Heterosigma akashiwo in Patagonian Fjords: Genetics, Growth, Pigment Signature and Role of PUFA and ROS in Ichthyotoxicity
Abstract
:1. Introduction
2. Results
2.1. Molecular Identification
2.2. Pigments Signature
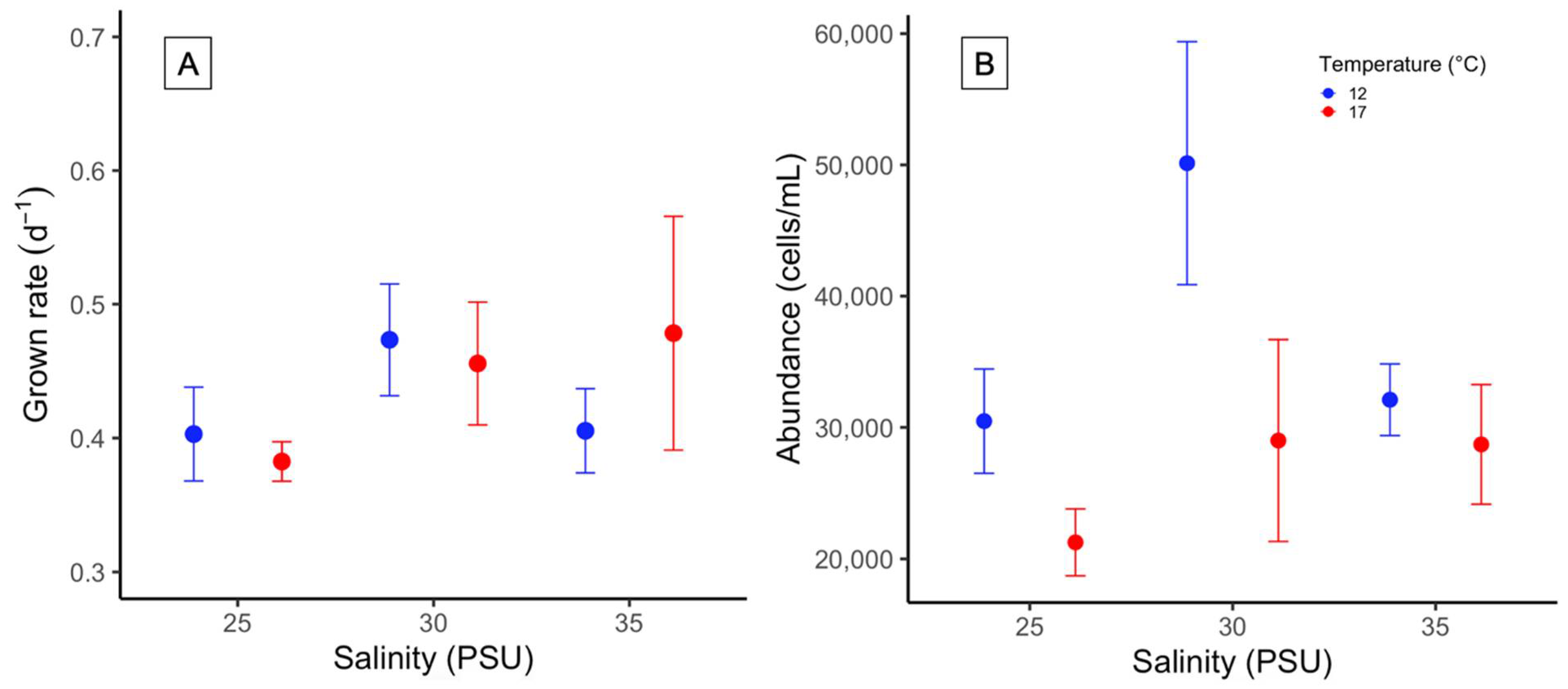
2.3. Growth Rate and Cell Density
2.4. Fatty Acid Profile and Superoxide Production
2.5. Intra- and Extracellular Ichthyotoxicity
3. Discussion
4. Materials and Methods
4.1. Microalgal Culture Conditions
4.2. DNA Extraction, Amplification, Sequencing, and Phylogeny
4.3. HPLC Pigment Analysis
4.4. Lipid Extraction and Analysis
4.5. Production of Superoxide Anion by H. akashiwo
4.6. RTgill-W1 Cell Line Maintenance
4.7. Gill Cell Assay with H. akashiwo
4.8. H. akashiwo Growth under Different Temperatures and Salinities
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamochi, S. Mechanisms for outbreak of Heterosigma akashiwo red tide in Osaka Bay, Japan. Bull. Osaka Pref. Fish. Exp. Stat. 1989, 8, 1–110. [Google Scholar]
- Martínez, R.; Orive, E.; Laza-Martínez, A.; Seoane, S. Growth response of six strains of Heterosigma akashiwo to varying temperature, salinity and irradiance conditions. J. Plankton Res. 2010, 32, 529–538. [Google Scholar] [CrossRef]
- Mardones, J.; Clément, A.; Rojas, X. Monitoring potentially ichthyotoxic phytoflagellates in southern fjords of Chile. Harmful Algae News 2012, 45, 6–7. [Google Scholar]
- Strom, S.L.; Harvey, E.L.; Fredrickson, K.A.; Menden-Deuer, S. Broad salinity tolerance as a refuge from predation in the harmful raphidophyte alga Heterosigma akashiwo (Raphidophyceae). J. Phycol. 2013, 49, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Imai, I.; Itakura, S. Importance of cysts in the population dynamics of the red tide flagellate Heterosigma akashiwo (Raphidophyceae). Mar. Biol. 1999, 133, 755–762. [Google Scholar] [CrossRef]
