Venom Variation of Neonate and Adult Chinese Cobras in Captivity Concerning Their Foraging Strategies
Abstract
:1. Introduction
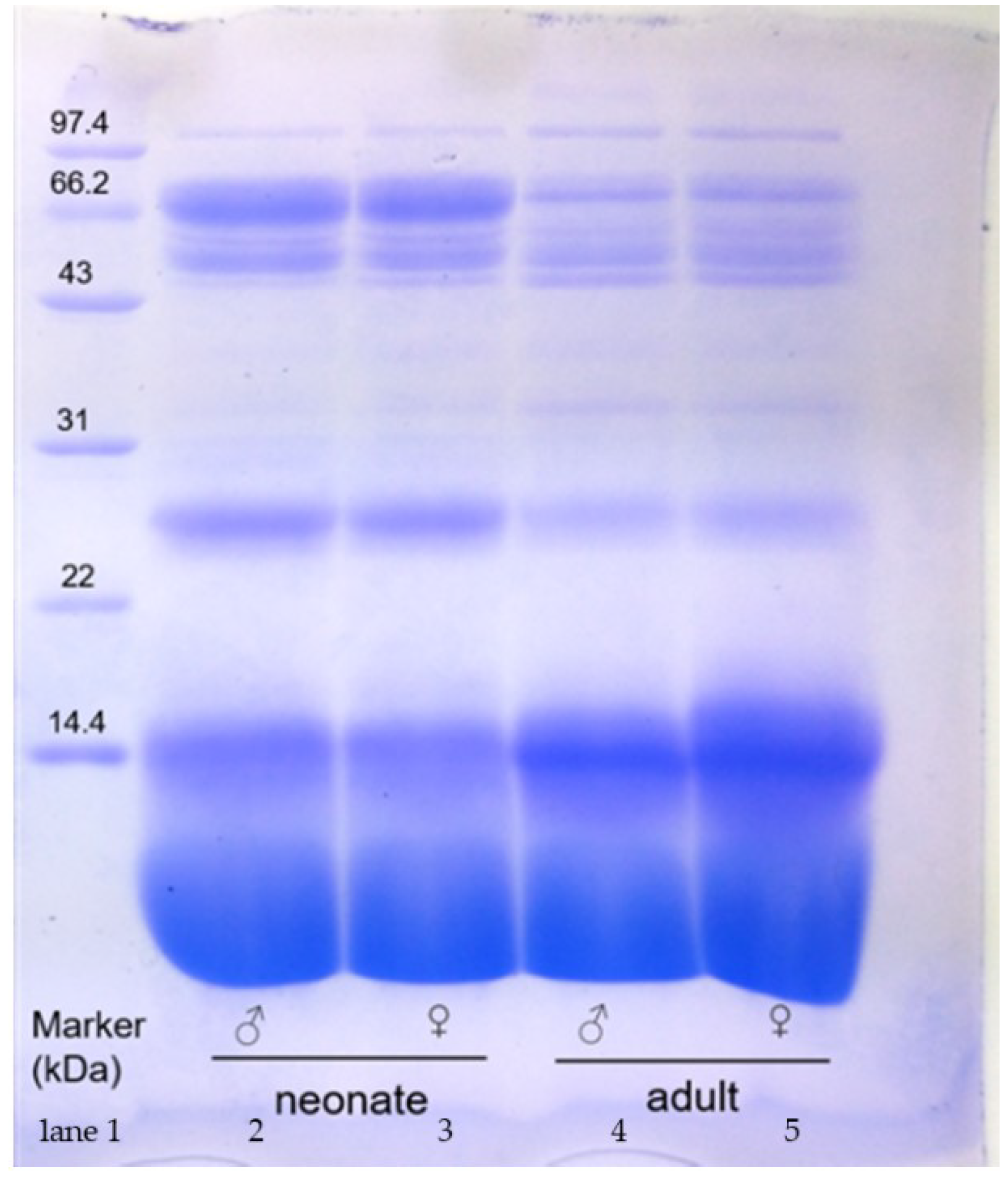
2. Results
2.1. SDS-PAGE
2.2. Venom Protein Concentration and Captive Snake Growth Status
2.3. Venom Proteomics
2.4. Protein Sequence Alignment
2.5. Venom Gland Transcriptome
2.6. Cooperative Proteomic and Transcriptomic Analysis
2.7. Toxicological and Enzymatic Activity of N. atra Venom
3. Discussion
3.1. Ontogenetic Variation Predominates in the Snake Venom of Captive Species
3.2. Neonate and Adult Snakes have Different Strategies for Handling Prey
3.3. The Hatchery Breeding Process Rearranges Snake Venom
3.4. Asynchronous Snake Venom Synthesis Process
3.5. Widespread Distribution of Toxin Gene on Chromosomes
4. Conclusions
5. Materials and Methods
5.1. Animals Keeping and Feeding
5.2. Snake Venom and Gland
5.3. Library Preparation for Transcriptome Sequencing
5.4. cDNA Sequencing and Data Analysis
5.5. TMT Labeling of Peptides
5.6. Separation of Fractions
5.7. LC-MS/MS Analysis
5.8. The Identification and Quantitation of Protein
5.9. Protein and Transcript Alignment
5.10. Snake Protein Concentration Determination
5.11. SDS-PAGE
5.12. Median Lethal Dose (LD50) Determination
5.13. Enzyme Activity Assay
5.14. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Snakebite-Envenoming. Available online: https://www.who.int/zh/news-room/fact-sheets/detail/snakebite-envenoming (accessed on 8 April 2019).
