Six Feet under Microbiota: Microbiologic Contamination and Toxicity Profile in Three Urban Cemeteries from Lisbon, Portugal
Abstract
:1. Introduction
2. Results
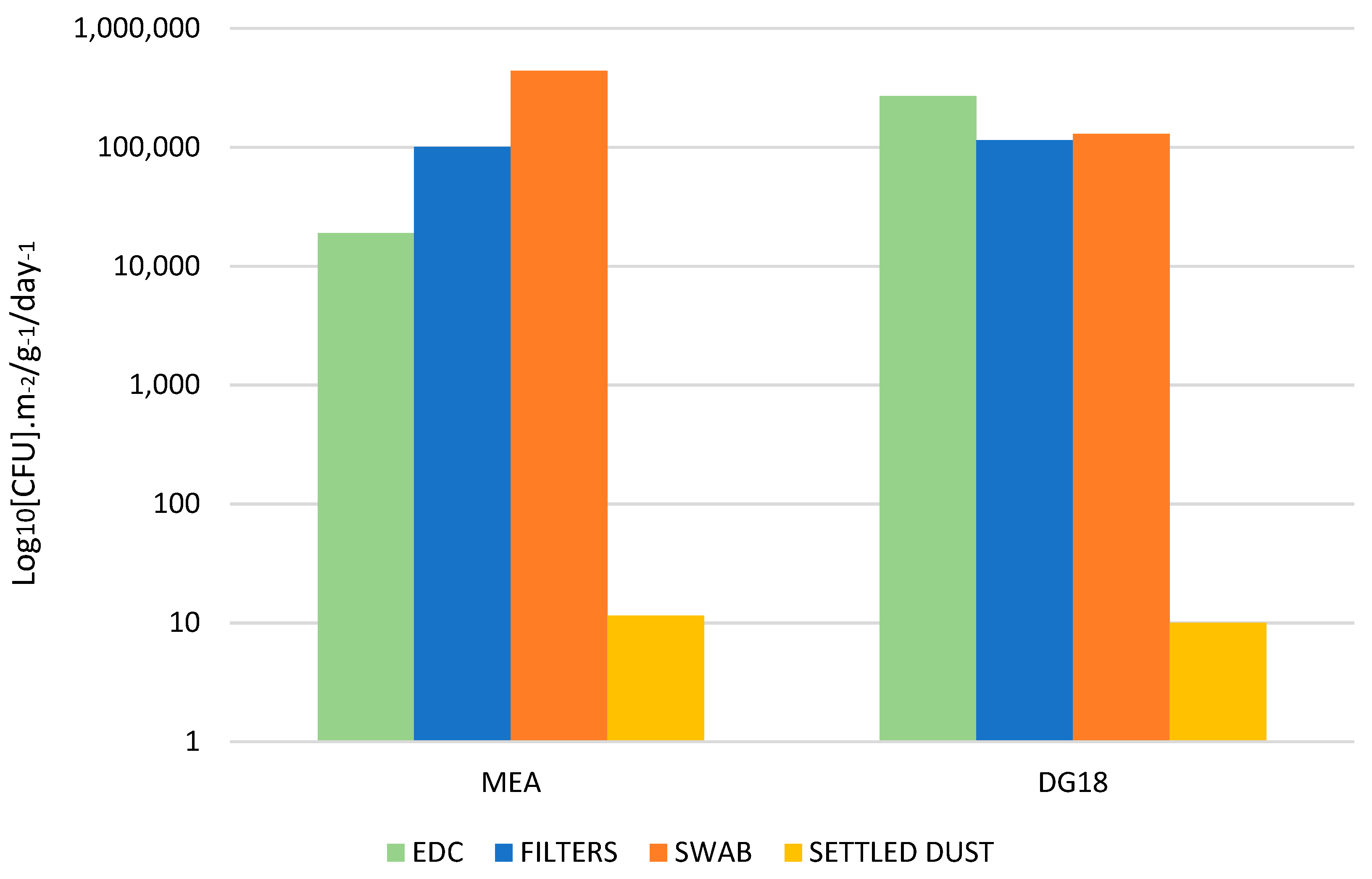
2.1. Viable Bacterial Contamination
2.2. Viable Fungal Contamination
2.3. Azole Resistance Profile
2.4. Detection of SARS-CoV-2 and the Targeted Fungal Sections
2.5. Mycotoxins Results
2.6. Cytotoxicity Evaluation
2.7. Correlation and Comparison Analysis
2.8. Correlation and Comparison Analysis
3. Discussion
4. Conclusions
5. Materials and Methods
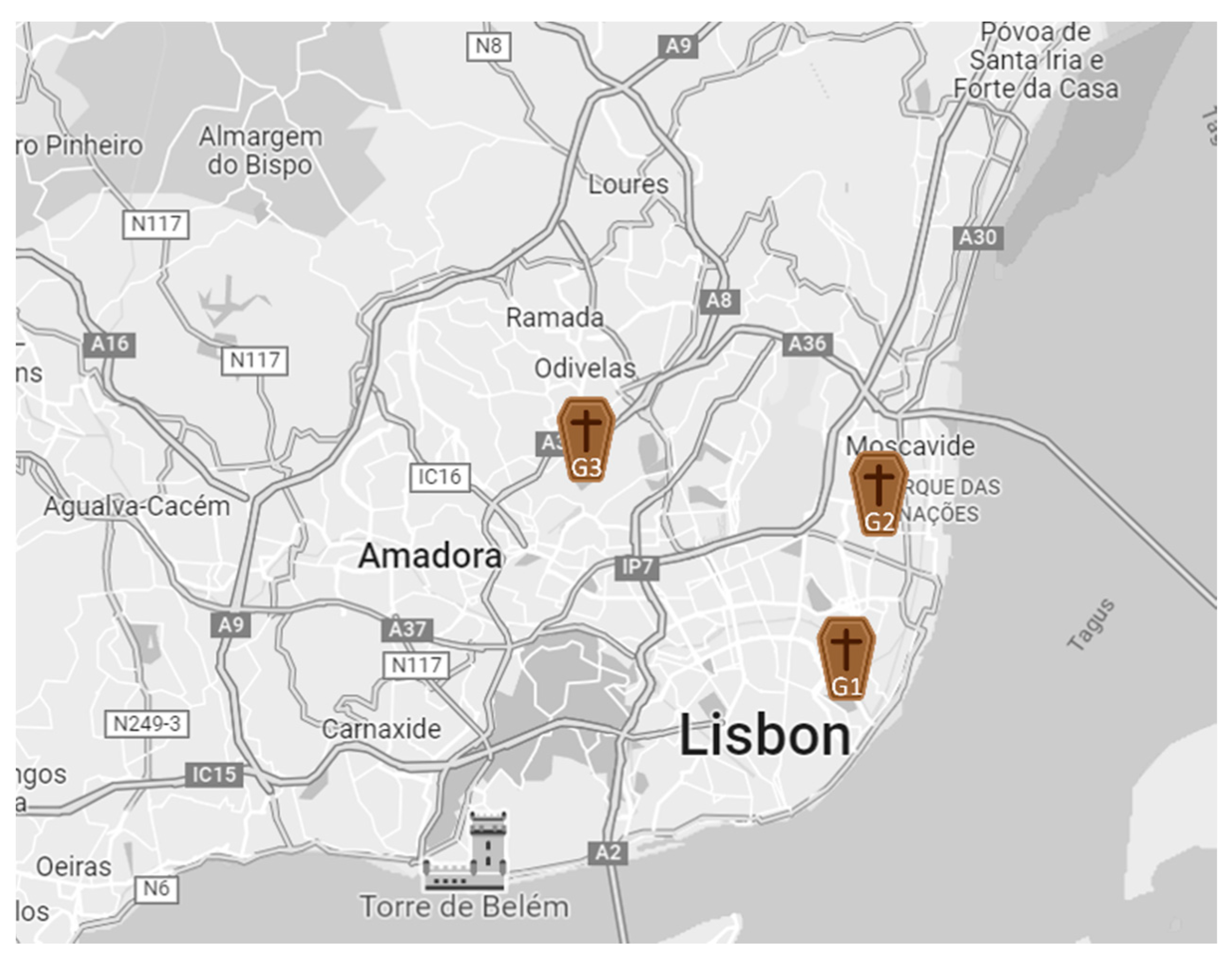
5.1. Graveyards Assessed
5.2. Sampling Approach and Characterization through Culture Dependent-Methods
5.3. Azole Resistance Screening
5.4. Sampling and Molecular Detection of SARS-CoV-2 and Targeted Aspergillus Sections
5.5. Mycotoxins Analysis
5.6. Cytotoxicity Analyses
5.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Uslu, A.; Bari, E.; Erdo, E. Ecological concerns over cemeteries. Afr. J. Agric. Res. 2009, 4, 1505–1511. [Google Scholar]
- Całkosiński, I.; Płoneczka-Janeczko, K.; Ostapska, M.; Dudek, K.; Gamian, A.; Rypuła, K. Microbiological Analysis of Necrosols Collected from Urban Cemeteries in Poland. Biomed Res. Int. 2015, 2015, 169573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Üçisik, A.S.; Rushbrook, P. The Impact of Cemeteries on the Environment and Public Health an Introductory Briefing; EUR/HFA target 23, EUR/ICP/EHNA 010401(A); WHO Regional Office for Europe, Nancy Project Office: Copenhagen, Denmark, 1998. [Google Scholar]
- Rodrigues, L.; Pacheco, A. Groundwater contamination from cemeteries cases of study. In Proceedings of the Environmental 2010: Situation and Perspectives for the European Union, Porto, Portugal, 6–10 May 2003; pp. 1–6. [Google Scholar]
- Directive 89/391/EEC; Council Directive of 12 June 1989 on the Introduction of Measures to Encourage Improvements in the Safety and Health of Workers at Work. Official Journal of the European Communities: Brussels, Belgium, 1989. Available online: https://eur-lex.europa.eu/legalcontent/EN/TXT/PDF/?uri=CELEX:31989L0391&from=IT (accessed on 9 April 2022).