- Itakura, S.; Nagasaki, K.; Yamaguchi, M.; Imai, I. Cyst formation in the red tide flagellate Heterosigma akashiwo (Raphidophyceae). J. Plankton Res. 1996, 18, 1975–1979. [Google Scholar] [CrossRef]
- Branco, S.; Menezes, M.; Alves-de-Souza, C.; Domingos, P.; Schramm, M.A.; Proença, L.A.O. Recurrent blooms of Heterosigma akashiwo (Raphidophyceae) in the Piraquê Channel, rodrigo de freitas Lagoon, southeast Brazil. Braz. J. Biol. 2014, 74, 529–537. [Google Scholar] [CrossRef] [Green Version]
- Engesmo, A.; Eikrem, W.; Seoane, S.; Smith, K.; Edvardsen, B.; Hofgaard, A.; Tomas, C.R. New insights into the morphology and phylogeny of Heterosigma akashiwo (Raphidophyceae), with the description of Heterosigma minor sp. nov. Phycologia 2016, 55, 279–294. [Google Scholar] [CrossRef]
- Hallegraeff, G.M.; Hara, Y. Taxonomy of harmful marine raphidophytes. In Manual on Harmful Marine Microalgae; UNESCO Digital Library: Paris, France, 2003; pp. 511–522. [Google Scholar]
- Powers, L.; Creed, I.F.; Trick, C.G. Sinking of Heterosigma akashiwo results in increased toxicity of this harmful algal bloom species. Harmful Algae 2012, 13, 95–104. [Google Scholar] [CrossRef]
- Salmon Expert. Heterosigma Akashiwo Y Karenia: Las Dos Microalgas Detectadas En Fiordo Comau. Available online: https://www.salmonexpert.cl/article/heterosigma-akashiwo-y-karenia-las-dos-microalgas-detectadas-en-fiordo-comau/ (accessed on 29 June 2022).
