Transcriptional Analysis of Cotton Bollworm Strains with Different Genetic Mechanisms of Resistance and Their Response to Bacillus thuringiensis Cry1Ac Toxin
Abstract
:1. Introduction
2. Results
2.1. Overview of the Midgut Transcriptomes of Four Cotton Bollworm Strains with or without Cry1Ac Exposure
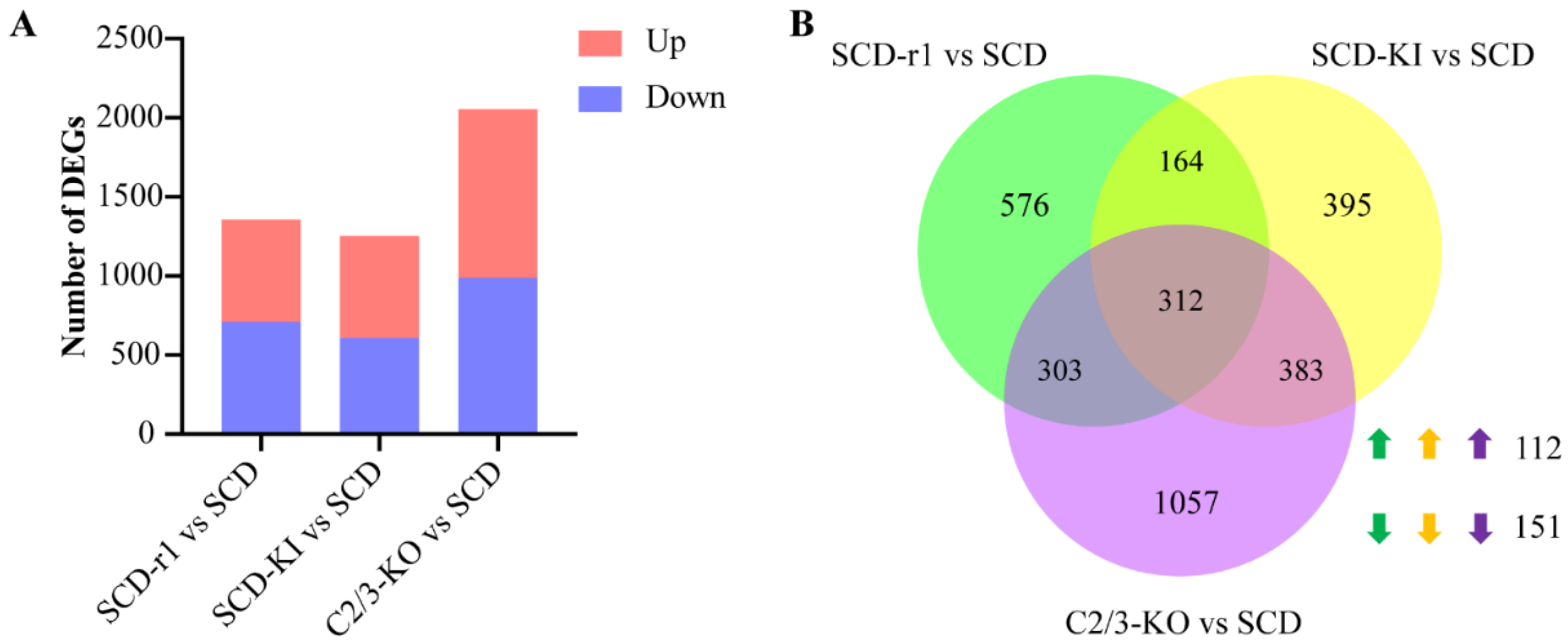
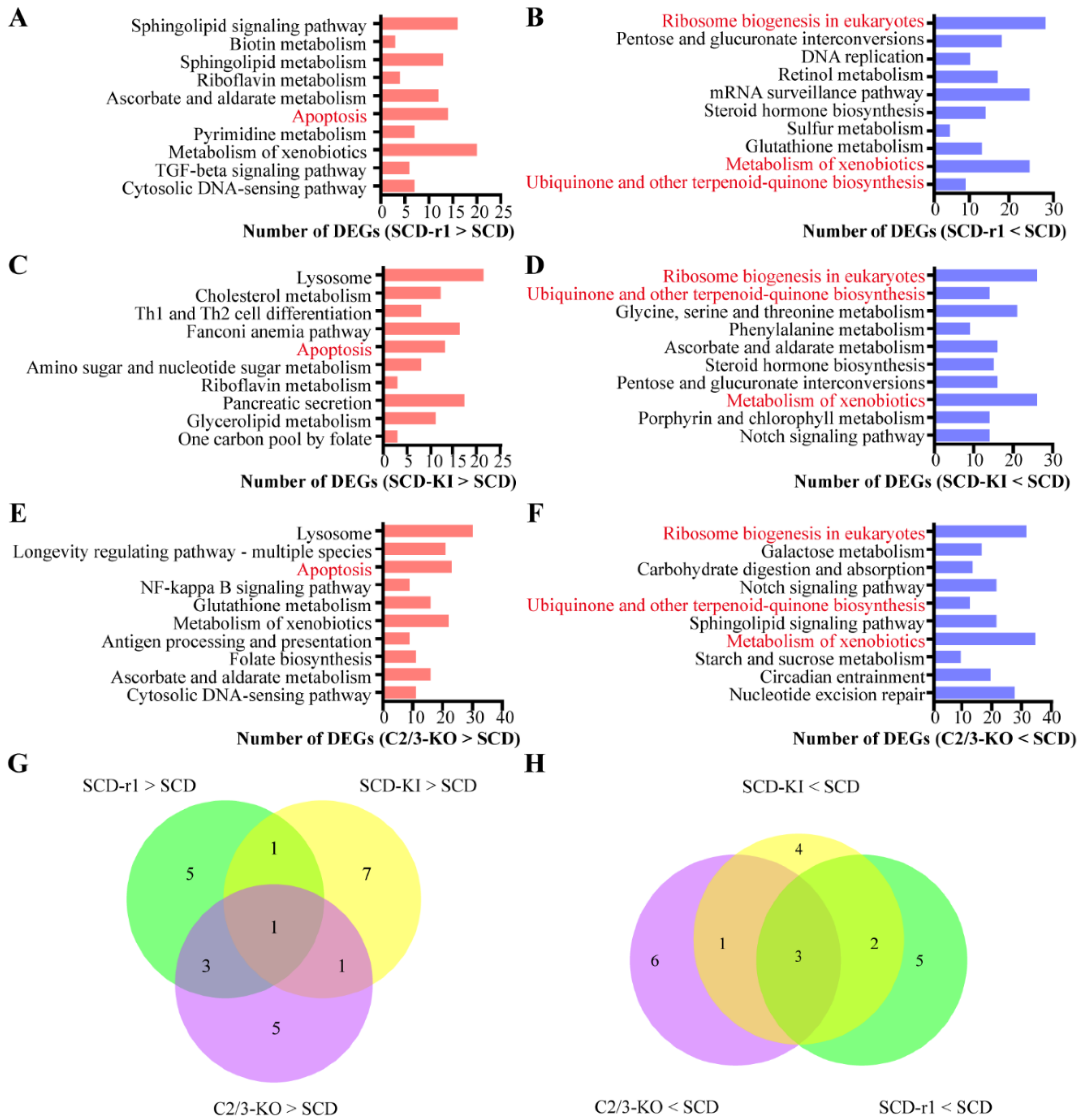
2.2. Effects of Different Genetic Mutations on Midgut Transcriptomes of Cotton Bollworms without Cry1Ac Exposure
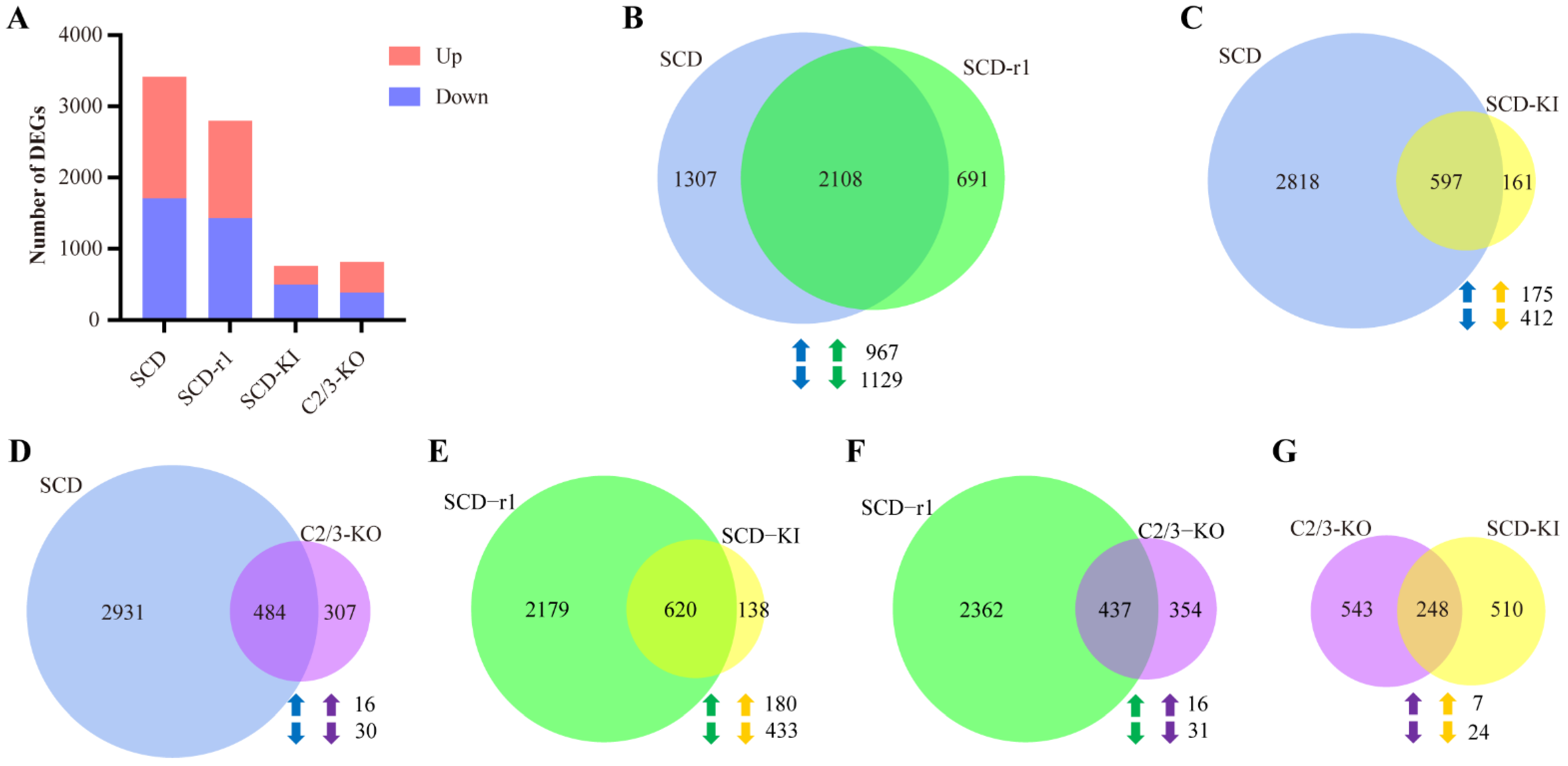
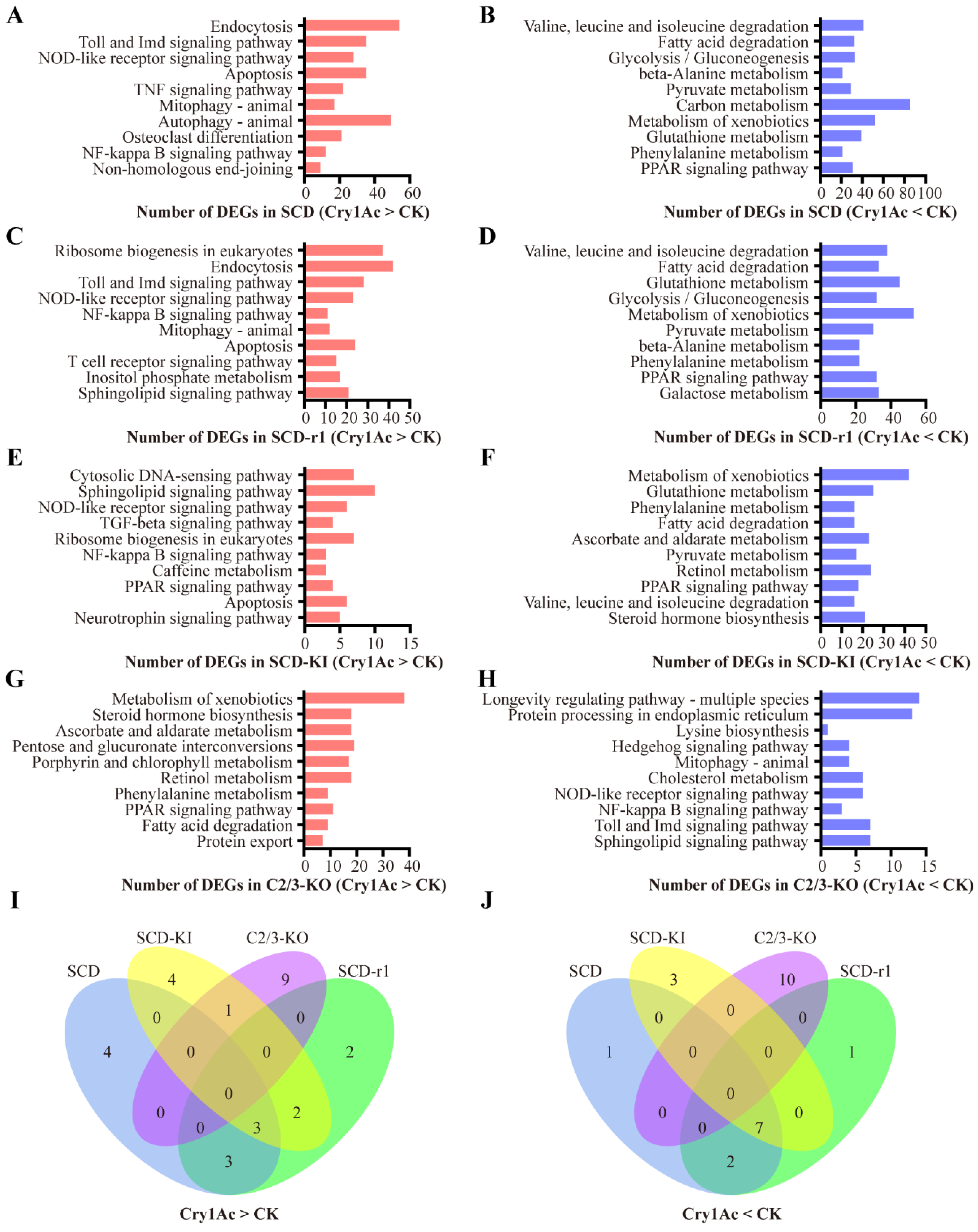
2.3. Effects of Cry1Ac on Midgut Transcriptomes of Susceptible and Resistant Cotton Bollworms
2.4. Expression Profiles of Midgut Epithelium Healing Genes
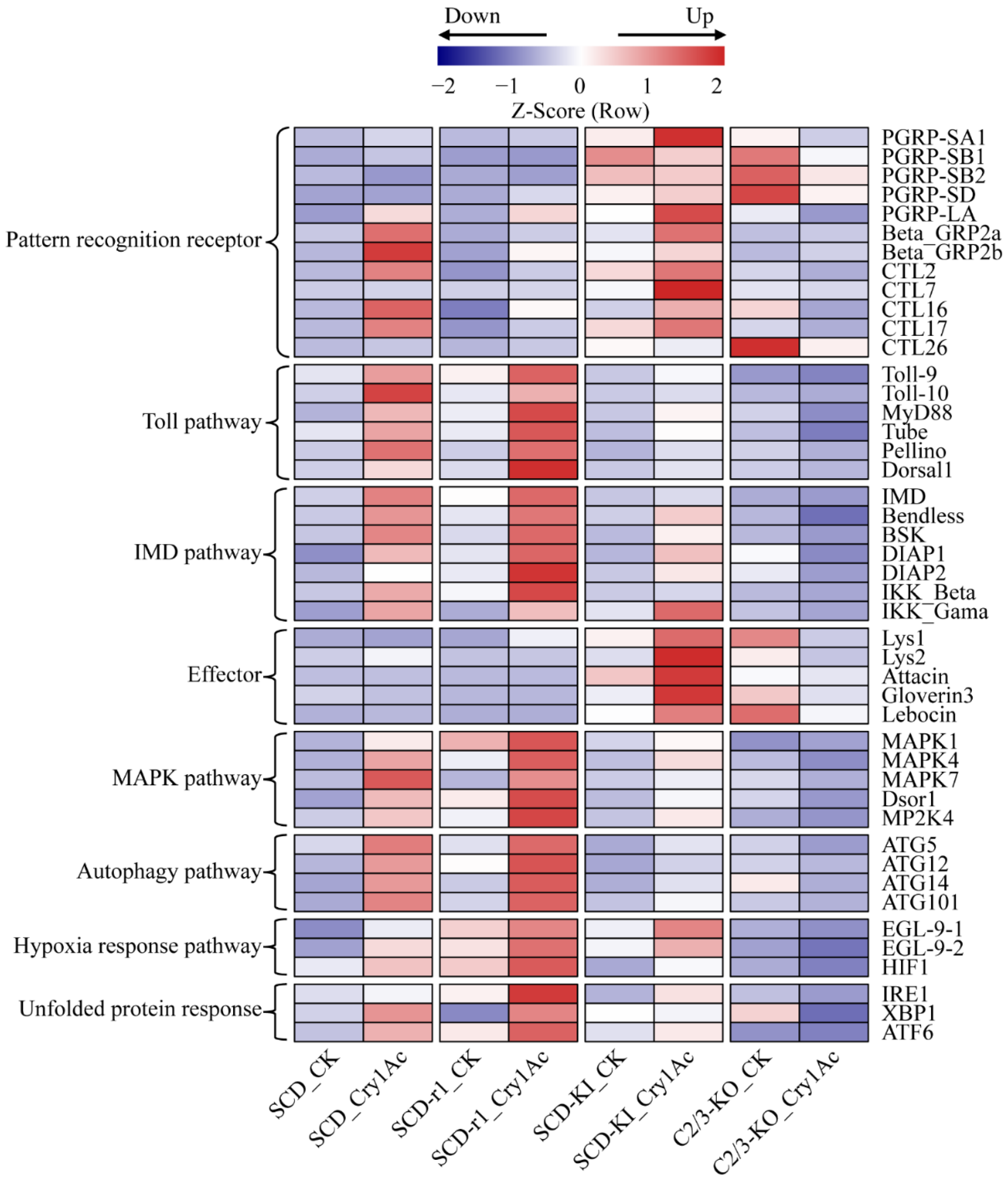
2.5. Expression Profiles of Defense Related Genes
3. Discussion
4. Materials and Methods
4.1. Insect Strains and Rearing
4.2. Dissection of Midgut and Extraction of RNA
4.3. mRNA Library Construction
4.4. RNA-Seq Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanahuja, G.; Banakar, R.; Twyman, R.M.; Capell, T.; Christou, P. Bacillus thuringiensis: A century of research, development and commercial applications. Plant Biotechnol. J. 2011, 9, 283–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardo-López, L.; Soberón, M.; Bravo, A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 2013, 37, 3–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendelsohn, M.; Kough, J.; Vaituzis, Z.; Matthews, K. Are Bt crops safe? Nat. Biotechnol. 2003, 21, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Comas, C.; Lumbierres, B.; Pons, X.; Albajes, R. No effects of Bacillus thuringiensis maize on nontarget organisms in the field in southern Europe: A meta-analysis of 26 arthropod taxa. Transgenic Res. 2014, 23, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Nicolia, A.; Manzo, A.; Veronesi, F.; Rosellini, D. An overview of the last 10 years of genetically engineered crop safety research. Crit. Rev. Biotechnol. 2014, 34, 77–88. [Google Scholar] [CrossRef]
- International Service for the Acquisition of Agri-Biotech Applications (ISAAA). Global Status of Commercialized Biotech/GM Crops in 2019; Brief 55; ISAAA: Ithaca, NY, USA, 2019. [Google Scholar]