- Domingo, J.L.; Nadal, M. Domestic waste composting facilities: A review of human health risks. Env. Inter. 2009, 35, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, O.; Huyard, A. Bioaerosols in composting plants: Occupational exposure and health. Environ. Risques Santé. 2008, 7, 37–45. [Google Scholar] [CrossRef]
- Viegas, C.; Gomes, B.; Pimenta, R.; Dias, M.; Cervantes, R.; Caetano, L.A.; Carolino, E.; Twarużek, M.; Soszczyńska, E.; Kosicki, R.; et al. Microbial contamination in firefighter Headquarters’: A neglected occupational exposure scenario. Build. Environ. 2022, 213, 108862. [Google Scholar] [CrossRef]
- Gonçalves, P.; Melo, A.; Dias, M.; Almeida, B.; Caetano, L.A.; Veríssimo, C.; Viegas, C.; Sabino, R. Azole-Resistant Aspergillus fumigatus Harboring the TR34/L98H Mutation: First Report in Portugal in Environmental Samples. Microorganisms 2021, 9, 57. [Google Scholar] [CrossRef]
- Abia, A.L.K.; Alisoltani, A.; Ubomba-Jaswa, E.; Dippenaar, M.A. Microbial life beyond the grave: 16S rRNA gene-based metagenomic analysis of bacteria diversity and their functional profiles in cemetery environments. Sci. Total Environ. 2019, 655, 831–841. [Google Scholar] [CrossRef]
- Łukaszuk, C.; Krajewska-Kułak, E.; Guzowski, A.; Kraszyńska, B.; Grassmann, M.; Dobrowolski, R. Analysis of the incidence fungi in a crypt cemetery. J. Air Waste Manag. Assoc. 2015, 65, 1141–1147. [Google Scholar] [CrossRef] [Green Version]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.; Meis, J.F. Azole Resistance in Aspergillus fumigatus: Can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef] [Green Version]
- Burks, C.; Darby, A.; Gómez Londoño, L.; Momany, M.; Brewer, M.T. Azole-resistant Aspergillus fumigatus in the environment: Identifying key reservoirs and hotspots of antifungal resistance. PLoS Pathog. 2021, 17, e1009711. [Google Scholar] [CrossRef]
- Bart, F.; Sarah, A.; Steve, H.; Andy, M.; John, L. The Multi-Fungicide Resistance Status of Aspergillus fumigatus Populations in Arable Soils and the Wider European Environment. Front. Microbiol. 2020, 11, 599233. [Google Scholar] [CrossRef]
- Pena, P.; Morais, J.; Caetano, L.A.; Viegas, C. Screening of fungal azole resistance in different environmental samples. In Encyclopedia of Mycology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 150–158. [Google Scholar] [CrossRef]