- Mardones, J. Screening of Chilean fish-killing microalgae using a gill cell-based assay. Lat. Am. J. Aquat. Res. 2020, 48, 329–335. [Google Scholar] [CrossRef]
- Sandoval-Sanhueza, A.; Aguilera-Belmonte, A.; Basti, L.; Figueroa, R.I.; Molinet, C.; Álvarez, G.; Oyanedel, S.; Riobó, P.; Mancilla-Gutiérrez, G.; Díaz, P.A. Interactive effects of temperature and salinity on the growth and cytotoxicity of the fish-killing microalgal species Heterosigma akashiwo and Pseudochattonella verruculosa. Mar. Pollut. Bull. 2022, 174, 113234. [Google Scholar] [CrossRef] [PubMed]
- Astuya, A.; Ramírez, A.E.; Aballay, A.; Araya, J.; Silva, J.; Ulloa, V.; Fuentealba, J. Neurotoxin-like compounds from the ichthyotoxic red tide alga Heterosigma akashiwo induce a TTX-like synaptic silencing in mammalian neurons. Harmful Algae 2015, 47, 1–8. [Google Scholar] [CrossRef]
- Gallardo-Rodríguez, J.; Astuya-Villalón, A.; Avello, V.; Llanos-Rivera, A.; Krock, B.; Agurto-Muñoz, C.; Sánchez-Mirón, A.; García-Camacho, F. Production of extracts with anaesthetic activity from the culture of Heterosigma akashiwo in pilot-scale photobioreactors. Algal Res. 2020, 45, 101760. [Google Scholar] [CrossRef]
- Astuya, A.; Rivera, A.; Vega-Drake, K.; Aburto, C.; Cruzat, F.; Ulloa, V.; Caprile, T.; Gallardo-Rodríguez, J.J. Study of the ichthyotoxic microalga Heterosigma akashiwo by transcriptional activation of sublethal marker Hsp70b in Transwell co-culture assays. PLoS ONE 2018, 13, e0201438. [Google Scholar]
- Gómez, P.I.; Inostroza, I.; Castro-Varela, P.; Silva, J.; Clément, A.; Rojas, G.; Belmonte, A.A. Comparison of a Chilean strain of the ichthyotoxic phytoflagellate Heterosigma akashiwo (Raphidophyceae) with strains from France, Spain and New Zealand. Phycologia 2022, 61, 7–15. [Google Scholar] [CrossRef]
- Dorantes-Aranda, J.J.; Seger, A.; Mardones, J.I.; Nichols, P.D.; Hallegraeff, G.M. Progress in understanding algal bloom-mediated fish kills: The role of superoxide radicals, phycotoxins and fatty acids. PLoS ONE 2015, 10, e0133549. [Google Scholar] [CrossRef]
- Yamasaki, Y.; Shikata, T.; Nukata, A.; Ichiki, S.; Nagasoe, S.; Matsubara, T.; Shimasaki, Y.; Nakao, M.; Yamaguchi, K.; Oshima, Y.; et al. Extracellular polysaccharide-protein complexes of a harmful alga mediate the allelopathic control it exerts within the phytoplankton community. ISME J. 2009, 3, 808–817. [Google Scholar] [CrossRef] [Green Version]
