In Silico Approach Gives Insights into Ig-like Fold Containing Proteins in Vibrio parahaemolyticus: A Focus on the Fibrillar Adhesins
Abstract
:1. Introduction
2. Results
2.1. Distribution of Ig-like Fold Containing Proteins
2.2. Orthogroup Analysis
2.3. Identification of Fibrillar Adhesin Like Proteins
2.4. Three-Dimensional Structure and Protein Interface Analysis
2.5. Molecular Docking
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Definition of Profiles for Ig-like Fold Proteins
5.2. Orthogroup Inference of Ig-Fold Containing Proteins
5.3. Identification and Characterization of Fibrillar Adhesin-like Proteins
5.4. Structure Prediction and Evaluation of Fibrillar Adhesin-like Proteins
5.5. Molecular Docking of Virulence Inhibitors with Fibrillar Adhesion Proteins
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, L.H.; Teh, C.S.J.; Yap, K.P.; Thong, K.L. Diagnostic approaches and contribution of next-generation sequencing technologies in genomic investigation of Vibrio parahaemolyticus that caused Acute Hepatopancreatic Necrosis Disease (AHPND). Aquac. Int. 2020, 28, 2547–2559. [Google Scholar] [CrossRef]
- Ma, R.; Wang, Y.; Huang, L.; Ying, N.; Ling, H.; Fang, W. Complete genome sequence of highly pathogenic Vibrio parahaemolyticus isolated from mariculture Penaeus vannamei reveals virulence factor genes. Aquac. Res. 2021, 52, 1401–1413. [Google Scholar] [CrossRef]
- Yang, J.; Deng, D.; Brandt, B.W.; Nazmi, K.; Wu, Y.; Crielaard, W.; Ligtenberg, A.J. Diversity of SpaP in genetic and salivary agglutinin mediated adherence among Streptococcus mutans strains. Sci. Rep. 2019, 9, 19943. [Google Scholar] [CrossRef]
- Monzon, V.; Lafita, A.; Bateman, A. Discovery of fibrillar adhesins across bacterial species. BMC Genom. 2021, 22, 550. [Google Scholar] [CrossRef]
- Sikora, A.E.; Zielke, R.A.; Lawrence, D.A.; Andrews, P.C.; Sandkvist, M. Proteomic Analysis of the Vibrio cholerae Type II Secretome Reveals New Proteins, Including Three Related Serine Proteases. J. Biol. Chem. 2011, 286, 16555–16566. [Google Scholar] [CrossRef] [Green Version]
- Gadwal, S.; Korotkov, K.V.; Delarosa, J.R.; Hol, W.G.J.; Sandkvist, M. Functional and structural characterization of Vibrio cholerae extracellular serine protease B, VesB. J. Biol. Chem. 2014, 289, 8288–8298. [Google Scholar] [CrossRef] [Green Version]
- Schleicher, T.R. Proteomic Characterization of Host-Microbe Interactions in the Euprymna scolopes Light Organ Symbiosis with Vibrio fischeri. 2014. Available online: https://opencommons.uconn.edu/dissertations/481/ (accessed on 2 December 2021).