- Rocchi, S.; Ponçot, M.; Morin-Crini, N.; Laboissière, A.; Valot, B.; Godeau, C.; Léchenault-Bergerot, C.; Reboux, G.; Crini, G.; Millon, L. Determination of azole fungal residues in soils and detection of Aspergillus fumigatus-resistant strains in market gardens of Eastern France. Environ. Sci. Pollut. Res. Int. 2018, 25, 32015–32023. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dong, F.; Zhao, J.; Fan, H.; Qin, C.; Li, R.; Verweij, P.E.; Zheng, Y.; Han, L. High Azole Resistance in Aspergillus fumigatus Isolates from Strawberry Fields, China, 2018. Emerg. Infect. Dis. 2020, 26, 81–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viegas, S.; Viegas, C.; Oppliger, A. Occupational exposure to mycotoxins: Current knowledge and prospects. Ann. Work Expo. Health 2018, 62, 923–941. [Google Scholar] [CrossRef]
- WHO. World Health Organisation guidelines for indoor air quality: Dampness and mould. World Health Organisation Regional Office for Europe, 2009. Available online: https://www.who.int/publications/i/item/9789289041683 (accessed on 9 April 2022).
- Viegas, C.; Caetano, L.A.; Viegas, S. Occupational exposure to Aspergillus section Fumigati: Tackling the knowledge gap in Portugal. Environ. Res. 2021, 194, 110674. [Google Scholar] [CrossRef]
- Bergwall, C.; Stehn, B. Comparison of selective mycological agar media for the isolation and enumeration of xerophilic moulds and osmotolerant yeasts in granulated white sugar. Zuckerindustrie 2002, 127, 259–264. [Google Scholar]
- Caetano, L.A.; Almeida, B.; Viegas, C. Assessment of Azole Resistance in Clinical Settings by Passive Sampling. In Health and Social Care Systems of the Future: Demographic Changes, Digital Age and Human Factors; Cotrim, T., Serranheira, F., Sousa, P., Hignett, S., Albolino, S., Tartaglia, R., Eds.; Springer: Berlin/Heidelberg, Germany; HEPS: Lisbon, Portugal, 2019; Volume 1012. [Google Scholar]
- Viegas, C.; Monteiro, A.; Ribeiro, E.; Caetano, L.A.; Carolino, E.; Assunção, R.; Viegas, S. Organic Dust Exposure in Veterinary Clinics: A Case Study of a Small-Animal Practice in Portugal. Arch. Ind. Hyg. Toxicol. 2018, 69, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Viegas, C.; Dias, M.; Almeida, B.; Carolino, E.; Viegas, S. Aspergillus spp. presence on mechanical protection gloves from the waste sorting industry. J. Occup. Environ. Hyg. 2020, 17, 523–530. [Google Scholar] [CrossRef]