- Branco, S.; Almeida, L.L.; Alves-De-Souza, C.; Oliveira, M.M.; Proença, L.A.; Menezes, M. Morphological and genetic characterization of bloom-forming Raphidophyceae from Brazilian coast. Phycol. Res. 2019, 67, 279–290. [Google Scholar] [CrossRef]
- Connell, L.B. Nuclear ITS region of the alga Heterosigma akashiwo (Chromophyta: Raphidophyceae) is identical in isolates from Atlantic and Pacific basins. Mar. Biol. 2000, 136, 953–960. [Google Scholar] [CrossRef]
- Bowers, H.A.; Tomas, C.; Tengs, T.; Kempton, J.W.; Lewitus, A.J.; Oldach, D.W. Raphidophyceae [Chaedefaud ex silva] Systematics and rapid identification: Sequence analyses and real-time PCR assays. J. Phycol. 2006, 42, 1333–1348. [Google Scholar] [CrossRef] [Green Version]
- Fredrickson, K.A.; Strom, S.L.; Crim, R.; Coyne, K.J. Interstrain variability in physiology and genetics of Heterosigma akashiwo (Raphidophyceae) from the west coast of north America. J. Phycol. 2011, 47, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Marzetz, V.; Spijkerman, E.; Striebel, M.; Wacker, A. Phytoplankton community responses to interactions between light intensity, light variations, and phosphorus supply. Front. Environ. Sci. 2020, 8, 539733. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Adams, W.W.; Ebbert, V.; Logan, B.A. Ecophysiology of the xanthophyll cycle. In The Photochemistry of Carotenoids; Springer: Dordrecht, The Netherlands, 1999; Volume 8, Chapter 14; pp. 245–269. [Google Scholar]
- Jacob-Lopes, E.; Queiroz, M.I.; Zepka, L.Q. Pigments from Microalgae Handbook; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Butrón, A.; Madariaga, I.; Orive, E. Tolerance to high irradiance levels as a determinant of the bloom-forming Heterosigma akashiwo success in estuarine waters in summer. Estuar. Coast. Shelf Sci. 2012, 107, 141–149. [Google Scholar] [CrossRef]
- Jeong, H.J. Mixotrophy in red tide algae raphidophytes. J. Eukaryot. Microbiol. 2011, 58, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Villanova, V.; Spetea, C. Mixotrophy in diatoms: Molecular mechanism and industrial potential. Physiol. Plant. 2021, 173, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.; Jeong, H.J.; Kim, S.; Kim, G.; Kang, J. Bacterivory by co-occurring red-tide algae, heterotrophic nanoflagellates, and ciliates. Mar. Ecol. Prog. Ser. 2006, 322, 85–97. [Google Scholar] [CrossRef]
