Susceptibility of Sea Bream (Sparus aurata) to AIP56, an AB-Type Toxin Secreted by Photobacterium damselae subsp. piscicida
Abstract
:1. Introduction
2. Results
2.1. aip56 Is Ubiquitous among Sea Bream and Sea Bass Phdp Isolates
2.2. Sea Bream and Sea Bass Phdp Isolates Secrete Identical AIP56 Toxins
2.3. AIP56 Does Not Induce Apoptosis or Other Signs of Toxicity in Sea Bream Peritoneal Leukocytes Ex Vivo
2.4. AIP56 Displays Proteolytic Activity against Sea Bream NF-kB p65
2.5. The Internalization of AIP56-488 in Sea Bream Peritoneal Leukocytes Is Inefficient
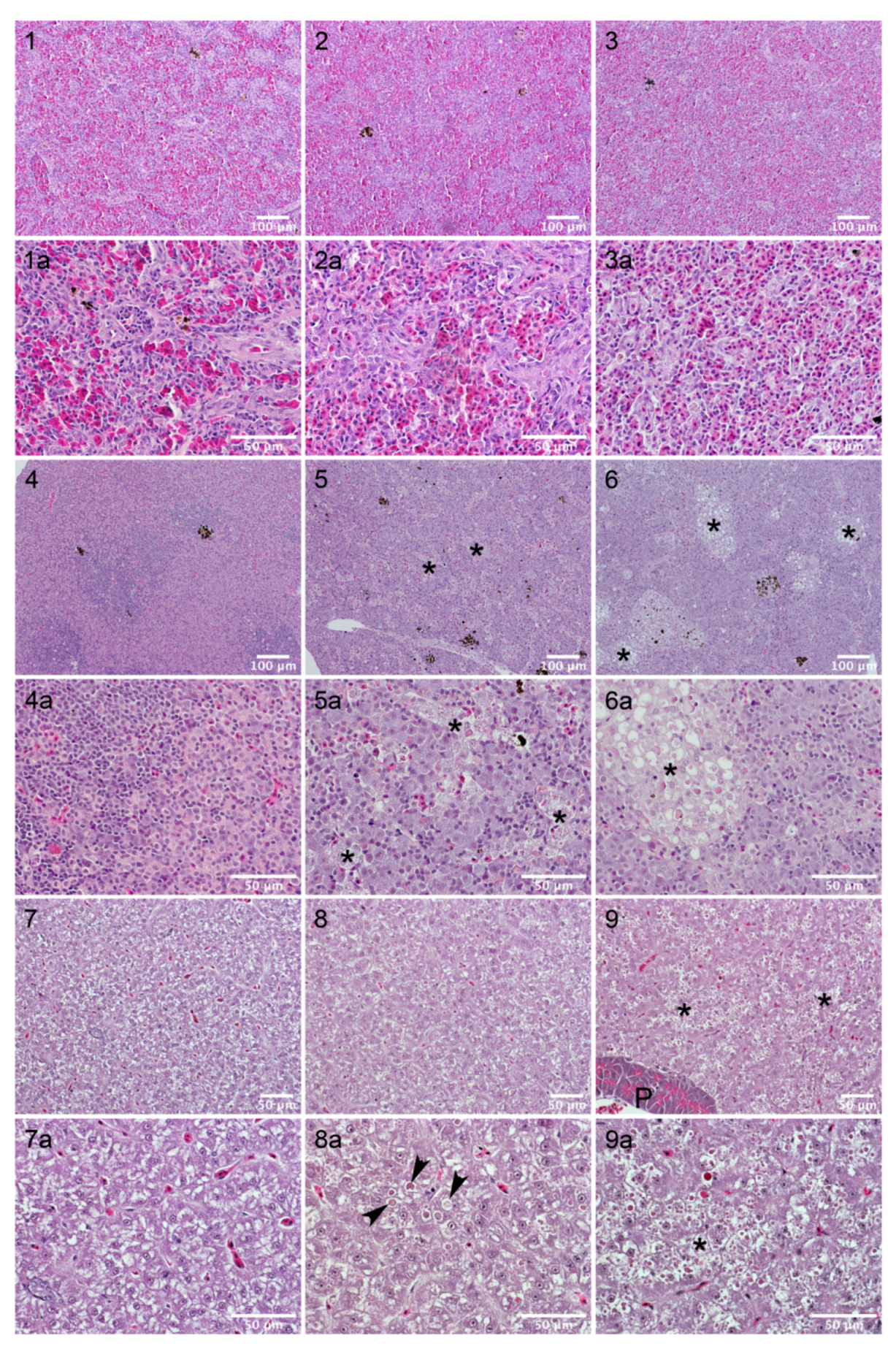
2.6. Sea Bream Are Highly Resistant to AIP56 Toxicity
3. Discussion
4. Materials and Methods
4.1. Production and Fluorescence Labeling of Recombinant Proteins
4.2. Determination of Recombinant Protein Concentration
4.3. Fish
4.4. Cells
4.5. Cell Intoxication Assays
4.6. AIP56 Internalization Assays
4.7. Detection of aip56 in Phdp Field Isolates
4.8. Comparative Analysis of AIP56 from Different Phdp Strains
4.9. Cloning Sea Bream NF-kB p65
4.10. NF-kB Cleavage Assay
4.11. In Vivo Toxicity Tests
4.12. GenBank Accession Numbers
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Do Vale, A.; Silva, M.T.; dos Santos, N.M.S.; Nascimento, D.S.; Reis-Rodrigues, P.; Costa-Ramos, C.; Ellis, A.E.; Azevedo, J.E. AIP56, a novel plasmid-encoded virulence factor of Photobacterium damselae subsp. piscicida with apoptogenic activity against sea bass macrophages and neutrophils. Mol. Microbiol. 2005, 58, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Andreoni, F.; Magnani, M. Photobacteriosis: Prevention and diagnosis. J. Immunol. Res. 2014, 2014, 793817. [Google Scholar] [CrossRef] [Green Version]
- Colloca, F.; Cerasi, S. Sparus aurata. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/sparus_aurata/en (accessed on 14 December 2021).