- Viegas, C.; Dias, M.; Carolino, E.; Sabino, R. Culture Media and Sampling Collection Method for Aspergillus spp. Assessment: Tackling the Gap between Recommendations and the Scientific Evidence. Atmosphere 2021, 12, 23. [Google Scholar] [CrossRef]
- Varga, J.; Baranyi, N.; Chandrasekaran, M.; Vágvölgyi, C.; Kocsubé, S. Mycotoxin producers in the Aspergillus genus: An update. Acta Biol. Szeged. 2015, 59, 151–167. [Google Scholar]
- Sabino, R.; Veríssimo, C.; Viegas, C.; Viegas, S.; Brandão, J.; Alves-Correia, M.; Borrego, L.M.; Clemons, K.V.; Stevens, D.A.; Richardson, M. The role of occupational Aspergillus exposure in the development of diseases. Med. Mycol. 2019, 57, S196–S205. [Google Scholar] [CrossRef] [PubMed]
- Matos, J.; Brantes, J.; Cunha, A.M. Qualidade do Ar em Espaços Interiores Um Guia Técnico 2010. In Agência Port; do Ambient: Lisbon, Portugal, 2010. [Google Scholar]
- MacNeil, L.; Kauri, T.; Robertson, W. Molecular techniques and their potential application in monitoring the microbiological quality of indoor air. Can. J. Microbiol. 1995, 41, 657–665. [Google Scholar] [CrossRef]
- Madsen, A.M.; Frederiksen, M.W.; Jacobsen, M.H.; Tendal, K. Towards a risk evaluation of workers’ exposure to handborne and airborne microbial species as exemplified with waste collection workers. Environ. Res. 2020, 183, 109177. [Google Scholar] [CrossRef] [PubMed]
- Viegas, S.; Assunção, R.; Nunes, C.; Osteresch, B.; Twarużek, M.; Kosicki, R.; Grajewski, J.; Martins, C.; Alvito, P.; Almeida, A.; et al. Exposure assessment to mycotoxins in a Portuguese fresh bread dough company by using a multi-biomarker approach. Toxins. 2018, 10, 342. [Google Scholar] [CrossRef] [Green Version]
- Vaali, K.; Tuomela, M.; Mannerström, M.; Heinonen, T.; Tuuminen, T. Toxic Indoor Air Is aPotential Risk of ausing Immuno Suppression and Morbidity—A Pilot Study. J. Fungi 2022, 8, 104. [Google Scholar] [CrossRef]
- Swain, R.J.; Kemp, S.J.; Goldstraw, P.; Tetley, T.D.; Stevens, M.M. Assessment of Cell Line Models of Primary Human Cells by Raman Spectral Phenotyping. Biophys. J. 2010, 98, 1703–1711. [Google Scholar] [CrossRef] [Green Version]