- Mardones, J.I.; Norambuena, L.; Paredes, J.; Fuenzalida, G.; Dorantes-Aranda, J.J.; Chang, K.J.L.; Guzmán, L.; Krock, B.; Hallegraeff, G. Unraveling the Karenia selliformis complex with the description of a non-gymnodimine producing Patagonian phylo-type. Harmful Algae 2020, 98, 101892. [Google Scholar] [CrossRef]
- Mardones, J.I.; Dorantes-Aranda, J.J.; Nichols, P.D.; Hallegraeff, G.M. Fish gill damage by the dinoflagellate Alexandrium catenella from Chilean fjords: Synergistic action of ROS and PUFA. Harmful Algae 2015, 49, 40–49. [Google Scholar] [CrossRef]
- Marshall, J.A.; Nichols, P.D.; Hamilton, B.; Lewis, R.J.; Hallegraeff, G.M. Ichthyotoxicity of Chattonella marina (Raphidophyceae) to damselfish (Acanthochromis polycanthus): The synergistic role of reactive oxygen species and free fatty acids. Harmful Algae 2003, 2, 273–281. [Google Scholar] [CrossRef]
- Scholin, C.A.; Herzog, M.; Sogin, M.; Anderson, D.M. Identification of group-and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). ii. sequence analysis of a fragment of the LSU rRNA gene. J. Phycol. 1994, 30, 999–1011. [Google Scholar] [CrossRef]
- Edvardsen, B.; Shalchian-Tabrizi, K.; Jakobsen, K.S.; Medlin, L.K.; Dahl, E.; Brubak, S.; Paasche, E. Genetic variability and molecular phylogeny of Dinophysis species (DINOPHYCEAE) from Norwegian waters inferred from single cell analyses of rDNA. J. Phycol. 2003, 39, 395–408. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Sanz, N.; García-Blanco, A.; Gavalás-Olea, A.; Loures, P.; Garrido, J.L. Phytoplankton pigment biomarkers: HPLC separation using a pentafluorophenyloctadecyl silica column. Methods Ecol. Evol. 2015, 6, 1199–1209. [Google Scholar] [CrossRef] [Green Version]
- Mooney, B.D.; Nichols, P.D.; de Salas, M.F.; Hallegraeff, G.M. Lipid, fatty acid and sterol composition of eight species of Kareniaceae (Dinophyta): Chemotaxonomy and putative lipid phycotoxins. J. Phycol. 2007, 43, 101–111. [Google Scholar] [CrossRef]
- Godrant, A.; Rose, A.L.; Sarthou, G.; Waite, T.D. New method for the determination of extracellular production of superoxide by marine phytoplankton using the chemiluminescence probes MCLA and red-CLA. Limnol. Oceanogr. Methods 2009, 7, 682–692. [Google Scholar] [CrossRef]
- Bols, N.C.; Barlian, A.; Chirino-Trejo, M.; Caldwell, S.J.; Goegan, P.; Lee, L.E.J. Development of a cell line from primary cultures of rainbow trout, Oncorhynchus mykiss (Walbaum), gills. J. Fish Dis. 1994, 17, 601–611. [Google Scholar] [CrossRef]