- Bagni, M. Dicentrarchus labrax. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/dicentrarchus_labrax/en (accessed on 14 December 2021).
- Colen, R.; Ramalho, A.; Rocha, F.; Dinis, M.T. Solea solea. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/solea_spp/en (accessed on 14 December 2021).
- Dhirendra, P.T. Seriola quiqueradiata. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/seriola_quinqueradiata/en (accessed on 14 December 2021).
- Kaiser, J.B.; Holt, J.G. Rachycentron canadum. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/rachycentron_canadum/en (accessed on 14 December 2021).
- do Vale, A.; Costa-Ramos, C.; Silva, A.; Silva, D.S.; Gartner, F.; dos Santos, N.M.; Silva, M.T. Systemic macrophage and neutrophil destruction by secondary necrosis induced by a bacterial exotoxin in a Gram-negative septicaemia. Cell. Microbiol. 2007, 9, 988–1003. [Google Scholar] [CrossRef]
- Tung, M.-C.T.; Tsai, S.S.; Ho, L.-F.; Huang, S.T.; Chen, S.C. An acute septicemic infection of Pasteurella organism in pond-cultured Formosa snake-head fish (Channa maculata Lacepeda) in Taiwan. Fish Pathol. 1985, 20, 143–148. [Google Scholar] [CrossRef] [Green Version]
- Hawke, J.P.; Plakas, S.M.; Minton, R.V.; McPhearson, R.M.; Snider, T.G.; Guarino, A.M. Fish pasteurellosis of cultured striped bass (Morone saxatilis) in coastal Alabama. Aquaculture 1987, 65, 193–204. [Google Scholar] [CrossRef]
- Noya, M.; Magarinos, B.; Toranzo, A.E.; Lamas, J. Sequential pathology of experimental pasteurellosis in gilthead seabream Sparus aurata. A light- and electron-microscopic study. Dis. Aquat. Org. 1995, 21, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Magarinos, B.; Toranzo, A.E.; Romalde, J.L. Phenotypic and pathobiological characteristics of Pasteurella piscicida. Annu. Rev. Fish Dis. 1996, 6, 41–64. [Google Scholar] [CrossRef]
- do Vale, A.; Pereira, C.; Osorio, C.R.; dos Santos, N.M.S. The Apoptogenic Toxin AIP56 Is Secreted by the Type II Secretion System of Photobacterium damselae subsp. piscicida. Toxins 2017, 9, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, M.T.; do Vale, A.; dos Santos, N.M. Secondary necrosis in multicellular animals: An outcome of apoptosis with pathogenic implications. Apoptosis 2008, 13, 463–482. [Google Scholar] [CrossRef]
- Pereira, L.M.; Pinto, R.D.; Silva, D.S.; Moreira, A.R.; Beitzinger, C.; Oliveira, P.; Sampaio, P.; Benz, R.; Azevedo, J.E.; dos Santos, N.M.S.; et al. Intracellular trafficking of AIP56, an NF-kB cleaving toxin from Photobacterium damselae subsp. piscicida. Infect. Immun. 2014, 82, 5270–5285. [Google Scholar] [CrossRef] [Green Version]
- Silva, D.S.; Pereira, L.M.G.; Moreira, A.R.; Ferreira-da-Silva, F.; Brito, R.M.; Faria, T.Q.; Zornetta, I.; Montecucco, C.; Oliveira, P.; Azevedo, J.E.; et al. The Apoptogenic Toxin AIP56 is a Metalloprotease A-B Toxin that Cleaves NF-κb P65. PLoS Pathog. 2013, 9, e1003128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, I.S.; Pereira, L.M.G.; Lisboa, J.; Pereira, C.; Oliveira, P.; Dos Santos, N.M.S.; do Vale, A. Involvement of Hsp90 and cyclophilins in intoxication by AIP56, a metalloprotease toxin from Photobacterium damselae subsp. piscicida. Sci. Rep. 2019, 9, 9019. [Google Scholar] [CrossRef]
- Abushattal, S.; Vences, A.; Osorio, C.R. A virulence gene typing scheme for Photobacterium damselae subsp. piscicida, the causative agent of fish photobacteriosis, reveals a high prevalence of plasmid-encoded virulence factors and of type III secretion system genes. Aquaculture 2020, 521, 735057. [Google Scholar] [CrossRef]
- Magarinos, B.; Romalde, J.L.; Bandin, I.; Fouz, B.; Toranzo, A.E. Phenotypic, antigenic, and molecular characterization of Pasteurella piscicida strains isolated from fish. Appl. Environ. Microbiol. 1992, 58, 3316–3322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- do Vale, A.; Marques, F.; Silva, M.T. Apoptosis of sea bass (Dicentrarchus labrax L.) neutrophils and macrophages induced by experimental infection with Photobacterium damselae subsp. piscicida. Fish Shellfish Immunol. 2003, 15, 129–144. [Google Scholar] [CrossRef]
- Draft Assembly of Dicentrarchus labrax Genome (dicLab v1.0c). Available online: http://seabass.mpipz.mpg.de/ (accessed on 18 October 2021).
