Interindividual Differences in In Vitro Human Intestinal Microbial Conversion of 3-Acetyl-DON and 15-Acetyl-DON
Abstract
:1. Introduction
2. Results
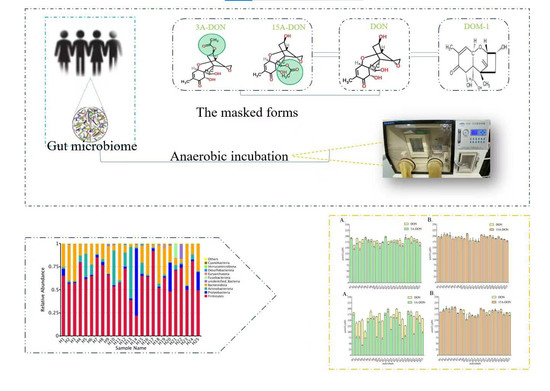
2.1. Quantitative Bacterial Taxonomy Profiling Analysis
2.2. Interindividual Differences of Human Gut Microbial Conversion of 3-Ac-DON and 15-Ac-DON
2.3. Estimation of Microbial Transformation of 3-Ac-DON and 15-Ac-DON by Human Individuals In Vivo
2.4. Correlation Analysis between Intestinal Microbial Profile and Metabolite Formation
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Solutions
5.2. Human Fecal Slurry Preparation
5.3. Fecal Batch Culture Incubations
5.4. HPLC Analysis of Fecal Incubation Samples
5.5. Microbial Taxonomic Profiling
5.6. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DON | Deoxynivalenol |
| 3-Ac-DON | 3-Acetyldeoxynivalenol |
| 15-Ac-DON | 15-Acetyldeoxynivalenol |
| DOM-1 | Deepoxydeoxynivalenol |
| DON-3-Glc | Deoxynivalenol-3-β-d-glucopyranoside |
References
- Guo, H.; Ji, J.; Wang, J.; Sun, X. Deoxynivalenol: Masked Forms, Fate during Food Processing, and Potential Biological Remedies. Compr. Rev. Food Sci. Food Saf. 2020, 19, 895–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payros, D.; Dobrindt, U.; Martin, P.; Secher, T.; Bracarense, A.P.F.L.; Boury, M.; Laffitte, J.; Pinton, P.; Oswald, E.; Oswald, I.P. The Food Contaminant Deoxynivalenol Exacerbates the Genotoxicity of Gut Microbiota. mBio 2017, 8, e00007–e00017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.-R.; He, K.; Landgraf, J.; Pan, X.; Pestka, J.J. Direct Activation of Ribosome-Associated Double-Stranded RNA-Dependent Protein Kinase (PKR) by Deoxynivalenol, Anisomycin and Ricin: A New Model for Ribotoxic Stress Response Induction. Toxins 2014, 6, 3406–3425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buszewska-Forajta, M. Mycotoxins, Invisible Danger of Feedstuff with Toxic Effect on Animals. Toxicon 2020, 182, 34–53. [Google Scholar] [CrossRef]
- EFSA Panel on Cantanimants in the Food Chain (CONTAM); Knutsen, H.K.; Alexander, J.; Barregård, L.; Bignami, M.; Brüschweiler, B.; Ceccatelli, S.; Cottrill, B.; DiNovi, M.; Grasl-Kraupp, B.; et al. Risks to Human and Animal Health Related to the Presence of Deoxynivalenol and Its Acetylated and Modified Forms in Food and Feed. EFSA J. 2017, 15, e04718. [Google Scholar] [CrossRef]
- Mruczyk, K.; Cisek-Woźniak, A.; Mizgier, M.; Wójciak, R.W. Natural Occurrence of Deoxynivalenol in Cereal-Based Baby Foods for Infants from Western Poland. Toxins 2021, 13, 777. [Google Scholar] [CrossRef]