- Gniadek, A.; Krzy´sciak, P.; Twaruzek, M.; Macura, A.B. Occurrence of fungi and cytotoxicity of the species: Aspergillus ochraceus, Aspergillus niger and Aspergillus flavus isolated from the air of hospital wards. Int. J. Occup. Med. Environ. Health 2017, 30, 231–239. [Google Scholar] [CrossRef]
- Kamei, K.; Watanabe, A.; Nishimura, K.; Miyaji, M. Cytotoxicity of Aspergillus fumigatus culture filtrate against macrophages. Nihon Ishinkin Gakkai Zasshi 2002, 43, 37–41. [Google Scholar] [CrossRef] [Green Version]
- Gniadek, A.; Macura, A.B.; Twarużek, M.; Grajewski, J. Cytotoxicity of Aspergillus strains isolated from the neonatal intensive care unit environment. Adv. Med. Sci. 2010, 55, 242–249. [Google Scholar] [CrossRef]
- Brown, R.; Priest, E.; Naglik, J.R.; Richardson, J.P. Fungal Toxins and Host Immune Responses. Front. Microbiol. 2021, 12, 643639. [Google Scholar] [CrossRef]
- Bräse, S.; Encinas, A.; Keck, J.; Nising, C. Chemistry and biology of mycotoxins and related fungal metabolites. Chem. Rev. 2009, 109, 3903–3990. [Google Scholar] [CrossRef] [PubMed]
- Gniadek, A. Cytotoxicity of Aspergillus Fungi as a Potential Infectious Threat. In Insight and Control of Infectious Disease in Global Scenario; Roy, P.K., Ed.; IntechOpen: London, UK, 2012. [Google Scholar]
- Liu, L.; Liu, R.; Basnet, B.B.; Bao, L.; Han, J.; Wang, L.; Liu, H. New phenolic bisabolane sesquiterpenoid derivatives with cytotoxicity from Aspergillus tennesseensis. J. Antibiot. 2018, 71, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Sparer, E.H.; Prendergast, D.P.; Apell, J.N.; Bartzak, M.R.; Wagner, G.R.; Adamkiewicz, G.; Hart, J.E.; Sorensen, G. Assessment of Ambient Exposures Firefighters Encounter while at the Fire Station: An Exploratory Study. J. Occup. Environ. Med. 2017, 59, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Viegas, C.; Sousa, P.; Dias, M.; Aranha Caetano, L.; Ribeiro, E.; Carolino, E.; Twarużek, M.; Kosicki, R.; Viegas, S. Bioburden contamination and Staphylococcus aureus colonization associated with firefighter’s ambulances. Environ. Res. 2021, 197, 111125. [Google Scholar] [CrossRef]
- ISO 18593; Microbiology of Food and Animal Feeding Stuffs, Horizontal Methods for Sampling Techniques from Surfaces Using Contact Plates and Swabs. American National Standards Institute (ANSI): Washington, DC, USA, 2004.