- Dorantes-Aranda, J.J.; Waite, T.D.; Godrant, A.; Rose, A.L.; Tovar, C.D.; Woods, G.M.; Hallegraeff, G.M. Novel application of a fish gill cell line assay to assess ichthyotoxicity of harmful marine microalgae. Harmful Algae 2011, 10, 366–373. [Google Scholar] [CrossRef]
- Page, B.; Page, M.; Noel, C. A new fluorometric assay for cytotoxicity measurements in vitro. Int. J. Oncol. 1993, 3, 473–476. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]

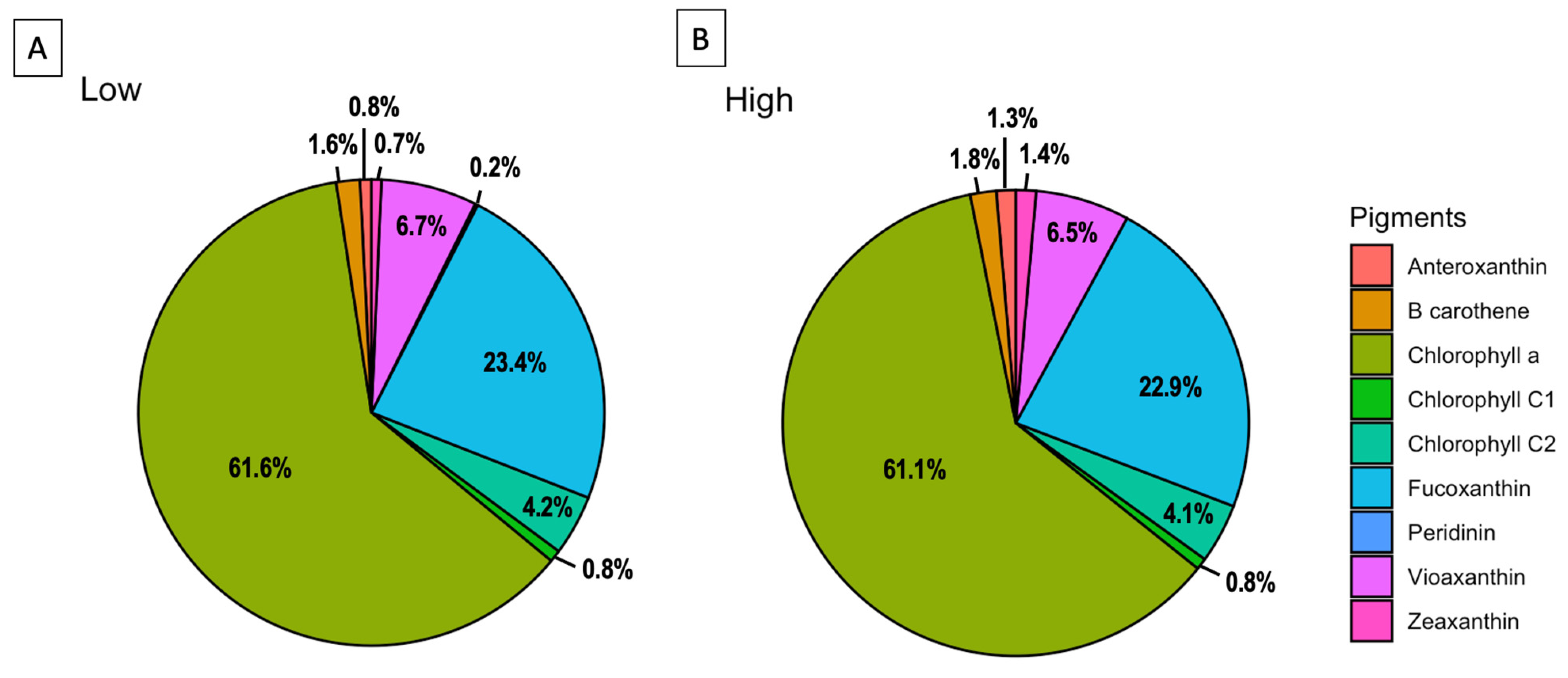

| Pigments | Sample | Bloom H. akashiwo | CREAN_ HA03 | ||
|---|---|---|---|---|---|
| Year | 2021 | 2020 | 2022 | ||
| Light Intensity | 1430 | 100 | 50 | 150 | |
| Peridinin | ND | ND | 0.20 | ND | |
| Chlorophyll C1 | 7.24 | 2.73 | 0.84 | 0.80 | |
| Chlorophyll C2 | 15.09 | 18.32 | 4.19 | 4.11 | |
| Fucoxanthin | 48.78 | 48.43 | 23.41 | 22.84 | |
| Violaxanthin | 14.73 | 9.77 | 6.67 | 6.77 | |
| Antheraxanthin | 1.78 | 3.80 | 0.80 | 1.34 | |
| Zeaxanthin | 1.60 | 2.25 | 0.68 | 1.34 | |
| Chlorophyll a | 9.80 | 14.70 | 61.59 | 60.95 | |
| β-Carotene | ND | ND | 1.63 | 1.84 | |
| Chlorophyllide a | 0.98 | 0.00 | ND | ND | |
| Ratio Fuco/Chl a | 4.98 | 3.29 | 0.38 | 0.37 | |
| Fatty Acid | Mean | SD | |
|---|---|---|---|
| SFA | |||
| 13:0 | 0.02 | ± | 0.02 |
| 14:0 | 5.47 | ± | 0.08 |
| 15:0 | 0.83 | ± | 0.04 |
| 16:0 PA | 20.94 | ± | 0.14 |
| 17:0 | 0.25 | ± | 0.01 |
| 18:0 | 2.77 | ± | 0.09 |
| 20:0 | 0.07 | ± | 0 |
| 22:0 | 0.16 | ± | 0.13 |
| 24:0 | 0.02 | ± | 0.02 |
| Hexadecenal | 0.04 | ± | 0.05 |
| Octadecenal | 0.15 | ± | 0 |
| Branched | |||
| a15:0 | 0.01 | ± | 0.01 |
| i14:0 | 0.02 | ± | 0.02 |
| i15:0 | 0.63 | ± | 0.01 |
| i16:0 | 0.22 | ± | 0.01 |
| i17:0 | 0.03 | ± | 0 |
| i18:0 | 0.16 | ± | 0.02 |
| MUFA | |||
| 17:1ω8c + a17:0 | 0.15 | ± | 0.03 |
| 14:01 | 0.06 | ± | 0.02 |
| 16:1ω9c | 0.33 | ± | 0.29 |
| 16:1ω7c | 8.60 | ± | 0.09 |
| 16:1ω7t | 0.02 | ± | 0.03 |
| 16:1ω5c | 2.54 | ± | 0.02 |
| 16:1ω13t | 1.05 | ± | 0.91 |
| 18:1ω9c | 1.36 | ± | 0.04 |
| 18:1ω7c | 7.09 | ± | 0.09 |
| 18:1ω7t | 0.05 | ± | 0.05 |
| 18:1 a | 0.10 | ± | 0.08 |
| 18:1 b | 0.04 | ± | 0.07 |
| 18:1 c | 0.04 | ± | 0.04 |
| 19:1 a | 0.01 | ± | 0.01 |
| 19:1 b | 0.02 | ± | 0.02 |
| 20:1ω11c | 0.40 | ± | 0.01 |
| 20:1ω9c | 0.12 | ± | 0 |
| 20:1ω7c | 0.14 | ± | 0 |
| 22:1ω11c | 0.05 | ± | 0.04 |
| 22:1ω9c | 0.10 | ± | 0.07 |
| PUFA | |||