- He, X.; Yu, M.; Wu, Y.; Ran, L.; Liu, W.; Zhang, X.-H. Two Highly Similar Chitinases from Marine Vibrio Species have Different Enzymatic Properties. Mar. Drugs 2020, 18, 139. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.S.; Yu, Z.; Maxwell, K.L.; Davidson, A.R. Ig-Like Domains on Bacteriophages: A Tale of Promiscuity and Deceit. J. Mol. Biol. 2006, 359, 496–507. [Google Scholar] [CrossRef]
- Solís-Sánchez, A.; Hernández-Chiñas, U.; Navarro-Ocaña, A.; De la Mora, J.; Xicohtencatl-Cortes, J.; Eslava-Campos, C. Genetic characterization of ØVC8 lytic phage for Vibrio cholerae O1. Virol. J. 2016, 13, 47. [Google Scholar] [CrossRef] [Green Version]
- Bodelón, G.; Palomino, C.; Fernández, L.Á. Immunoglobulin domains in Escherichia coli and other Enterobacteria: From pathogenesis to applications in antibody technologies. FEMS Microbiol. Rev. 2013, 37, 204–250. [Google Scholar] [CrossRef] [Green Version]
- Buchinger, E.; Knudsen, D.H.; Behrens, M.A.; Pedersen, J.S.; Aarstad, O.A.; Tøndervik, A.; Valla, S.; Skjåk-Bræk, G.; Wimmer, R.; Aachmann, F.L. Structural and functional characterization of the R-modules in alginate C-5 epimerases AlgE4 and AlgE6 from Azotobacter vinelandii. J. Biol. Chem. 2014, 289, 31382–31396. [Google Scholar] [CrossRef] [Green Version]
- Kong, W.-J.; Zhao, Y.-L.; Xiao, X.-H.; Wang, J.-B.; Li, H.-B.; Li, Z.-L.; Jin, C.; Liu, Y. Spectrum–Effect relationships between ultra performance liquid chromatography fingerprints and anti-bacterial activities of Rhizoma coptidis. Anal. Chim. Acta 2009, 634, 279–285. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Hwang, E.-K.; Baek, Y.-M.; Kim, H.-D. Deodorizing function and antibacterial activity of fabrics dyed with gallnut (Galla Chinensis) extract. Text. Res. J. 2015, 85, 1045–1054. [Google Scholar] [CrossRef]
- Nowak, A.; Zakłos-Szyda, M.; Błasiak, J.; Nowak, A.; Zhang, Z.; Zhang, B. Potential of Schisandra chinensis (Turcz.) Baill. in human health and nutrition: A review of current knowledge and therapeutic perspectives. Nutrients 2019, 11, 333. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Stein, B.D. Antimicrobial activity of a combination of Mume fructus, Schizandrae fructus, and Coptidis rhizoma on enterohemorrhagic Escherichia coli O26, O111, and O157 and its effect on Shiga toxin releases. Foodborne Pathog. Dis. 2011, 8, 643–646. [Google Scholar] [CrossRef]
- Jakubovics, N.S.; Brittan, J.L.; Dutton, L.C.; Jenkinson, H.F. Multiple adhesin proteins on the cell surface of Streptococcus gordonii are involved in adhesion to human fibronectin. Microbiology 2009, 155, 3572. [Google Scholar] [CrossRef] [Green Version]
- Back, C.R.; Sztukowska, M.N.; Till, M.; Lamont, R.J.; Jenkinson, H.F.; Nobbs, A.H.; Race, P.R. The Streptococcus gordonii Adhesin CshA Protein Binds Host Fibronectin via a Catch-Clamp Mechanism. J. Biol. Chem. 2017, 292, 1538–1549. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.-H.; Jiang, Y.-L.; Zhang, J.; Wang, L.; Bai, X.-H.; Zhang, S.-J.; Ren, Y.-M.; Li, N.; Zhang, Y.-H.; Zhang, Z.; et al. Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells. PLOS Pathog. 2014, 10, e1004169. [Google Scholar] [CrossRef]