- Wu, K.M.; Lu, Y.H.; Feng, H.Q.; Jiang, Y.Y.; Zhao, J.Z. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 2008, 321, 1676–1678. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Wu, K.; Jiang, Y.; Guo, Y.; Desneux, N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 2012, 487, 362–365. [Google Scholar] [CrossRef]
- Carpenter, J.E. Peer-reviewed surveys indicate positive impact of commercialized GM crops. Nat. Biotechnol. 2010, 28, 319–321. [Google Scholar] [CrossRef]
- Hutchison, W.D.; Burkness, E.C.; Mitchell, P.D.; Moon, R.D.; Leslie, T.W.; Fleischer, S.J.; Abrahamson, M.; Hamilton, K.L.; Steffey, K.L.; Gray, M.E.; et al. Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science 2010, 330, 222–225. [Google Scholar] [CrossRef] [Green Version]
- Dively, G.P.; Venugopal, P.D.; Bean, D.; Whalen, J.; Holmstrom, K.; Kuhar, T.P.; Doughty, H.B.; Patton, T.; Cissel, W.; Hutchison, W.D. Regional pest suppression associated with widespread Bt maize adoption benefits vegetable growers. Proc. Natl. Acad. Sci. USA 2018, 115, 3320–3325. [Google Scholar] [CrossRef] [Green Version]
- Tabashnik, B.E.; Carrière, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 2017, 35, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Jurat-Fuentes, J.L.; Heckel, D.G.; Ferré, J. Mechanisms of resistance to insecticidal proteins from Bacillus thuringiensis. Annu. Rev. Entomol. 2021, 66, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Carrière, Y. Global patterns of resistance to Bt crops highlighting pink bollworm in the United States, China, and India. J. Econ. Entomol. 2019, 112, 2513–2523. [Google Scholar] [CrossRef] [PubMed]
- Heckel, D.G. How do toxins from Bacillus thuringiensis kill insects? An evolutionary perspective. Arch. Insect Biochem. Physiol. 2020, 104, e21673. [Google Scholar] [CrossRef] [Green Version]
- Endo, H.; Azuma, M.; Adegawa, S.; Kikuta, S.; Sato, R. Water influx via aquaporin directly determines necrotic cell death induced by the Bacillus thuringiensis Cry toxin. FEBS Lett. 2017, 591, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y. Detection and mechanisms of resistance evolved in insects to Cry toxins from Bacillus thuringiensis. Adv. Insect Phys. 2014, 47, 297–342. [Google Scholar]