- Dong, F.; Qiu, J.; Xu, J.; Yu, M.; Wang, S.; Sun, Y.; Zhang, G.; Shi, J. Effect of Environmental Factors on Fusarium Population and Associated Trichothecenes in Wheat Grain Grown in Jiangsu Province, China. Int. J. Food Microbiol. 2016, 230, 58–63. [Google Scholar] [CrossRef]
- Maresca, M. From the Gut to the Brain: Journey and Pathophysiological Effects of the Food-Associated Trichothecene Mycotoxin Deoxynivalenol. Toxins 2013, 5, 784–820. [Google Scholar] [CrossRef]
- Sobrova, P.; Adam, V.; Vasatkova, A.; Beklova, M.; Zeman, L.; Kizek, R. Deoxynivalenol and Its Toxicity. Interdiscip. Toxicol. 2010, 3, 94–99. [Google Scholar] [CrossRef]
- Broekaert, N.; Devreese, M.; De Baere, S.; De Backer, P.; Croubels, S. Modified Fusarium Mycotoxins Unmasked: From Occurrence in Cereals to Animal and Human Excretion. Food Chem. Toxicol. 2015, 80, 17–31. [Google Scholar] [CrossRef]
- Jin, J.; Spenkelink, A.; Beekmann, K.; Baccaro, M.; Xing, F.; Rietjens, I.M.C.M. Species Differences in in Vitro and Estimated in Vivo Kinetics for Intestinal Microbiota Mediated Metabolism of Acetyl-Deoxynivalenols. Mol. Nutr. Food Res. 2021, 65, 2001085. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C.; Zhou, T.; Yu, H.; Zhu, H.; Gong, J. Degradation of Trichothecene Mycotoxins by Chicken Intestinal Microbes. Food Chem. Toxicol. 2007, 45, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.W.; Duncan, G.; Richardson, A.J. The Human Fecal Microbiota Metabolizes Deoxynivalenol and Deoxynivalenol-3-Glucoside and May Be Responsible for Urinary Deepoxy-Deoxynivalenol. Appl. Environ. Microbiol. 2013, 79, 1821–1825. [Google Scholar] [CrossRef] [Green Version]
- Eriksen, G.S.; Pettersson, H. Lack of De-Epoxidation of Type B Trichothecenes in Incubates with Human Faeces. Food Addit. Contam. 2003, 20, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Rose, C.; Parker, A.; Jefferson, B.; Cartmell, E. The Characterization of Feces and Urine: A Review of the Literature to Inform Advanced Treatment Technology. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1827–1879. [Google Scholar] [CrossRef] [Green Version]
- Kararli, T.T. Comparison of the Gastrointestinal Anatomy, Physiology, and Biochemistry of Humans and Commonly Used Laboratory Animals. Biopharm. Drug Dispos. 1995, 16, 351–380. [Google Scholar] [CrossRef] [PubMed]
- King, R.R.; McQueen, R.E.; Levesque, D.; Greenhalgh, R. Transformation of Deoxynivalenol (Vomitoxin) by Rumen Microorganisms. J. Agric. Food Chem. 1984, 32, 1181–1183. [Google Scholar] [CrossRef]
- Hillman, E.T.; Lu, H.; Yao, T.; Nakatsu, C.H. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017, 32, 300–313. [Google Scholar] [CrossRef] [Green Version]