- De Hoog, D.; Guarro, J.; Gene, G.; Figueras, M. Atlas of Clinical Fungi—The Ultimate Benchtool for Diagnosis; Utr Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2016; Volume 4. [Google Scholar]
- Arendrup, M.C.; Rodriguez-Tudela, J.L.; Lass-Flörl, C.; Cuenca-Estrella, M.; Donnelly, J.P.; Hope, W. EUCAST technical note on anidulafungin. Clin. Microbiol. Infect. 2013, 19, 278–280. [Google Scholar] [CrossRef] [Green Version]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Routine and Extended Internal Quality Control for MIC Determination and Agar Dilution for Yeasts, Moulds and Dermatophytes as Recommended by EUCAST. Version 5.0. 2020. Available online: http://www.eucast.org (accessed on 12 September 2021).
- Viegas, C.; Almeida, B.; Aranha Caetano, L.; Afanou, A.; Straumfors, A.; Veríssimo, C.; Gonçalves, P.; Sabino, R. Algorithm to assess the presence of Aspergillus fumigatus resistant strains: The case of Norwegian sawmills. Int. J. Environ. Res. 2020, 32, 963–971. [Google Scholar] [CrossRef]
- Hanelt, M.; Gareis, M.; Kollarczik, B. Cytotoxicity of mycotoxins evaluated by the MTT-cell culture assay. Mycopathologia 1994, 128, 167–174. [Google Scholar] [CrossRef]


| SDA | ITZ | VCZ | PSZ | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Matrice | Species | CFU·m−2·Day−1/g−1/m2 | % | CFU·m−2·Day−1/g−1/m−2 | % | CFU·m−2·Day−1/g−1/m−2 | % | CFU·m−2·Day−1/g−1/m−2 | % |
| EDC | A. section Nidulantes | 1.06 × 102 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| A. section Nigri | 1.06 × 102 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Cladosporium sp. | 5.20 × 104 | 54.4 | 7.01 × 104 | 36.9 | 1.46 × 105 | 64.1 | 1.00 × 104 | 16.5 | |
| Chrysosporium sp. | 0.0 | 0.0 | 0.0 | 0.0 | 2.00 × 104 | 8.8 | 0.0 | 0.0 | |
| C. sitophila | 2.06 × 104 | 21.6 | 6.01 × 104 | 31.6 | 5.03 × 104 | 22.0 | 5.03 × 104 | 83.3 | |
| Penicillium sp. | 2.17 × 104 | 22.7 | 6.00 × 104 | 31.5 | 0.0 | 0.0 | 1.06 × 104 | 0.2 | |
| Other species | 1.06 × 103 | 1.1 | 0.0 | 0.0 | 1.16 × 104 | 5.1 | 0.0 | 0.0 | |
| Total | 9.56 × 104 | 100.0 | 1.90 × 105 | 100.0 | 2.28 × 105 | 100.0 | 6.04 × 104 | 100.0 | |
| FILTERS | Cladosporium sp. | 1.05 × 104 | 33.9 | 5.00 × 102 | 33.3 | 1.00 × 104 | 50.0 | 0.0 | 0.0 |
| C. sitophila | 1.00 × 103 | 3.2 | 5.00 × 102 | 33.3 | 5.00 × 102 | 25.0 | 6.00 × 103 | 92.3 | |
| Fusarium verticilloides | 0.0 | 0.0 | 5.00 × 102 | 33.3 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Penicillium sp. | 1.90 × 104 | 61.3 | 0.0 | 0.0 | 5.00 × 102 | 25.0 | 5.00 × 102 | 7.7 | |
| Other species | 5.00 × 102 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Total | 3.10 × 104 | 100.0 | 1.50 × 103 | 100.0 | 2.00 × 103 | 100.0 | 6.50 × 103 | 100.0 | |