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- do Vale, A.; Afonso, A.; Silva, M.T. The professional phagocytes of sea bass (Dicentrarchus labrax L.): Cytochemical characterisation of neutrophils and macrophages in the normal and inflamed peritoneal cavity. Fish Shellfish Immunol. 2002, 13, 183–198. [Google Scholar] [CrossRef]
- Toranzo, A.E.; Barreiro, S.; Casal, J.F.; Figueras, A.; Magarinos, B.; Barja, J.L. Pasteurellosis in cultured gilthead seabream (Sparus aurata): First report in Spain. Aquaculture 1991, 99, 1–15. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Muñoz, P.; López-Muñoz, A.; Pelegrín, P.; García Ayala, A.; Mulero, V.; Meseguer, J. Early innate immune response and redistribution of inflammatory cells in the bony fish gilthead seabream experimentally infected with Vibrio anguillarum. Cell Tissue Res. 2005, 320, 61–68. [Google Scholar] [CrossRef]
- Sepulcre, M.; Pelegrín, P.; Mulero, V.; Meseguer, J. Characterisation of gilthead seabream acidophilic granulocytes by a monoclonal antibody unequivocally points to their involvement in fish phagocytic response. Cell Tissue Res. 2002, 308, 97–102. [Google Scholar] [CrossRef]
- Sepulcre, M.P.; Lopez-Castejon, G.; Meseguer, J.; Mulero, V. The activation of gilthead seabream professional phagocytes by different PAMPs underlines the behavioural diversity of the main innate immune cells of bony fish. Mol. Immunol. 2007, 44, 2019–2026. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, A.; Wan, F. Interference with nuclear factor kappaB signaling pathway by pathogen-encoded proteases: Global and selective inhibition. Mol. Microbiol. 2016, 99, 439–452. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.H.; Cheng, T.-C.; Wang, P.-C.; Chen, S.-C. Genotypic diversity, and molecular and pathogenic characterization of Photobacterium damselae subsp. piscicida isolated from different fish species in Taiwan. J. Fish Dis. 2020, 43, 757–774. [Google Scholar] [CrossRef]
- Baseggio, L.; Silayeva, O.; Buller, N.; Landos, M.; Englestädter, J.; Barnes, A.C. Complete, closed and curated genome sequences of Photobacterium damselae subsp. piscicida isolates from Australia indicate mobilome-driven localized evolution and novel pathogenicity determinants. Microb. Genom. 2021, 7, 000562. [Google Scholar] [CrossRef]
- ProtParam Tool on the Expasy Server. Available online: http://www.expasy.org/tools/protparam.html (accessed on 18 October 2021).
- Afonso, A.; Ellis, A.E.; Silva, M.T. The leucocyte population of the unstimulated peritoneal cavity of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 1997, 7, 335–348. [Google Scholar] [CrossRef]
- Costa-Ramos, C.; do Vale, A.; Ludovico, P.; dos Santos, N.M.S.; Silva, M.T. The bacterial exotoxin AIP56 induces fish macrophage and neutrophil apoptosis using mechanisms of the extrinsic and intrinsic pathways. Fish Shellfish Immunol. 2011, 30, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Translate Tool on the Expasy Server. Available online: https://web.expasy.org/translate/ (accessed on 18 October 2021).