- Sakai, H.; Naito, K.; Takahashi, Y.; Sato, T.; Yamamoto, T. The Vigna Genome Server, ‘VigGS’: A Genomic Knowledge Base of the Genus Vigna Based on High-Quality, Annotated Genome Sequence of the Azuki Bean, Vigna Angularis (Willd) Ohwi Ohashi. Plant Cell Physiol. 2016, 57, e2. [Google Scholar] [CrossRef] [Green Version]
- Mendez-Catala, D.M.; Spenkelink, A.; Rietjens, I.M.; Beekmann, K. An In Vitro model to Quantify Interspecies Differences in Kinetics for Intestinal Microbial Bioactivation and Detoxification of Zearalenone. Toxicol. Rep. 2020, 7, 938–946. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, G.; Dai, Y.; Wang, Y.; Lee, Y.-W.; Shi, J.; Xu, J. Biodegradation of Deoxynivalenol by a Novel Microbial Consortium. Front. Microbiol. 2020, 10, 2964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ajandouz, E.H.; Berdah, S.; Moutardier, V.; Bege, T.; Birnbaum, D.J.; Perrier, J.; Di Pasquale, E.; Maresca, M. Hydrolytic Fate of 3/15-Acetyldeoxynivalenol in Humans: Specific Deacetylation by the Small Intestine and Liver Revealed Using in Vitro and ex Vivo Approaches. Toxins 2016, 8, 232. [Google Scholar] [CrossRef] [PubMed]
- Koppel, N.; Rekdal, V.M.; Balskus, E.P. Chemical Transformation of Xenobiotics by the Human Gut Microbiota. Science 2017, 356, 6344. [Google Scholar] [CrossRef]
- Daud, N.; Currie, V.; Duncan, G.; Busman, M.; Gratz, S.W. Intestinal Hydrolysis and Microbial Biotransformation of Diacetoxyscirpenol-α-Glucoside, HT-2-β-Glucoside and N-(1-Deoxy-d-Fructos-1-yl) Fumonisin B1 by Human Gut Microbiota in Vitro. Int. J. Food Sci. Nutr. 2019, 71, 540–548. [Google Scholar] [CrossRef] [PubMed]








| 3-Ac-DON | 15-Ac-DON | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individuals | DON Concentration (μM) | Transformation Percentage (%) | Transformation Rate (In Vitro) a | Calculated Transformation Time (min) | Reported Fecal Transit Time (min) | Conversion Rate (%) of Total Parent Compounds | DON Concentration (μM) | Transformation Percentage (%) | Transformation Rate (In Vitro) a | Calculated Transformation Time (min) | Reported Fecal Transit Time (min) | Conversion Rate (%) of Total Parent Compounds | |
| 1 | 3.54 ± 0.20 | 1.8 | 0.44 | 452 | 1848 | 100 | 0.00 | / | / | / | / | 1848 | / |
| 2 | 18.24 ± 0.61 | 9.1 | 2.28 | 88 | 100 | 2.18 ± 0.38 | 1.1 | 0.27 | 734 | 100 | |||
| 3 | 23.56 ± 0.75 | 11.8 | 2.94 | 68 | 100 | 3.56 * ± 1.13 | 1.8 | 0.07 | 2698 | 68.5 | |||
| 4 | 35.40 ± 1.14 | 17.7 | 4.42 | 45 | 100 | 5.40 ± 1.97 | 2.7 | 0.67 | 296 | 100 | |||
| 5 | 16.49 ± 0.56 | 8.2 | 2.06 | 97 | 100 | 5.32 ± 0.89 | 2.7 | 0.66 | 301 | 100 | |||
| 6 | 6.52 ± 0.76 | 3.3 | 0.81 | 245 | 100 | 3.36 * ± 1.39 | 1.7 | 0.07 | 2859 | 64.6 | |||