| SWABS | Cladosporium sp. | 1.10 × 105 | 37.9 | 0.0 | 0.0 | 4.00 × 104 | 57.1 | 0.0 | 0.0 |
| C. sitophila | 1.00 × 105 | 34.5 | 3.00 × 104 | 60.0 | 0.0 | 0.0 | 3.00 × 104 | 75.0 | |
| Penicillium sp. | 5.00 × 104 | 17.2 | 1.00 × 104 | 20.0 | 2.00 × 104 | 28.6 | 0.0 | 0.0 | |
| Rhizopus sp. | 0.0 | 0.0 | 0.0 | 0.0 | 1.00 × 104 | 14.3 | 0.0 | 0.0 | |
| Trichoderma sp. | 0.0 | 0.0 | 1.00 × 104 | 20.0 | 0.0 | 0.0 | 1.00 × 104 | 25.0 | |
| Other species | 3.00 × 104 | 10.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Total | 2.90 × 105 | 100.0 | 5.00 × 104 | 100.0 | 7.00 × 104 | 100.0 | 4.00 × 104 | 100.0 | |
| SETTLED DUST | Aureobasidium sp. | 4.00 | 7.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| A. section Nigri | 1.00 | 1.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Cladosporium sp. | 0.0 | 0.0 | 4.00 | 57.1 | 1.06 × 102 | 92.2 | 0.0 | 0.0 | |
| Chrysosporium sp. | 0.0 | 0.0 | 0.0 | 0.0 | 2.00 | 1.7 | 0.0 | 0.0 | |
| C. sitophila | 1.50 | 2.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Geotrichum sp. | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.00 | 25.0 | |
| Penicillium sp. | 4.45 × 101 | 82.4 | 3.00 | 42.9 | 6.00 | 5.2 | 3.00 | 75.0 | |
| Other species | 3.00 | 5.6 | 0.0 | 0.0 | 1.00 | 0.9 | 0.0 | 0.0 | |
| Total | 5.40 × 101 | 100.0 | 7.00 | 100.0 | 1.15 × 102 | 100.0 | 4.00 | 100.0 | |
| Dilution Step | Filters | EDC | ||||
|---|---|---|---|---|---|---|
| IC50 | A549 | HepG2 | IC50 | A549 | HepG2 | |
| 1 | 1 | 13 | 6 | 0.016 | 1 | 1 |
| 2 | 0.5 | 3 | 1 | 0.008 | 0 | 0 |
| 3 | 0.25 | 1 | 0 | 0.004 | 0 | 0 |
| 4 | 0.125 | 0 | 2 | 0.002 | 0 | 0 |
| (-) | 1 | 9 | 17 | 17 | ||
| Method | Media | Fungi (CFU·m−2/m−2·Day−1) | Fungal Resistance (CFU·m−2/m−2·Day−1) | ||||
|---|---|---|---|---|---|---|---|
| DG18 | SDA | ITZ | VCZ | PSZ | |||
| EDC | Fungi (CFU·m−2·day−1) | MEA | 0.242 | 0.105 | 0.284 | 0.340 | −0.035 |
| DG18 | 0.606 ** | 0.510 * | 0.692 ** | 0.345 | |||
| Fungal resistance (CFU·m−2·day−1) | SDA | 0.446 | 0.514 * | −0.034 | |||
| ITZ | 0.628 ** | 0.261 | |||||
| VCZ | 0.411 | ||||||
| Filters | Fungi (CFU·m−2) | MEA | 0.598 ** | 0.507 * | 0.188 | 0.675 ** | 0.162 |
| DG18 | 0.271 | 0.132 | 0.460 | −0.257 | |||
| Fungal resistance (CFU·m−2) | SDA | 0.378 | 0.623 ** | 0.238 | |||
| ITZ | 0.478 * | 0.452 | |||||
| VCZ | 0.225 | ||||||
| Swabs | Fungi (CFU·m−2) | MEA | 0.221 | 0.105 | −0.387 | −0.166 | −0.183 |
| DG18 | 0.646 ** | 0.228 | 0.405 | 0.257 | |||
| Fungal resistance (CFU·m−2) | SDA | 0.283 | 0.074 | 0.045 | |||
| ITZ | 0.214 | 0.037 | |||||
| VCZ | 0.399 | ||||||