- Madeira, F.; Park, Y.m.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
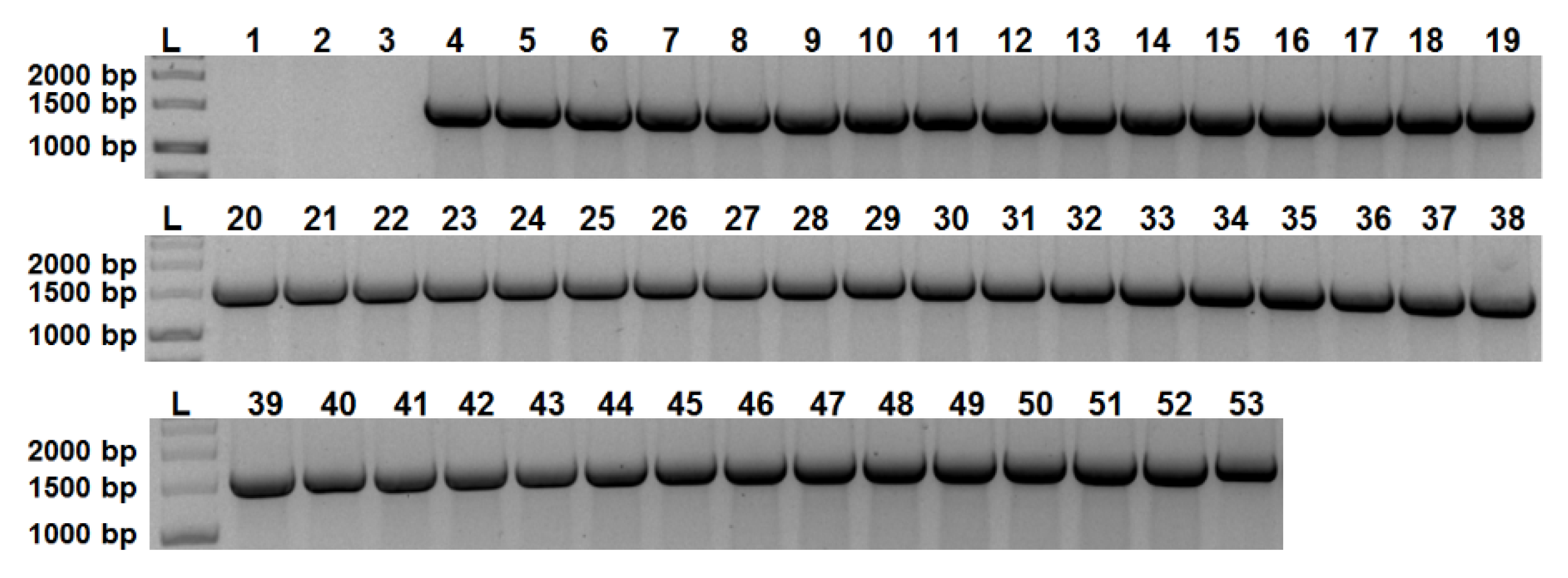
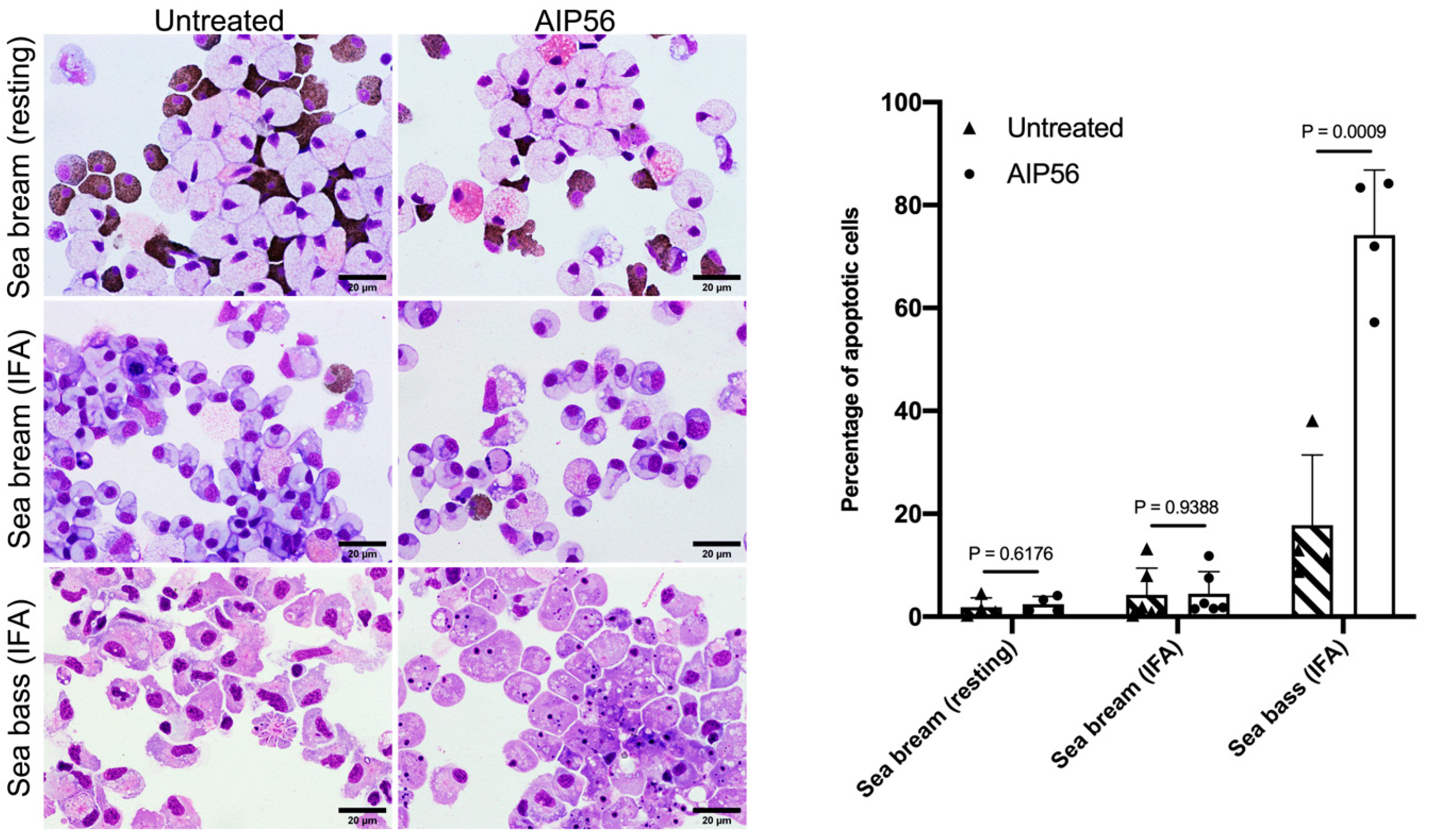

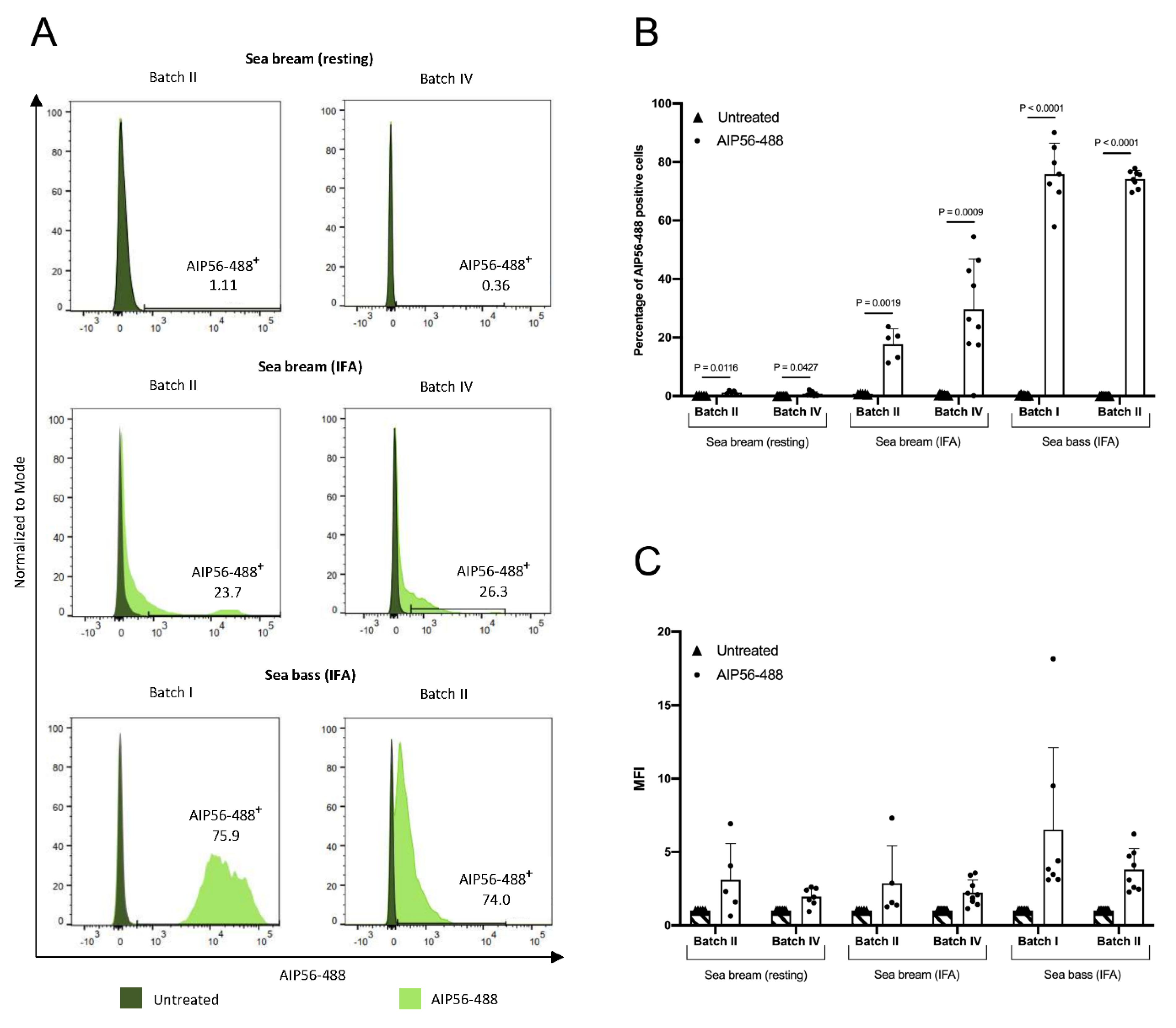
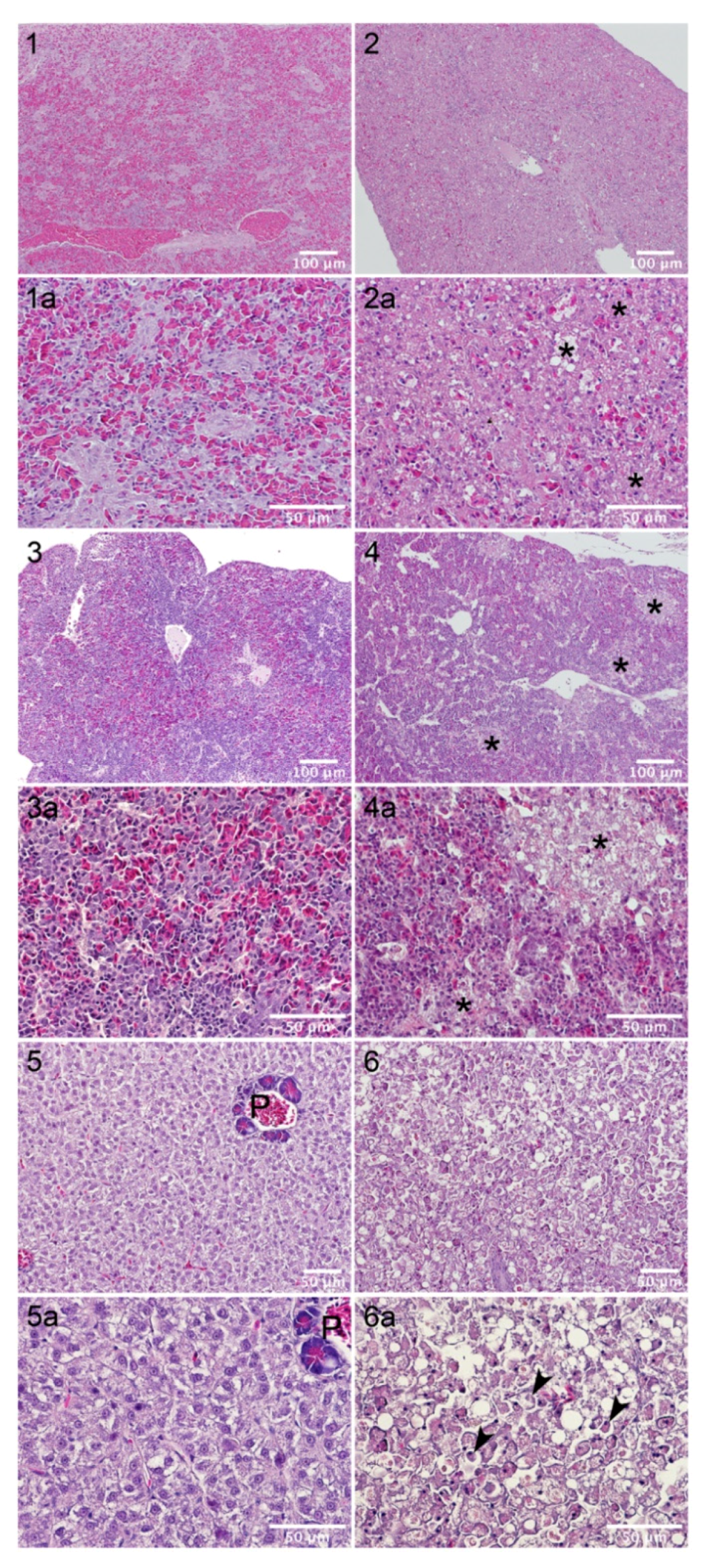
| Strain | Host Species | Year | Geographical Origin | |
|---|---|---|---|---|
| 1 | ATCC29690 | Seriola quinqueradiata | 1972 | Japan |
| 2 | EPOY8803-II | Epinephelus akaara | 1988 | Japan |
| 3 | DI21 | Sparus aurata | 1990 | Spain |
| 4 | MT1415 | Dicentrarchus labrax | unknown | Italy |
| 5 | SPSA14-1 | Sparus aurata | 2014 | Spain |
| 6 | SPDL15-1 | Dicentrarchus labrax | 2015 | Spain |
| 7 | SPSA16-1 | Sparus aurata | 2016 | Spain (Mediterranean) |
| 8 | SPDL17-1 | Dicentrarchus labrax | 2017 | Spain (Atlantic) |
| 9 | SPDL17-3 | Dicentrarchus labrax | 2017 | Spain (Atlantic) |
| 10 | SPDL17-5 | Dicentrarchus labrax | 2017 | Spain (Atlantic) |
| 11 | SPDL17-2 | Dicentrarchus labrax | 2017 | Spain (Canary Islands) |
| 12 | SPDL17-4 | Dicentrarchus labrax | 2017 | Spain (Mediterranean) |
| 13 | GRDL17-1 | Dicentrarchus labrax | 2017 | Greece |
| 14 | SPSA18-1 | Sparus aurata | 2018 | Spain (Canary Islands) |
| 15 | SPSA18-2 | Sparus aurata | 2018 | Spain (Mediterranean) |
| 16 | SPDL18-1 | Dicentrarchus labrax | 2018 | Spain (Atlantic) |
| 17 | SPDL18-2 | Dicentrarchus labrax | 2018 | Spain (Mediterranean) |
| 18 | SPDL18-3 | Dicentrarchus labrax | 2018 | Spain (Mediterranean) |
| 19 | SPDL18-4 | Dicentrarchus labrax | 2018 | Spain (Mediterranean) |
| 20 | SPDL18-5 | Dicentrarchus labrax | 2018 | Spain (Mediterranean) |