- Oleastro, M.; Ménard, A. The Role of Helicobacter pylori Outer Membrane Proteins in Adherence and Pathogenesis. Biology 2013, 2, 1110–1134. [Google Scholar] [CrossRef] [Green Version]
- van ‘t Wout, E.F.; van Schadewijk, A.; van Boxtel, R.; Dalton, L.E.; Clarke, H.J.; Tommassen, J.; Marciniak, S.J.; Hiemstra, P.S. Virulence factors of Pseudomonas aeruginosa induce both the unfolded protein and integrated stress responses in airway epithelial cells. PLoS Pathog. 2015, 11, e1004946. [Google Scholar] [CrossRef] [Green Version]
- Quintieri, L.; Fanelli, F.; Zühlke, D.; Caputo, L.; Logrieco, A.F.; Albrecht, D.; Riedel, K. Biofilm and Pathogenesis-Related Proteins in the Foodborne P. fluorescens ITEM 17298 With Distinctive Phenotypes During Cold Storage. Front. Microbiol. 2020, 11, 991. [Google Scholar] [CrossRef]
- King, M.M.; Kayastha, B.B.; Franklin, M.J.; Patrauchan, M.A. Calcium Regulation of Bacterial Virulence. In Calcium Signaling; Islam, M.S., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 827–855. [Google Scholar]
- Guo, S.; Stevens, C.A.; Vance, T.D.R.; Olijve, L.L.C.; Graham, L.A.; Campbell, R.L.; Yazdi, S.R.; Escobedo, C.; Bar-Dolev, M.; Yashunsky, V.; et al. Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice. Sci. Adv. 2017, 3, e1701440. [Google Scholar] [CrossRef] [Green Version]
- Tiruvayipati, S.; Bhassu, S. Host, pathogen and the environment: The case of Macrobrachium rosenbergii, Vibrio parahaemolyticus and magnesium. Gut Pathog. 2016, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Choy, H.A.; Kelley, M.M.; Chen, T.L.; Møller, A.K.; Matsunaga, J.; Haake, D.A. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 2007, 75, 2441–2450. [Google Scholar] [CrossRef] [Green Version]
- Zavala-Alvarado, C.; Huete, S.G.; Vincent, A.T.; Sismeiro, O.; Legendre, R.; Varet, H.; Bussotti, G.; Lorioux, C.; Lechat, P.; Coppée, J.-Y.; et al. The oxidative stress response of pathogenic Leptospira is controlled by two peroxide stress regulators which putatively cooperate in controlling virulence. PLoS Pathog. 2021, 17, e1009087. [Google Scholar] [CrossRef]
- Riangrungroj, P.; Polizzi, K.M. BeQuIK (Biosensor Engineered Quorum Induced Killing): Designer bacteria for destroying recalcitrant biofilms. Microb. Biotechnol. 2020, 13, 311–314. [Google Scholar] [CrossRef] [Green Version]
- Hüttener, M.; Prieto, A.; Aznar, S.; Bernabeu, M.; Glaría, E.; Valledor, A.F.; Paytubi, S.; Merino, S.; Tomás, J.; Juárez, A. Expression of a novel class of bacterial Ig-like proteins is required for IncHI plasmid conjugation. PLoS Genet. 2019, 15, e1008399. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Wang, B.; Wu, Y. Structural characterization and function prediction of immunoglobulin-like fold in cell adhesion and cell signaling. J. Chem. Inf. Modeling 2018, 58, 532–542. [Google Scholar] [CrossRef]