| Sampling Method | Ranks | Test Statistics | Kruskal–Wallis Multiple Comparisons Test | |||||
|---|---|---|---|---|---|---|---|---|
| n | Mean Rank | Kruskal–Wallis H | df | p | ||||
| Fungi (CFU·m−2/m−2·day−1) | MEA | EDC | 18 | 19.58 | 6.943 | 2 | 0.031 * | EDC ≠ Filter (p = 0.048) |
| Filter | 18 | 32.17 | ||||||
| Swabs | 18 | 30.75 | ||||||
| Total | 54 | |||||||
| DG18 | EDC | 18 | 31.42 | 4.948 | 2 | 0.084 | ||
| Filter | 18 | 30.17 | ||||||
| Swabs | 18 | 20.92 | ||||||
| Total | 54 | |||||||
| Fungal resistance (CFU·m−2/m−2·day−1) | SDA | EDC | 18 | 24.67 | 16.003 | 2 | 0.000 * | EDC ≠ Swabs (p = 0.017) |
| Filter | 18 | 18.81 | Filter ≠ Swabs (p = 0.000) | |||||
| Swabs | 18 | 39.03 | ||||||
| Total | 54 | |||||||
| ITZ | EDC | 18 | 36.58 | 12.915 | 2 | 0.002 * | EDC ≠ Filter (p = 0.002) | |
| Filter | 18 | 20.92 | EDC ≠ Swabs (p = 0.031) | |||||
| Swabs | 18 | 25.00 | ||||||
| Total | 54 | |||||||
| VCZ | EDC | 18 | 36.94 | 12.729 | 2 | 0.002 * | EDC ≠ Filter (p = 0.002) | |
| Filter | 18 | 20.78 | EDC ≠ Swabs (p = 0.030) | |||||
| Swabs | 18 | 24.78 | ||||||
| Total | 54 | |||||||
| PSZ | EDC | 18 | 30.17 | 5.591 | 2 | 0.061 | ||
| Filter | 18 | 31.33 | ||||||
| Swabs | 18 | 21.00 | ||||||
| Total | 54 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viegas, C.; Cervantes, R.; Dias, M.; Gomes, B.; Pena, P.; Carolino, E.; Twarużek, M.; Kosicki, R.; Soszczyńska, E.; Viegas, S.; et al. Six Feet under Microbiota: Microbiologic Contamination and Toxicity Profile in Three Urban Cemeteries from Lisbon, Portugal. Toxins 2022, 14, 348. https://doi.org/10.3390/toxins14050348
Viegas C, Cervantes R, Dias M, Gomes B, Pena P, Carolino E, Twarużek M, Kosicki R, Soszczyńska E, Viegas S, et al. Six Feet under Microbiota: Microbiologic Contamination and Toxicity Profile in Three Urban Cemeteries from Lisbon, Portugal. Toxins. 2022; 14(5):348. https://doi.org/10.3390/toxins14050348
Chicago/Turabian StyleViegas, Carla, Renata Cervantes, Marta Dias, Bianca Gomes, Pedro Pena, Elisabete Carolino, Magdalena Twarużek, Robert Kosicki, Ewelina Soszczyńska, Susana Viegas, and et al. 2022. "Six Feet under Microbiota: Microbiologic Contamination and Toxicity Profile in Three Urban Cemeteries from Lisbon, Portugal" Toxins 14, no. 5: 348. https://doi.org/10.3390/toxins14050348
APA StyleViegas, C., Cervantes, R., Dias, M., Gomes, B., Pena, P., Carolino, E., Twarużek, M., Kosicki, R., Soszczyńska, E., Viegas, S., & Caetano, L. A. (2022). Six Feet under Microbiota: Microbiologic Contamination and Toxicity Profile in Three Urban Cemeteries from Lisbon, Portugal. Toxins, 14(5), 348. https://doi.org/10.3390/toxins14050348