| 21 | SPDL18-6 | Dicentrarchus labrax | 2018 | Spain (Mediterranean) |
| 22 | SPDL18-7 | Dicentrarchus labrax | 2018 | Spain (Mediterranean) |
| 23 | ITDL18-1 | Dicentrarchus labrax | 2018 | Italy |
| 24 | SPSA19-1 | Sparus aurata | 2019 | Spain |
| 25 | SPSA19-2 | Sparus aurata | 2019 | Spain (Canary Islands) |
| 26 | GRSA19-1 | Sparus aurata | 2019 | Greece |
| 27 | SPDL19-2 | Dicentrarchus labrax | 2019 | Spain |
| 28 | SPDL19-3 | Dicentrarchus labrax | 2019 | Spain (Atlantic) |
| 29 | SPDL19-4 | Dicentrarchus labrax | 2019 | Spain (Canary Islands) |
| 30 | SPDL19-1 | Dicentrarchus labrax | 2019 | Spain (Mediterranean) |
| 31 | SPDL19-5 | Dicentrarchus labrax | 2019 | Spain (Mediterranean) |
| 32 | SPDL19-6 | Dicentrarchus labrax | 2019 | Spain (Mediterranean) |
| 33 | SPDL19-7 | Dicentrarchus labrax | 2019 | Spain (Mediterranean) |
| 34 | SPDL19-8 | Dicentrarchus labrax | 2019 | Spain (Mediterranean) |
| 35 | SPDL19-9 | Dicentrarchus labrax | 2019 | Spain (Mediterranean) |
| 36 | SPDL19-10 | Dicentrarchus labrax | 2019 | Spain (Mediterranean) |
| 37 | GRDL19-1 | Dicentrarchus labrax | 2019 | Greece |
| 38 | GRDL19-2 | Dicentrarchus labrax | 2019 | Greece |
| 39 | ITDL19-1 | Dicentrarchus labrax | 2019 | Italy |
| 40 | SPSA20-1 | Sparus aurata | 2020 | Spain (Mediterranean) |
| 41 | SPSA20-2 | Sparus aurata | 2020 | Spain (Mediterranean) |
| 42 | GRSA20-1 | Sparus aurata | 2020 | Greece |
| 43 | SPDL20-1 | Dicentrarchus labrax | 2020 | Spain (Atlantic) |
| 44 | SPDL20-2 | Dicentrarchus labrax | 2020 | Spain (Atlantic) |