- Gahan, L.J.; Gould, F.; Heckel, D.G. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 2001, 293, 857–860. [Google Scholar] [CrossRef]
- Gahan, L.J.; Pauchet, Y.; Vogel, H.; Heckel, D.G. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010, 6, e1001248. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ma, H.; Zhao, S.; Huang, J.; Yang, Y.; Tabashnik, B.E.; Wu, Y. Functional redundancy of two ABC transporter proteins in mediating toxicity of Bacillus thuringiensis to cotton bollworm. PLoS Pathog. 2020, 16, e1008427. [Google Scholar] [CrossRef]
- Zhang, H.; Tian, W.; Zhao, J.; Jin, L.; Yang, J.; Liu, C.; Yang, Y.; Wu, S.; Wu, K.; Cui, J.; et al. Diverse genetic basis of field-evolved resistance to Bt cotton in cotton bollworm from China. Proc. Natl. Acad. Sci. USA 2012, 109, 10275–10280. [Google Scholar] [CrossRef] [Green Version]
- Jin, L.; Wang, J.; Guan, F.; Zhang, J.; Yu, S.; Liu, S.; Xue, Y.; Li, L.; Wu, S.; Wang, X.; et al. Dominant point mutation in a tetraspanin gene associated with field-evolved resistance of cotton bollworm to transgenic Bt cotton. Proc. Natl. Acad. Sci. USA 2018, 115, 11760–11765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, W.T.; Soria, M.F.; Walsh, T.; Thomazoni, D.; Silvie, P.; Behere, G.T.; Anderson, C.; Downes, S. A brave new world for an old world pest: Helicoverpa armigera (Lepidoptera: Noctuidae) in Brazil. PLoS ONE 2013, 8, e80134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, L.; Zhang, H.; Lu, Y.; Yang, Y.; Wu, K.; Tabashnik, B.E.; Wu, Y. Large-scale test of the natural refuge strategy for delaying insect resistance to transgenic Bt crops. Nat. Biotechnol. 2015, 33, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xiao, Y.; Chen, W.; Lu, Y.; Wu, K. Field monitoring of Helicoverpa armigera (Lepidoptera: Noctuidae) Cry1Ac insecticidal protein resistance in China (2005–2017). Pest Manag. Sci. 2019, 75, 753–759. [Google Scholar]