| 18:3ω6 + 18:5ω3 | 3.63 | ± | 0.05 |
| 18:4ω3 | 10.44 | ± | 0.13 |
| 18:2ω6 | 3.73 | ± | 0.06 |
| 18:3ω3 | 2.54 | ± | 0.05 |
| 20:4ω6 | 1.09 | ± | 0.01 |
| 20:5ω3 | 13.04 | ± | 0.15 |
| 20:3ω6 | 0.66 | ± | 0.02 |
| 20:4ω3 | 2.93 | ± | 0.03 |
| 20:2ω6 | 0.11 | ± | 0.01 |
| 21:5ω3 | 0.04 | ± | 0.03 |
| 22:5ω6 | 0.16 | ± | 0.02 |
| 22:6ω3 DHA | 1.70 | ± | 0.02 |
| 22:5ω3 | 0.34 | ± | 0.06 |
| FALD | |||
| 16:0 FALD | 5.11 | ± | 0.06 |
| 18:0 FALD | 0.26 | ± | 0.04 |
| CLA | |||
| CLAa | 0.04 | ± | 0.07 |
| CLAb | 0.01 | ± | 0.02 |
| CLAc | 0.12 | ± | 0.1 |
| Sum SFA | 30.73 | ||
| Sum Branched | 1.06 | ||
| Sum MUFA | 22.27 | ||
| Sum PUFA | 40.41 | ||
| Sum FALD | 5.37 | ||
| Sum CLA | 0.17 |
| Species | Strain | Isolator | Origin | Whole Cells (pmol O2– cell−1 h−1) | Lysed Cells (pmol O2– cell−1 h−1) | ||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||||
| H. akashiwo | CREAN_HA03 | J.I. Mardones, 2019 | Chile, Los Lagos | 0.308 | 0.03 | 0.293 | 0.013 |
| K. selliformis | CREAN_KS02 | J.I. Mardones, 2018 | Chile, Aysén | 0.311 | 0.467 | 0.861 | 0.535 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores-Leñero, A.; Vargas-Torres, V.; Paredes-Mella, J.; Norambuena, L.; Fuenzalida, G.; Lee-Chang, K.; Mardones, J.I. Heterosigma akashiwo in Patagonian Fjords: Genetics, Growth, Pigment Signature and Role of PUFA and ROS in Ichthyotoxicity. Toxins 2022, 14, 577. https://doi.org/10.3390/toxins14090577
Flores-Leñero A, Vargas-Torres V, Paredes-Mella J, Norambuena L, Fuenzalida G, Lee-Chang K, Mardones JI. Heterosigma akashiwo in Patagonian Fjords: Genetics, Growth, Pigment Signature and Role of PUFA and ROS in Ichthyotoxicity. Toxins. 2022; 14(9):577. https://doi.org/10.3390/toxins14090577
Chicago/Turabian StyleFlores-Leñero, Ana, Valentina Vargas-Torres, Javier Paredes-Mella, Luis Norambuena, Gonzalo Fuenzalida, Kim Lee-Chang, and Jorge I. Mardones. 2022. "Heterosigma akashiwo in Patagonian Fjords: Genetics, Growth, Pigment Signature and Role of PUFA and ROS in Ichthyotoxicity" Toxins 14, no. 9: 577. https://doi.org/10.3390/toxins14090577
APA StyleFlores-Leñero, A., Vargas-Torres, V., Paredes-Mella, J., Norambuena, L., Fuenzalida, G., Lee-Chang, K., & Mardones, J. I. (2022). Heterosigma akashiwo in Patagonian Fjords: Genetics, Growth, Pigment Signature and Role of PUFA and ROS in Ichthyotoxicity. Toxins, 14(9), 577. https://doi.org/10.3390/toxins14090577