| 45 | SPDL20-5 | Dicentrarchus labrax | 2020 | Spain (Atlantic) |
| 46 | SPDL20-6 | Dicentrarchus labrax | 2020 | Spain (Atlantic) |
| 47 | SPDL20-3 | Dicentrarchus labrax | 2020 | Spain (Mediterranean) |
| 48 | SPDL20-4 | Dicentrarchus labrax | 2020 | Spain (Mediterranean) |
| 49 | SPDL20-7 | Dicentrarchus labrax | 2020 | Spain (Mediterranean) |
| 50 | SPDL20-8 | Dicentrarchus labrax | 2020 | Spain (Mediterranean) |
| 51 | GRDL20-1 | Dicentrarchus labrax | 2020 | Greece |
| 52 | GRDL20-2 | Dicentrarchus labrax | 2020 | Greece |
| 53 | PTDL20-1 | Dicentrarchus labrax | 2020 | Portugal |
| Experiment | Species | Average Weight (g) | Batch | Treatment | Dose (µg/g Body Weight) | Mortality |
|---|---|---|---|---|---|---|
| 1 | Sparus aurata | 11.4 ± 2.6 | I | AIP56 | 0.9 | 0% (0/10) |
| AIP56 | 0.09 | 0% (0/10) | ||||
| Dicentrarchus labrax | 9.6 ± 2.1 | I | AIP56 | 1 | 100% (10/10) | |
| AIP56 | 0.1 | 60% (6/10) | ||||
| 2 | Sparus aurata | 12.0 ± 2.0 | I | AIP56 | 4.2 | 0% (0/10) |
| AIP56 | 0.8 | 0% (0/10) | ||||
| Dicentrarchus labrax | 10.4 ± 2.1 | I | AIP56 | 1 | 100% (10/10) | |
| AIP56 | 0.1 | 90% (9/10) | ||||
| 3 | Sparus aurata | 16.0 ± 3.8 | I | AIP56 | 3.1 | 0% (0/10) |
| Dicentrarchus labrax | 14.8 ± 6.6 | I | AIP56 | 0.7 | 100% (10/10) | |
| AIP56 | 0.07 | 70% (7/10) | ||||
| Vehicle | x | 0% (0/10) | ||||
| 4 | Sparus aurata | 11.6 ± 2.8 | II | AIP56 | 2.2 | 100% (10/10) |
| AIP56 | 0.2 | 30% (3/10) | ||||
| AIP56AAIVAA | 2.2 | 0% (0/10) | ||||
| Vehicle | x | 0% (0/10) | ||||