- Xu, X.; Yu, L.; Wu, Y. Disruption of a cadherin gene associated with resistance to Cry1Ac delta-endotoxin of Bacillus thuringiensis in Helicoverpa armigera. Appl. Environ. Microbiol. 2005, 71, 948–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.H.; Yang, Y.J.; Gao, W.Y.; Guo, J.J.; Wu, Y.H.; Wu, Y.D. Introgression of a disrupted cadherin gene enables susceptible Helicoverpa armigera to obtain resistance to Bacillus thuringiensis toxin Cry1Ac. Bull. Entomol. Res. 2009, 99, 175–181. [Google Scholar] [CrossRef]
- Cancino-Rodezno, A.; Porta, H.; Soberon, M.; Bravo, A. Defense and death responses to pore forming toxins. Biotechnol. Genet. Eng. Rev. 2010, 26, 65–82. [Google Scholar] [CrossRef] [Green Version]
- Pinos, D.; Andrés-Garrido, A.; Ferré, J.; Hernández-Martínez, P. Response mechanisms of invertebrates to Bacillus thuringiensis and Its pesticidal proteins. Microbiol. Mol. Biol. Rev. 2021, 85, e00007-20. [Google Scholar] [CrossRef]
- Blackburn, M.B.; Loeb, M.J.; Clark, E.; Jaffe, H. Stimulation of midgut stem cell proliferation by Manduca sexta alpha-arylphorin. Arch. Insect Biochem. Physiol. 2004, 55, 26–32. [Google Scholar] [CrossRef]
- Hakim, R.S.; Blackburn, M.B.; Corti, P.; Gelman, D.B.; Goodman, C.; Elsen, K.; Loeb, M.J.; Lynn, D.; Soin, T.; Smagghe, G. Growth and mitogenic effects of arylphorin in vivo and in vitro. Arch. Insect Biochem. Physiol. 2007, 64, 63–73. [Google Scholar] [CrossRef]
- Castagnola, A.; Jackson, J.; Perera, O.P.; Oppert, C.; Eda, S.; Jurat-Fuentes, J.L. Alpha-arylphorin is a mitogen in the Heliothis virescens midgut cell secretome upon Cry1Ac intoxication. PeerJ 2017, 5, e3886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Patel, P.H.; Kohlmaier, A.; Grenley, M.O.; McEwen, D.G.; Edgar, B.A. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 2009, 137, 1343–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Edgar, B.A. Intestinal stem cells in the adult Drosophila midgut. Exp. Cell Res. 2011, 317, 2780–2788. [Google Scholar] [CrossRef] [PubMed]