- Pownraj, B.; Ramu, M.; Haque, E. Important Insight of Hypothetical Proteins from E. faecium Strain 13-022, the Drug Targeted Virulent Big-1 Adhesins: An in-silico Approach. Int. J. Pharm. Investig. 2021, 11, 32–40. [Google Scholar] [CrossRef]
| Pfam ID | Pfam | Escherichia coli K-12 MG1655 | Vibrio cholerae NCTC 9420 | Vibrio parahaemolyticus CHN25 | Vibrio parahaemolyticus RIMD 2210633 |
|---|---|---|---|---|---|
| PF00028 | Cadherin | 0 | 1 | 0 | 0 |
| PF00041 | fn3 | 0 | 1 | 0 | 0 |
| PF00207 | A2M | 1 | 0 | 0 | 0 |
| PF00630 | Filamin | 1 | 0 | 0 | 0 |
| PF00703 | Glyco_hydro_2 | 3 | 1 | 2 | 2 |
| PF00801 | PKD | 0 | 1 | 1 | 1 |
| PF00932 | LTD | 0 | 1 | 1 | 1 |
| PF01345 | DUF11 | 0 | 0 | 1 | 1 |
| PF01835 | MG2 | 1 | 0 | 0 | 1 |
| PF02010 | REJ | 0 | 0 | 1 | 1 |
| PF02368 | Big_2 | 0 | 1 | 1 | 1 |
| PF02369 | Big_1 | 1 | 0 | 0 | 0 |
| PF02753 | PapD_C | 10 | 0 | 0 | 0 |
| PF02903 | Alpha-amylase_N | 1 | 0 | 1 | 1 |
| PF02927 | CelD_N | 0 | 1 | 1 | 1 |
| PF03067 | LPMO_10 | 0 | 2 | 2 | 2 |
| PF03160 | Calx-beta | 0 | 1 | 2 | 1 |
| PF03174 | CHB_HEX_C | 0 | 1 | 1 | 1 |
| PF03404 | Mo-co_dimer | 0 | 0 | 1 | 1 |
| PF04234 | CopC | 1 | 0 | 0 | 0 |
| PF04379 | DUF525 | 1 | 1 | 1 | 1 |
| PF05345 | He_PIG | 0 | 0 | 1 | 1 |
| PF05753 | TRAP_beta | 0 | 0 | 1 | 1 |
| PF06832 | BiPBP_C | 1 | 0 | 0 | 0 |
| PF07233 | DUF1425 | 1 | 1 | 1 | 1 |
| PF07495 | Y_Y_Y | 0 | 1 | 0 | 0 |
| PF07703 | A2M_BRD | 1 | 0 | 0 | 0 |
| PF08329 | ChitinaseA_N | 0 | 1 | 1 | 1 |
| PF09134 | Invasin_D3 | 1 | 0 | 0 | 0 |
| PF09619 | YscW | 1 | 1 | 2 | 2 |
| PF10029 | DUF2271 | 0 | 0 | 1 | 1 |
| PF10610 | Tafi-CsgC | 1 | 0 | 0 | 0 |
| PF10633 | NPCBM_assoc | 0 | 0 | 2 | 2 |
| PF11412 | DsbC | 1 | 1 | 2 | 2 |
| PF11614 | FixG_C | 0 | 1 | 1 | 1 |
| PF11806 | Enterochelin_N | 1 | 0 | 0 | 0 |
| PF11940 | DUF3458 | 1 | 1 | 1 | 1 |
| PF11974 | bMG3 | 1 | 0 | 0 | 0 |
| PF12262 | Lipase_bact_N | 0 | 1 | 1 | 1 |
| PF13584 | BatD | 0 | 1 | 2 | 2 |
| PF13629 | T2SS-T3SS_pil_N | 0 | 0 | 2 | 2 |
| PF13860 | FlgD_ig | 1 | 1 | 3 | 3 |
| PF14310 | Fn3-like | 1 | 0 | 0 | 0 |
| PF14467 | DUF4426 | 0 | 1 | 1 | 1 |
| PF16184 | Cadherin_3 | 0 | 0 | 1 | 1 |
| PF16353 | LacZ_4 | 2 | 1 | 1 | 1 |
| PF16640 | Big_3_5 | 1 | 0 | 0 | 0 |
| PF16655 | PhoD_N | 0 | 0 | 1 | 1 |
| PF16967 | TcfC | 1 | 0 | 0 | 0 |
| PF17753 | Ig_mannosidase | 0 | 0 | 1 | 1 |
| PF17756 | RET_CLD1 | 0 | 1 | 0 | 0 |
| PF17786 | Mannosidase_ig | 0 | 0 | 1 | 1 |
| PF17789 | MG4 | 1 | 0 | 0 | 0 |
| PF17803 | Cadherin_4 | 0 | 1 | 4 | 4 |
| PF17892 | Cadherin_5 | 0 | 1 | 5 | 5 |
| PF17957 | Big_7 | 0 | 2 | 2 | 2 |
| PF17962 | bMG6 | 1 | 0 | 0 | 0 |
| PF17963 | Big_9 | 0 | 1 | 5 | 5 |
| PF17967 | Pullulanase_N2 | 0 | 0 | 1 | 1 |
| PF17970 | bMG1 | 1 | 0 | 0 | 0 |
| PF17972 | bMG5 | 1 | 0 | 0 | 0 |
| PF17973 | bMG10 | 1 | 0 | 0 | 0 |
| PF18200 | Big_11 | 0 | 1 | 0 | 0 |
| PF18565 | Glyco_hydro2_C5 | 1 | 0 | 0 | 0 |
| PF18911 | PKD_4 | 0 | 2 | 2 | 3 |
| PF19076 | CshA_repeat | 0 | 0 | 1 | 1 |
| Orthogroup | V. parahaemolyticus RIMD 2210633 | V. parahaemolyticus CHN25 | E. coli K-12 MG1655 | V. cholerae NCTC 9420 |
|---|---|---|---|---|