| 7 | 8.48 ± 0.99 | 4.2 | 1.06 | 189 | 100 | 2.00 * ± 0.15 | 1.0 | 0.04 | 4805 | 38.5 | |||
| 8 | 10.56 ± 0.47 | 5.3 | 1.32 | 152 | 100 | 2.74 * ± 0.20 | 1.4 | 0.06 | 3508 | 52.7 | |||
| 9 | 25.97 ± 0.14 | 13.0 | 3.24 | 62 | 100 | 9.26 ± 0.00 | 4.6 | 1.16 | 173 | 100 | |||
| 10 | 10.52 ± 0.31 | 5.3 | 1.31 | 152 | 100 | 3.60 * ± 0.27 | 1.8 | 0.08 | 2665 | 69.3 | |||
| 11 | 21.29 ± 0.82 | 10.6 | 2.66 | 75 | 100 | 7.11 ± 0.80 | 3.6 | 0.89 | 225 | 100 | |||
| 12 | 3.71 ± 0.11 | 1.9 | 0.46 | 431 | 100 | 2.11 * ± 0.13 | 1.1 | 0.04 | 4546 | 40.6 | |||
| 13 | 5.42 ± 0.21 | 2.7 | 0.68 | 295 | 100 | 4.99 * ± 3.24 | 2.5 | 0.10 | 1925 | 96.0 | |||
| 14 | 13.87 ± 0.33 | 6.9 | 1.73 | 115 | 100 | 2.74 ± 0.10 | 1.4 | 0.34 | 584 | 100 | |||
| 15 | 6.53 ± 0.00 | 3.3 | 0.82 | 245 | 100 | 2.70 ± 0.59 | 1.3 | 0.34 | 593 | 100 | |||
| 16 | 9.23 ± 0.50 | 4.6 | 1.15 | 173 | 100 | 2.27 ± 0.16 | 1.1 | 0.28 | 705 | 100 | |||
| 17 | 13.87 ± 0.33 | 6.9 | 1.73 | 115 | 100 | 4.24 ± 0.03 | 2.1 | 0.53 | 377 | 100 | |||
| 18 | 27.96 ± 7.92 | 14.0 | 3.49 | 57 | 100 | 5.66 ± 0.41 | 2.8 | 0.71 | 283 | 100 | |||
| 19 | 12.97 ± 0.11 | 6.5 | 1.62 | 123 | 100 | 3.67 ± 0.23 | 1.8 | 0.46 | 437 | 100 | |||
| 20 | 6.68 ± 1.69 | 3.3 | 0.83 | 240 | 100 | 3.52 ± 0.24 | 1.8 | 0.44 | 455 | 100 | |||
| 21 | 3.71 ± 0.09 | 1.9 | 0.46 | 431 | 100 | 2.97 ± 0.24 | 1.5 | 0.37 | 540 | 100 | |||
| 22 | 27.27 ± 3.70 | 13.6 | 3.41 | 59 | 100 | 5.98 ± 0.27 | 3.0 | 0.75 | 268 | 100 | |||
| 23 | 8.64 ± 2.19 | 4.3 | 1.08 | 185 | 100 | 2.64 ± 0.32 | 1.3 | 0.33 | 606 | 100 | |||
| 24 | 11.12 ± 1.58 | 5.6 | 1.39 | 144 | 100 | 2.20 ± 0.01 | 1.1 | 0.27 | 728 | 100 | |||
| 25 | 14.42 ± 1.18 | 7.2 | 1.80 | 111 | 100 | 4.70 ± 0.87 | 2.4 | 0.59 | 341 | 100 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Jin, J.; Rietjens, I.M.C.M.; Xing, F. Interindividual Differences in In Vitro Human Intestinal Microbial Conversion of 3-Acetyl-DON and 15-Acetyl-DON. Toxins 2022, 14, 199. https://doi.org/10.3390/toxins14030199
Li F, Jin J, Rietjens IMCM, Xing F. Interindividual Differences in In Vitro Human Intestinal Microbial Conversion of 3-Acetyl-DON and 15-Acetyl-DON. Toxins. 2022; 14(3):199. https://doi.org/10.3390/toxins14030199
Chicago/Turabian StyleLi, Fangfang, Jing Jin, Ivonne M. C. M. Rietjens, and Fuguo Xing. 2022. "Interindividual Differences in In Vitro Human Intestinal Microbial Conversion of 3-Acetyl-DON and 15-Acetyl-DON" Toxins 14, no. 3: 199. https://doi.org/10.3390/toxins14030199
APA StyleLi, F., Jin, J., Rietjens, I. M. C. M., & Xing, F. (2022). Interindividual Differences in In Vitro Human Intestinal Microbial Conversion of 3-Acetyl-DON and 15-Acetyl-DON. Toxins, 14(3), 199. https://doi.org/10.3390/toxins14030199