- Arbouzova, N.I.; Zeidler, M.P. JAK/STAT signalling in Drosophila: Insights into conserved regulatory and cellular functions. Development 2006, 133, 2605–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brito, C.; Cabanes, D.; Sarmento Mesquita, F.; Sousa, S. Mechanisms protecting host cells against bacterial pore-forming toxins. Cell Mol. Life Sci. 2019, 76, 1319–1339. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, S.J.; Rappoport, J.Z. The role of vesicle trafficking in epithelial cell motility. Biochem. Soc. Trans. 2009, 37, 1072–1076. [Google Scholar] [CrossRef] [Green Version]
- Caccia, S.; Di Lelio, I.; La Storia, A.; Marinelli, A.; Varricchio, P.; Franzetti, E.; Banyuls, N.; Tettamanti, G.; Casartelli, M.; Giordana, B.; et al. Midgut microbiota and host immunocompetence underlie Bacillus thuringiensis killing mechanism. Proc. Natl. Acad. Sci. USA 2016, 113, 9486–9491. [Google Scholar] [CrossRef] [Green Version]
- Ma, G.; Roberts, H.; Sarjan, M.; Featherstone, N.; Lahnstein, J.; Akhurst, R.; Schmidt, O. Is the mature endotoxin Cry1Ac from Bacillus thuringiensis inactivated by a coagulation reaction in the gut lumen of resistant Helicoverpa armigera larvae? Insect Biochem. Mol. Biol. 2005, 35, 729–739. [Google Scholar] [CrossRef]
- Ferrandon, D.; Imler, J.L.; Hetru, C.; Hoffmann, J.A. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 2007, 7, 862–874. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, J.P.; Kanost, M.R.; Trenczek, T. Biological mediators of insect immunity. Annu. Rev. Entomol. 1997, 42, 611–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, H.; Wang, H.; Zhao, S.; Zuo, Y.; Yang, Y.; Wu, Y. Functional validation of cadherin as a receptor of Bt toxin Cry1Ac in Helicoverpa armigera utilizing the CRISPR/Cas9 system. Insect Biochem. Mol. Biol. 2016, 76, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Angst, B.D.; Marcozzi, C.; Magee, A.I. The cadherin superfamily: Diversity in form and function. J. Cell. Sci. 2001, 114, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, Y.; Wan, P.; Liu, K.; Xiao, Y.; Wang, J.; Cong, S.; Xu, D.; Wu, K.; Fabrick, J.A.; et al. Resistance to Bacillus thuringiensis linked with a cadherin transmembrane mutation affecting cellular trafficking in pink bollworm from China. Insect Biochem. Mol. Biol. 2018, 94, 28–35. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, H.; Wu, S.; Yang, Y.; Xu, X.; Wu, Y. Identification and molecular detection of a deletion mutation responsible for a truncated cadherin of Helicoverpa armigera. Insect Biochem. Mol. Biol. 2006, 36, 735–740. [Google Scholar] [CrossRef]