| Dicentrarchus labrax | 53.6 ± 12.5 | I | AIP56 | 0.2 | 90% (9/10) | |
| 5 | Sparus aurata | 34.4 ± 4.5 | III | AIP56 | 2.9 | 0% (0/6) |
| Dicentrarchus labrax | 177.8 ± 23.2 | II | AIP56 | 0.06 | 100% (5/5) | |
| 6 | Sparus aurata | 53.9 ± 14.5 | IV | AIP56 | 1.9 | 0% (0/5) |
| Dicentrarchus labrax | 183.4 ± 41.0 | II | AIP56 | 0.005 | 80% (4/5) |
| Experiment | Species | Average Weight (g) | Batch | Treatment | Dose (µg/g Body Weight) | Lethality | Histopathology |
|---|---|---|---|---|---|---|---|
| 1 | Sparus aurata | 11.4 ± 2.6 | I | AIP56 | 0.9 | Yes (n = 10) | No |
| AIP56 | 0.09 | Yes (n = 10) | No | ||||
| Dicentrarchus labrax | 9.6 ± 2.1 | I | AIP56 | 1 | Yes (n = 10) | No | |
| AIP56 | 0.1 | Yes (n = 10) | No | ||||
| 2 | Sparus aurata | 12.0 ± 2.0 | I | AIP56 | 4.2 | Yes (n = 10) | No |
| AIP56 | 0.8 | Yes (n = 10) | No | ||||
| Dicentrarchus labrax | 10.4 ± 2.1 | I | AIP56 | 1 | Yes (n = 10) | No | |
| AIP56 | 0.1 | Yes (n = 10) | No | ||||
| 3 | Sparus aurata | 16.0 ± 3.8 | I | AIP56 | 3.1 | Yes (n = 10) | Yes (n = 10) |
| Vehicle | x | No | Yes (n = 10) | ||||
| Dicentrarchus labrax | 14.8 ± 6.6 | I | AIP56 | 0.7 | Yes (n = 10) | Yes (n = 10) | |
| AIP56 | 0.07 | Yes (n = 10) | No | ||||
| Vehicle | x | Yes (n = 10) | Yes (n = 10) | ||||
| 4 | Sparus aurata | 11.6 ± 2.8 | II | AIP56 | 2.2 | Yes (n = 10) | Yes (n = 10) |
| AIP56 | 0.2 | Yes (n = 10) | No | ||||
| AIP56AAIVAA | 2.2 | Yes (n = 10) | No | ||||
| Vehicle | x | Yes (n = 10) | Yes (n = 10) | ||||
| Dicentrarchus labrax | 53.6 ± 12.5 | I | AIP56 | 0.2 | Yes (n = 10) | No | |
