [CrossRef] [Green Version]
- Ochoa-Reparaz, J.; Kasper, L.H. The Second Brain: Is the Gut Microbiota a Link Between Obesity and Central Nervous System Disorders? Curr. Obes. Rep. 2016, 5, 51–64. [Google Scholar] [CrossRef] [Green Version]
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. 2014, 13, 17–22. [Google Scholar]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, S.; Kimono, D.; Albadrani, M.; Seth, R.K.; Busbee, P.; Alghetaa, H.; Porter, D.E.; Scott, G.I.; Brooks, B.; Nagarkatti, M.; et al. Environmental microcystin targets the microbiome and increases the risk of intestinal inflammatory pathology via NOX2 in underlying murine model of Nonalcoholic Fatty Liver Disease. Sci. Rep. 2019, 9, 8742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, P.; Bose, D.; Stebliankin, V.; Cickovski, T.; Seth, R.K.; Porter, D.E.; Brooks, B.W.; Mathee, K.; Narasimhan, G.; Colwell, R.; et al. Prior exposure to microcystin alters host gut resistome and is associated with dysregulated immune homeostasis in translatable mouse models. Sci. Rep. 2022, 12, 11516. [Google Scholar] [CrossRef] [PubMed]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [Green Version]
- Byth, S. Palm Island mystery disease. Med. J. Aust. 1980, 2, 40–42. [Google Scholar] [CrossRef]
- Derrien, M.; Van Baarlen, P.; Hooiveld, G.; Norin, E.; Muller, M.; de Vos, W.M. Modulation of Mucosal Immune Response, Tolerance, and Proliferation in Mice Colonized by the Mucin-Degrader Akkermansia muciniphila. Front. Microbiol. 2011, 2, 166. [Google Scholar] [CrossRef] [Green Version]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Geerlings, S.Y.; Kostopoulos, I.; de Vos, W.M.; Belzer, C. Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef]
- Xu, J.; Bjursell, M.K.; Himrod, J.; Deng, S.; Carmichael, L.K.; Chiang, H.C.; Hooper, L.V.; Gordon, J.I. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 2003, 299, 2074–2076. [Google Scholar] [CrossRef] [Green Version]
- Hamajima, H.; Matsunaga, H.; Fujikawa, A.; Sato, T.; Mitsutake, S.; Yanagita, T.; Nagao, K.; Nakayama, J.; Kitagaki, H. Japanese traditional dietary fungus koji Aspergillus oryzae functions as a prebiotic for Blautia coccoides through glycosylceramide: Japanese dietary fungus koji is a new prebiotic. Springerplus 2016, 5, 1321. [Google Scholar] [CrossRef] [Green Version]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Hollander, D.; Kaunitz, J.D. The “Leaky Gut”: Tight Junctions but Loose Associations? Dig. Dis. Sci. 2020, 65, 1277–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuke, N.; Nagata, N.; Suganuma, H.; Ota, T. Regulation of Gut Microbiota and Metabolic Endotoxemia with Dietary Factors. Nutrients 2019, 11, 2277. [Google Scholar] [CrossRef] [Green Version]
- Mohammad, S.; Thiemermann, C. Role of Metabolic Endotoxemia in Systemic Inflammation and Potential Interventions. Front. Immunol. 2020, 11, 594150. [Google Scholar] [CrossRef] [PubMed]
- Biscetti, F.; Flex, A.; Alivernini, S.; Tolusso, B.; Gremese, E.; Ferraccioli, G. The Role of High-Mobility Group Box-1 and Its Crosstalk with Microbiome in Rheumatoid Arthritis. Mediat. Inflamm. 2017, 2017, 5230374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabo, G.; Petrasek, J. Inflammasome activation and function in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Kodama, Y.; Inokuchi, S.; Schnabl, B.; Aoyama, T.; Ohnishi, H.; Olefsky, J.M.; Brenner, D.A.; Seki, E. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology 2010, 139, 323–334 e327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dooley, S.; Ten Dijke, P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012, 347, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Nakao, A.; Imamura, T.; Souchelnytskyi, S.; Kawabata, M.; Ishisaki, A.; Oeda, E.; Tamaki, K.; Hanai, J.; Heldin, C.H.; Miyazono, K.; et al. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997, 16, 5353–5362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, C.; Galluzzi, L.; Kepp, O.; Kroemer, G. Decoding cell death signals in liver inflammation. J. Hepatol. 2013, 59, 583–594. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Hou, W.; Zhang, Q.; Kang, R.; Fan, X.G.; Tang, D. Emerging role of high-mobility group box 1 (HMGB1) in liver diseases. Mol. Med. 2013, 19, 357–366. [Google Scholar] [CrossRef]