- Walsh, T.; James, B.; Chakroun, M.; Ferré, J.; Downes, S. Isolating, characterising and identifying a Cry1Ac resistance mutation in field populations of Helicoverpa punctigera. Sci. Rep. 2018, 8, 2626. [Google Scholar] [CrossRef] [Green Version]
- Gómez, I.; Sánchez, J.; Miranda, R.; Bravo, A.; Soberón, M. Cadherin-like receptor binding facilitates proteolytic cleavage of helix alpha-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 2002, 513, 242–246. [Google Scholar] [CrossRef] [Green Version]
- Stevens, T.; Song, S.; Bruning, J.B.; Choo, A.; Baxter, S.W. Expressing a moth abcc2 gene in transgenic Drosophila causes susceptibility to Bt Cry1Ac without requiring a cadherin-like protein receptor. Insect Biochem. Mol. Biol. 2017, 80, 61–70. [Google Scholar] [CrossRef]
- Chauhan, V.K.; Dhania, N.K.; Chaitanya, R.K.; Senthilkumaran, B.; Dutta-Gupta, A. Larval midgut responses to sublethal dose of Cry toxin in lepidopteran pst Achaea janata. Front. Physiol. 2017, 8, 662. [Google Scholar] [CrossRef] [Green Version]
- Los, F.C.; Kao, C.Y.; Smitham, J.; McDonald, K.L.; Ha, C.; Peixoto, C.A.; Aroian, R.V. RAB-5- and RAB-11-dependent vesicle-trafficking pathways are required for plasma membrane repair after attack by bacterial pore-forming toxin. Cell Host Microbe 2011, 9, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Kang, S.; Chen, D.; Wu, Q.; Wang, S.; Xie, W.; Zhu, X.; Baxter, S.W.; Zhou, X.; Jurat-Fuentes, J.L.; et al. MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet. 2015, 11, e1005124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Z.; Kang, S.; Sun, D.; Gong, L.; Zhou, J.; Qin, J.; Guo, L.; Zhu, L.; Bai, Y.; Ye, F.; et al. MAPK-dependent hormonal signaling plasticity contributes to overcoming Bacillus thuringiensis toxin action in an insect host. Nat. Commun. 2020, 11, 3003. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, K.; Dawson, R.J.; Locher, K.P. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 2007, 17, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Chakrabarty, S.; Jin, M.; Liu, K.; Xiao, Y. Insect ATP-binding cassette (ABC) transporters: Roles in xenobiotic detoxification and Bt insecticidal activity. Int. J. Mol. Sci. 2019, 20, 2829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Li, C.Y.; Kong, A.N. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 2005, 28, 249–268. [Google Scholar] [CrossRef]
- Sato, R.; Adegawa, S.; Li, X.; Tanaka, S.; Endo, H. Function and role of ATP-binding cassette transporters as receptors for 3D-Cry toxins. Toxins 2019, 11, 124. [Google Scholar] [CrossRef] [Green Version]
- Tay, W.T.; Mahon, R.J.; Heckel, D.G.; Walsh, T.K.; Downes, S.; James, W.J.; Lee, S.F.; Reineke, A.; Williams, A.K.; Gordon, K.H. Insect resistance to Bacillus thuringiensis toxin Cry2Ab is conferred by mutations in an ABC transporter subfamily A protein. PLoS Genet. 2015, 11, e1005534. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Liu, S.; Liu, L.; Tay, W.T.; Walsh, T.K.; Yang, Y.; Wu, Y. CRISPR/Cas9 mediated genome editing of Helicoverpa armigera with mutations of an ABC transporter gene HaABCA2 confers resistance to Bacillus thuringiensis Cry2A toxins. Insect Biochem. Mol. Biol. 2017, 87, 147–153. [Google Scholar] [CrossRef]
- Pauchet, Y.; Bretschneider, A.; Augustin, S.; Heckel, D.G. A P-glycoprotein Is linked to resistance to the Bacillus thuringiensis Cry3Aa toxin in a leaf beetle. Toxins 2016, 8, 362. [Google Scholar] [CrossRef]
- Atsumi, S.; Miyamoto, K.; Yamamoto, K.; Narukawa, J.; Kawai, S.; Sezutsu, H.; Kobayashi, I.; Uchino, K.; Tamura, T.; Mita, K.; et al. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 2012, 109, E1591–E1598. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, S.; Endo, H.; Adegawa, S.; Kikuta, S.; Sato, R. Functional characterization of Bacillus thuringiensis Cry toxin receptors explains resistance in insects. FEBS J. 2016, 283, 4474–4490. [Google Scholar] [CrossRef] [PubMed]
- Florin, L.; Lang, T. Tetraspanin assemblies in virus infection. Front. Immunol. 2018, 9, 1140. [Google Scholar] [CrossRef] [PubMed]