| 5 | Sparus aurata | 34.4 ± 4.5 | III | AIP56 | 2.9 | Yes (n = 6) | No |
| Dicentrarchus labrax | 177.8 ± 23.2 | II | AIP56 | 0.06 | Yes (n = 5) | No | |
| 6 | Sparus aurata | 53.9 ± 14.5 | IV | AIP56 | 1.9 | Yes (n = 5) | No |
| Dicentrarchus labrax | 183.4 ± 41.0 | II | AIP56 | 0.005 | Yes (n = 5) | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freitas, I.L.; Teixeira, A.; Loureiro, I.; Lisboa, J.; Saraiva, A.; dos Santos, N.M.S.; do Vale, A. Susceptibility of Sea Bream (Sparus aurata) to AIP56, an AB-Type Toxin Secreted by Photobacterium damselae subsp. piscicida. Toxins 2022, 14, 119. https://doi.org/10.3390/toxins14020119
Freitas IL, Teixeira A, Loureiro I, Lisboa J, Saraiva A, dos Santos NMS, do Vale A. Susceptibility of Sea Bream (Sparus aurata) to AIP56, an AB-Type Toxin Secreted by Photobacterium damselae subsp. piscicida. Toxins. 2022; 14(2):119. https://doi.org/10.3390/toxins14020119
Chicago/Turabian StyleFreitas, Inês Lua, Alexandra Teixeira, Inês Loureiro, Johnny Lisboa, Aurélia Saraiva, Nuno Miguel Simões dos Santos, and Ana do Vale. 2022. "Susceptibility of Sea Bream (Sparus aurata) to AIP56, an AB-Type Toxin Secreted by Photobacterium damselae subsp. piscicida" Toxins 14, no. 2: 119. https://doi.org/10.3390/toxins14020119
APA StyleFreitas, I. L., Teixeira, A., Loureiro, I., Lisboa, J., Saraiva, A., dos Santos, N. M. S., & do Vale, A. (2022). Susceptibility of Sea Bream (Sparus aurata) to AIP56, an AB-Type Toxin Secreted by Photobacterium damselae subsp. piscicida. Toxins, 14(2), 119. https://doi.org/10.3390/toxins14020119