- Khambu, B.; Yan, S.; Huda, N.; Yin, X.M. Role of High-Mobility Group Box-1 in Liver Pathogenesis. Int. J. Mol. Sci. 2019, 20, 5314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Therap. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef] [Green Version]
- Aoun, A.; Darwish, F.; Hamod, N. The Influence of the Gut Microbiome on Obesity in Adults and the Role of Probiotics, Prebiotics, and Synbiotics for Weight Loss. Prev. Nutr. Food Sci. 2020, 25, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Hrncir, T.; Hrncirova, L.; Kverka, M.; Hromadka, R.; Machova, V.; Trckova, E.; Kostovcikova, K.; Kralickova, P.; Krejsek, J.; Tlaskalova-Hogenova, H. Gut Microbiota and NAFLD: Pathogenetic Mechanisms, Microbiota Signatures, and Therapeutic Interventions. Microorganisms 2021, 9, 957. [Google Scholar] [CrossRef] [PubMed]
- Menees, S.; Chey, W. The gut microbiome and irritable bowel syndrome. F1000Research 2018, 7, 1029. [Google Scholar] [CrossRef] [Green Version]
- Glassner, K.L.; Abraham, B.P.; Quigley, E.M.M. The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 2020, 145, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Li, G.; Huang, P.; Liu, Z.; Zhao, B. The Gut Microbiota and Alzheimer’s Disease. J. Alzheimers Dis. 2017, 58, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Savva, G.M.; Bedarf, J.R.; Charles, I.G.; Hildebrand, F.; Narbad, A. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Parkinsons Dis. 2021, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Bose, D.; Chatterjee, S.; Older, E.; Seth, R.; Janulewicz, P.; Saha, P.; Mondal, A.; Carlson, J.M.; Decho, A.W.; Sullivan, K.; et al. Host gut resistome in Gulf War chronic multisymptom illness correlates with persistent inflammation. Commun. Biol. 2022, 5, 552. [Google Scholar] [CrossRef]
- Saha, P.; Skidmore, P.T.; Holland, L.A.; Mondal, A.; Bose, D.; Seth, R.K.; Sullivan, K.; Janulewicz, P.A.; Horner, R.; Klimas, N.; et al. Andrographolide Attenuates Gut-Brain-Axis Associated Pathology in Gulf War Illness by Modulating Bacteriome-Virome Associated Inflammation and Microglia-Neuron Proinflammatory Crosstalk. Brain Sci. 2021, 11, 905. [Google Scholar] [CrossRef]
- Bose, D.; Saha, P.; Mondal, A.; Fanelli, B.; Seth, R.K.; Janulewicz, P.; Sullivan, K.; Lasley, S.; Horner, R.; Colwell, R.R.; et al. Obesity Worsens Gulf War Illness Symptom Persistence Pathology by Linking Altered Gut Microbiome Species to Long-Term Gastrointestinal, Hepatic, and Neuronal Inflammation in a Mouse Model. Nutrients 2020, 12, 2764. [Google Scholar] [CrossRef] [PubMed]
- Bose, D.; Mondal, A.; Saha, P.; Kimono, D.; Sarkar, S.; Seth, R.K.; Janulewicz, P.; Sullivan, K.; Horner, R.; Klimas, N.; et al. TLR Antagonism by Sparstolonin B Alters Microbial Signature and Modulates Gastrointestinal and Neuronal Inflammation in Gulf War Illness Preclinical Model. Brain Sci. 2020, 10, 532. [Google Scholar] [CrossRef]
- Seth, R.K.; Maqsood, R.; Mondal, A.; Bose, D.; Kimono, D.; Holland, L.A.; Janulewicz Lloyd, P.; Klimas, N.; Horner, R.D.; Sullivan, K.; et al. Gut DNA Virome Diversity and Its Association with Host Bacteria Regulate Inflammatory Phenotype and Neuronal Immunotoxicity in Experimental Gulf War Illness. Viruses 2019, 11, 968. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Stojanov, S.; Berlec, A.; Strukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Matsuoka, K.; Kanai, T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Carroll, I.M.; Ringel-Kulka, T.; Siddle, J.P.; Ringel, Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 2012, 24, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Schippa, S.; Iebba, V.; Barbato, M.; Di Nardo, G.; Totino, V.; Checchi, M.P.; Longhi, C.; Maiella, G.; Cucchiara, S.; Conte, M.P. A distinctive ‘microbial signature’ in celiac pediatric patients. BMC Microbiol. 2010, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyotylainen, T.; Hamalainen, A.M.; Peet, A.; Tillmann, V.; Poho, P.; Mattila, I.; et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [Green Version]
- Sonnenburg, J.L.; Xu, J.; Leip, D.D.; Chen, C.H.; Westover, B.P.; Weatherford, J.; Buhler, J.D.; Gordon, J.I. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 2005, 307, 1955–1959. [Google Scholar] [CrossRef] [Green Version]