- Charrin, S.; Jouannet, S.; Boucheix, C.; Rubinstein, E. Tetraspanins at a glance. J. Cell Sci. 2014, 127, 3641–3648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yáñez-Mó, M.; Barreiro, O.; Gordon-Alonso, M.; Sala-Valdés, M.; Sánchez-Madrid, F. Tetraspanin-enriched microdomains: A functional unit in cell plasma membranes. Trends Cell Biol. 2009, 19, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003, 19, 397–422. [Google Scholar] [CrossRef]
- Cannon, K.S.; Cresswell, P. Quality control of transmembrane domain assembly in the tetraspanin CD82. EMBO J. 2001, 20, 2443–2453. [Google Scholar] [CrossRef] [Green Version]
- Tarrant, J.M.; Robb, L.; van Spriel, A.B.; Wright, M.D. Tetraspanins: Molecular organisers of the leukocyte surface. Trends Immunol. 2003, 24, 610–617. [Google Scholar] [CrossRef]
- Zhuang, S.; Kelo, L.; Nardi, J.B.; Kanost, M.R. An integrin-tetraspanin interaction required for cellular innate immune responses of an insect, Manduca sexta. J. Biol. Chem. 2007, 282, 22563–22572. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Wang, C.; Li, K.; Yang, Y.; He, Y.-Z.; Wu, Y. Transcriptional Analysis of Cotton Bollworm Strains with Different Genetic Mechanisms of Resistance and Their Response to Bacillus thuringiensis Cry1Ac Toxin. Toxins 2022, 14, 366. https://doi.org/10.3390/toxins14060366
Yu S, Wang C, Li K, Yang Y, He Y-Z, Wu Y. Transcriptional Analysis of Cotton Bollworm Strains with Different Genetic Mechanisms of Resistance and Their Response to Bacillus thuringiensis Cry1Ac Toxin. Toxins. 2022; 14(6):366. https://doi.org/10.3390/toxins14060366
Chicago/Turabian StyleYu, Shan, Chenyang Wang, Kaixia Li, Yihua Yang, Ya-Zhou He, and Yidong Wu. 2022. "Transcriptional Analysis of Cotton Bollworm Strains with Different Genetic Mechanisms of Resistance and Their Response to Bacillus thuringiensis Cry1Ac Toxin" Toxins 14, no. 6: 366. https://doi.org/10.3390/toxins14060366
APA StyleYu, S., Wang, C., Li, K., Yang, Y., He, Y.-Z., & Wu, Y. (2022). Transcriptional Analysis of Cotton Bollworm Strains with Different Genetic Mechanisms of Resistance and Their Response to Bacillus thuringiensis Cry1Ac Toxin. Toxins, 14(6), 366. https://doi.org/10.3390/toxins14060366