| OG0000000 | NP_414682.1, NP_415064.1, NP_415245.1, NP_415459.1, NP_415464.4, NP_416613.1, NP_416839.1, NP_417519.4, NP_417612.1, NP_418736.3 | |||
| OG0000001 | WP_005482309.1 | WP_065870880.1 | NP_414878.1, NP_416134.1, YP_026199.1 | WP_001243585.1 |
| OG0000002 | WP_005456243.1 | WP_005456243.1 | NP_415622.1 | WP_001261381.1 |
| OG0000003 | WP_005459620.1 | WP_005459620.1 | NP_414592.1 | WP_000383338.1 |
| OG0000004 | WP_005461146.1 | WP_005461146.1 | NP_414987.3 | WP_000756880.1 |
| OG0000005 | WP_005462292.1 | WP_005462292.1 | NP_415593.1 | WP_000929365.1 |
| OG0000006 | WP_005478597.1 | WP_017449375.1 | NP_418559.1 | WP_001259538.1 |
| OG0000007 | WP_005480222.1, WP_005480358.1 | WP_065871343.1, WP_205390631.1 | ||
| OG0000008 | WP_005481930.1 | WP_065870668.1 | NP_415452.1 | WP_071919720.1 |
| OG0000009 | WP_005455369.1 | WP_065871306.1 | WP_000815658.1 | |
| OG0000010 | WP_005461703.1 | WP_005461703.1 | WP_001233676.1 | |
| OG0000011 | WP_005478358.1 | WP_162780945.1 | WP_000238825.1 | |
| OG0000012 | WP_005479039.1 | WP_065870865.1 | WP_071919742.1 | |
| OG0000013 | WP_005479087.1 | WP_065870903.1 | WP_000925616.1 | |
| OG0000014 | WP_005479129.1 | WP_025504563.1 | WP_071919697.1 | |
| OG0000015 | WP_005480168.1 | WP_005480168.1 | WP_000744635.1 | |
| OG0000016 | WP_005480704.1 | WP_065870579.1 | WP_000848953.1 | |
| OG0000017 | WP_005481163.1 | WP_015297519.1 | WP_000076153.1 | |
| OG0000018 | WP_005482420.1 | WP_065870820.1 | NP_414937.2 | |
| OG0000019 | WP_005482861.1 | WP_065871072.1 | WP_001894571.1 | |
| OG0000020 | WP_005482868.1 | WP_065871083.1 | WP_000873599.1 | |
| OG0000021 | WP_005488937.1 | WP_065870960.1 | WP_000426028.1 | |
| OG0000022 | WP_005489828.1 | WP_065870398.1 | WP_000914817.1 | |
| OG0000023 | WP_011105887.1 | WP_065870696.1 | WP_080488946.1 | |
| OG0000024 | WP_005454128.1 | WP_005454128.1 | ||
| OG0000025 | WP_005456668.1 | WP_065870884.1 | ||
| OG0000026 | WP_005462598.1 | WP_065870556.1 | ||
| OG0000027 | WP_005463605.1 | WP_065871223.1 | ||
| OG0000028 | WP_005463939.1 | WP_005463939.1 | ||
| OG0000029 | WP_005477556.1 | WP_079879954.1 | ||
| OG0000030 | WP_005477759.1 | WP_065870697.1 | ||
| OG0000031 | WP_005479027.1 | WP_065870427.1 | ||
| OG0000032 | WP_005479631.1 | WP_005479631.1 | ||
| OG0000033 | WP_005479868.1 | WP_065871168.1 | ||
| OG0000034 | WP_005481629.1 | WP_065870795.1 | ||
| OG0000035 | WP_005482097.1 | WP_065871538.1 | ||
| OG0000036 | WP_005489282.1 | WP_065871245.1 | ||
| OG0000037 | WP_005490731.1 | WP_205390630.1 | ||
| OG0000038 | WP_005492007.1 | WP_205390635.1 | ||
| OG0000039 | WP_011105908.1 | WP_065870717.1 |
| Protein Accession | Molecular Weight (MW) | Isoelectric Point (PI) | Grand Average of Hydropathicity (GRAVY) | Instability Index | Aliphatic Index |
|---|---|---|---|---|---|
| WP_005477759.1 | 338,024.95 | 3.72 | −0.242 | 19.69 | 87.55 |
| WP_005480168.1 | 53,630.31 | 4.58 | −0.464 | 29.84 | 70.70 |
| WP_005489282.1 | 638,384.33 | 3.58 | −0.097 | 19.27 | 92.16 |
| WP_005490731.1 | 75,993.93 | 4.5 | −0.27 | 36.66 | 88.21 |
| Protein Accession | Homologues | ||||||