- Durant, L.; Stentz, R.; Noble, A.; Brooks, J.; Gicheva, N.; Reddi, D.; O’Connor, M.J.; Hoyles, L.; McCartney, A.L.; Man, R.; et al. Bacteroides thetaiotaomicron-derived outer membrane vesicles promote regulatory dendritic cell responses in health but not in inflammatory bowel disease. Microbiome 2020, 8, 88. [Google Scholar] [CrossRef]
- Pan, N.; Imlay, J.A. How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron. Mol. Microbiol. 2001, 39, 1562–1571. [Google Scholar] [CrossRef]
- Derex, M.; Beugin, M.P.; Godelle, B.; Raymond, M. Experimental evidence for the influence of group size on cultural complexity. Nature 2013, 503, 389–391. [Google Scholar] [CrossRef]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef]
- Sehgal, K.; Khanna, S. Gut microbiome and Clostridioides difficile infection: A closer look at the microscopic interface. Ther. Adv. Gastroenterol. 2021, 14, 1756284821994736. [Google Scholar] [CrossRef]
- Moono, P.; Putsathit, P.; Knight, D.R.; Squire, M.M.; Hampson, D.J.; Foster, N.F.; Riley, T.V. Persistence of Clostridium difficile RT 237 infection in a Western Australian piggery. Anaerobe 2016, 37, 62–66. [Google Scholar] [CrossRef]
- Alhasson, F.; Das, S.; Seth, R.; Dattaroy, D.; Chandrashekaran, V.; Ryan, C.N.; Chan, L.S.; Testerman, T.; Burch, J.; Hofseth, L.J.; et al. Altered gut microbiome in a mouse model of Gulf War Illness causes neuroinflammation and intestinal injury via leaky gut and TLR4 activation. PLoS ONE 2017, 12, e0172914. [Google Scholar] [CrossRef] [Green Version]
- Jo, E.K.; Kim, J.K.; Shin, D.M.; Sasakawa, C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol. 2016, 13, 148–159. [Google Scholar] [CrossRef] [Green Version]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolios, G.; Valatas, V.; Kouroumalis, E. Role of Kupffer cells in the pathogenesis of liver disease. World J. Gastroenterol. 2006, 12, 7413–7420. [Google Scholar] [CrossRef] [PubMed]
- Humpage, A.R.; Falconer, I.R. Oral toxicity of the cyanobacterial toxin cylindrospermopsin in male Swiss albino mice: Determination of no observed adverse effect level for deriving a drinking water guideline value. Environ. Toxicol. 2003, 18, 94–103. [Google Scholar] [CrossRef] [PubMed]





| Genes | Primer Sequence (5′-3′ Orientation) |
|---|---|
| CD68 (Mus musculus) | Forward: GCTACATGGCGGTGGAGTACAA Reverse: ATGATGAGAGGCAGCAAGATGG |
| α-SMA (Mus musculus) | Forward: GGAGAAGCCCAGCCAGTCGC Reverse: ACCATTGTCGCACACCAGGGC |
| IL-1β (Mus musculus) | Forward: CCTCGGCCAAGACAGGTCGC Reverse: TGCCCATCAGAGGCAAGGAGGA |
| IL-6 (Mus musculus) | Forward: ACCAGAGGAAATTTTCAATAGGC Reverse: TGATGCACTTGCAGAAAACA |
| TGF-β (Mus musculus) | Forward: CTCACCGCGACTCCTGCTGC Reverse: TCGGAGAGCGGGAACCCTCG |
| 18S (Mus musculus) | Forward: TTCGAACGTCTGCCCTATCAA Reverse: ATGGTAGGCACGGCGATA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saha, P.; Upright, M.; Bose, D.; Roy, S.; Trivedi, A.; More, M.; Scott, G.I.; Brooks, B.W.; Chatterjee, S. Subchronic Oral Cylindrospermopsin Exposure Alters the Host Gut Microbiome and Is Associated with Progressive Hepatic Inflammation, Stellate Cell Activation, and Mild Fibrosis in a Preclinical Study. Toxins 2022, 14, 835. https://doi.org/10.3390/toxins14120835
Saha P, Upright M, Bose D, Roy S, Trivedi A, More M, Scott GI, Brooks BW, Chatterjee S. Subchronic Oral Cylindrospermopsin Exposure Alters the Host Gut Microbiome and Is Associated with Progressive Hepatic Inflammation, Stellate Cell Activation, and Mild Fibrosis in a Preclinical Study. Toxins. 2022; 14(12):835. https://doi.org/10.3390/toxins14120835
Chicago/Turabian StyleSaha, Punnag, Macayla Upright, Dipro Bose, Subhajit Roy, Ayushi Trivedi, Madhura More, Geoff I. Scott, Bryan W. Brooks, and Saurabh Chatterjee. 2022. "Subchronic Oral Cylindrospermopsin Exposure Alters the Host Gut Microbiome and Is Associated with Progressive Hepatic Inflammation, Stellate Cell Activation, and Mild Fibrosis in a Preclinical Study" Toxins 14, no. 12: 835. https://doi.org/10.3390/toxins14120835
APA StyleSaha, P., Upright, M., Bose, D., Roy, S., Trivedi, A., More, M., Scott, G. I., Brooks, B. W., & Chatterjee, S. (2022). Subchronic Oral Cylindrospermopsin Exposure Alters the Host Gut Microbiome and Is Associated with Progressive Hepatic Inflammation, Stellate Cell Activation, and Mild Fibrosis in a Preclinical Study. Toxins, 14(12), 835. https://doi.org/10.3390/toxins14120835