|---|---|---|---|---|---|---|---|
| 3D Structural Models | PDB ID | Organism | Protein Annotation | Unique Ligands | Chains | Protein Interface | |
| WP_005477759.1 |  | 1KAP | Pseudomonas aeruginosa | A two-domain protein AprA with a calcium binding parallel beta roll motif | ZN, CA | A [auth P] |  |
| B [auth I] | |||||||
 | 1AKL | P. aeruginosa | Alkaline protease | ZN, CA | A | 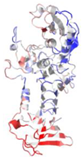 | |
 | 3VI1 | P. aeruginosa | Alkaline protease complexed with Substance P (1–6) | ZN, CA | C, D | 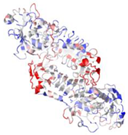 | |
| A, B | |||||||
| WP_005489282.1 | 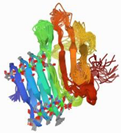 | 2ML1 | Azotobacter vinelandii | AlgE6R1 subunit from the Azotobacter vinelandii Mannuronan C5-epimerase AlgE6 | CA | A | 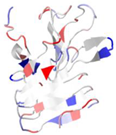 |
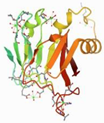 | 5JUH | Marinomonas primoryensis | C-terminal domain [12] of MpAFP, a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice | CA | A |  | |
| WP_005480168.1 |  | 2XWX | V. cholerae | Colonization factor GbpA | - | A, B | 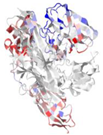 |
| WP_005490731.1 |  | 2N7S | Leptospira interrogans | Leptospiral immunoglobulin-like protein A (LigA), involved in the interaction of pathogenic Leptospira with mammalian host | - | A | 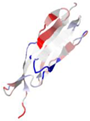 |
 | 2MH4 | L. interrogans | LigB-like protein | - | A |  | |
| WP_005477759.1 (1KAP) | WP_005489282.1 (2ML1) | WP_005480168.1 (2XWX) | WP_005490731.1 (2N7S) | |||||
|---|---|---|---|---|---|---|---|---|
| Ligand | Binding Affinity | Ligand | Binding Affinity | Ligand | Binding Affinity | Ligand | Binding Affinity | |
| 1 | Obacunone | −9.2 | Spinasterol | −8.5 | Cardenolide glycoside | −6.9 | Limonin | −9.9 |
| 2 | Pycnamine | −9.1 | Stigmasterin | −8.4 | Pycnamine | −6.9 | 8-Oxocoptisine | −9.7 |
| 3 | Rutin | −9.1 | Higenamine | −8.3 | 8-Oxocoptisine | −6.9 | Obacunone | −9.5 |
| 4 | Coptisine | −8.8 | Obacunone | −8.3 | Rutin | −6.8 | Ursolic Acid | −9.5 |
| 5 | Limonin | −8.8 | Sitosterol | −8.3 | Coptisine | −6.8 | Palmidin A | −9.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Wang, H. In Silico Approach Gives Insights into Ig-like Fold Containing Proteins in Vibrio parahaemolyticus: A Focus on the Fibrillar Adhesins. Toxins 2022, 14, 133. https://doi.org/10.3390/toxins14020133
Wang D, Wang H. In Silico Approach Gives Insights into Ig-like Fold Containing Proteins in Vibrio parahaemolyticus: A Focus on the Fibrillar Adhesins. Toxins. 2022; 14(2):133. https://doi.org/10.3390/toxins14020133
Chicago/Turabian StyleWang, Dan, and Haoran Wang. 2022. "In Silico Approach Gives Insights into Ig-like Fold Containing Proteins in Vibrio parahaemolyticus: A Focus on the Fibrillar Adhesins" Toxins 14, no. 2: 133. https://doi.org/10.3390/toxins14020133
APA StyleWang, D., & Wang, H. (2022). In Silico Approach Gives Insights into Ig-like Fold Containing Proteins in Vibrio parahaemolyticus: A Focus on the Fibrillar Adhesins. Toxins, 14(2), 133. https://doi.org/10.3390/toxins14020133