- Pin, L.; Wang, D.; Lutao, X.; Linjie, X. 2019 China Medical Rescue Association Animal Injury Treatment Branch Group Standard “Snake Bite Treatment Specification” Interpretation. In Proceedings of the 2020 China Animal Injury Diagnosis and Treatment Summit, Zhengzhou, China; 2020; p. 35. [Google Scholar] [CrossRef]
- Ying, H.; Jianfang, G.; Longhui, L.; Xiaomei, M.; Xiang, J. Age-related Variation in Snake Venom: Evidence from Two Snakes (Naja atra and Deinagkistrodon acutus) in Southeastern China. Asian Herpetol. Res. 2014, 5, 119–127. [Google Scholar] [CrossRef]
- Wang, W.; Chen, Q.-F.; Yin, R.-X.; Zhu, J.-J.; Li, Q.-B.; Chang, H.-H.; Wu, Y.-B.; Michelson, E. Clinical features and treatment experience: A review of 292 Chinese cobra snakebites. Environ. Toxicol. Pharmacol. 2014, 37, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Machado Braga, J.R.; de Morais-Zani, K.; Pereira, D.D.S.; Sant’Anna, S.S.; da Costa Galizio, N.; Tanaka-Azevedo, A.M.; Gomes Vilarinho, A.R.; Rodrigues, J.L.; da Rocha, M.M.T. Sexual and ontogenetic variation of Bothrops leucurus venom. Toxicon 2020, 184, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Amorim, F.G.; Costa, T.R.; Baiwir, D.; De Pauw, E.; Quinton, L.; Sampaio, S.V. Proteopeptidomic, Functional and Immunoreactivity Characterization of Bothrops moojeni Snake Venom: Influence of Snake Gender on Venom Composition. Toxins 2018, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Zelanis, A.; Menezes, M.C.; Kitano, E.S.; Liberato, T.; Tashima, A.K.; Pinto, A.F.; Sherman, N.E.; Ho, P.L.; Fox, J.W.; Serrano, S.M. Proteomic identification of gender molecular markers in Bothrops jararaca venom. J. Proteom. 2016, 139, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Leroy, J.; Mociño-Deloya, E.; Setser, K.; Bryson, R.W.; Saviola, A.J. Venom Ontogeny in the Mexican Lance-Headed Rattlesnake (Crotalus polystictus). Toxins 2018, 10, 271. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.N.; Lomonte, B.; del Carmen Gutierrez, M.; Alagon, A.; Gutierrez, J.M. Intraspecies variation in the venom of the rattlesnake Crotalus simus from Mexico: Different expression of crotoxin results in highly variable toxicity in the venoms of three subspecies. J. Proteom. 2013, 87, 103–121. [Google Scholar] [CrossRef]
- Zelanis, A.; Travaglia-Cardoso, S.R.; De Fátima Domingues Furtado, M. Ontogenetic changes in the venom of Bothrops insularis (Serpentes: Viperidae) and its biological implication. S. Am. J. Herpetol. 2008, 3, 43–50. [Google Scholar] [CrossRef]
- Cipriani, V.; Debono, J.; Goldenberg, J.; Jackson, T.N.W.; Arbuckle, K.; Dobson, J.; Koludarov, I.; Li, B.; Hay, C.; Dunstan, N.; et al. Correlation between ontogenetic dietary shifts and venom variation in Australian brown snakes (Pseudonaja). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2017, 197, 53–60. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, H.L.; Sanz, L.; Chiucchi, J.E.; Farrell, T.M.; Calvete, J.J. Proteomic analysis of ontogenetic and diet-related changes in venom composition of juvenile and adult Dusky Pigmy rattlesnakes (Sistrurus miliarius barbouri). J. Proteom. 2011, 74, 2169–2179. [Google Scholar] [CrossRef]
- Sasa, M. Diet and snake venom evolution: Can local selection alone explain intraspecific venom bvariation? Toxicon 1999, 37, 249–252. [Google Scholar] [PubMed]
- Daltry, J.C.; Wüster, W.; Thorpe, R.S. Diet and snake venom evolution. Nature 1996, 379, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Chanda, A.; Patra, A.; Kalita, B.; Mukherjee, A.K. Proteomics analysis to compare the venom composition between Naja naja and Naja kaouthia from the same geographical location of eastern India: Correlation with pathophysiology of envenomation and immunological cross-reactivity towards commercial polyantivenom. Expert Rev. Proteom. 2018, 15, 949–961. [Google Scholar] [CrossRef]
- Huang, H.-W.; Liu, B.-S.; Chien, K.-Y.; Chiang, L.-C.; Huang, S.-Y.; Sung, W.-C.; Wu, W.-G. Cobra venom proteome and glycome determined from individual snakes of Naja atra reveal medically important dynamic range and systematic geographic variation. J. Proteom. 2015, 128, 92–104. [Google Scholar] [CrossRef]
- Alape-Girón, A.; Sanz, L.; Escolano, J.; Flores-Díaz, M.; Madrigal, M.; Sasa, M.; Calvete, J.J. Snake venomics of the lancehead pitviper Bothrops asper geographic, individual, and ontogenetic variations. J. Proteome Res. 2008, 7, 3556–3571. [Google Scholar] [CrossRef]
- Warrell, D.A. Snake bite. Lancet 2010, 375, 77–88. [Google Scholar] [CrossRef]
- Alangode, A.; Rajan, K.; Nair, B.G. Snake antivenom: Challenges and alternate approaches. Biochem. Pharmacol. 2020, 181, 114135. [Google Scholar] [CrossRef]
- Liu, C.C.; You, C.H.; Wang, P.J.; Yu, J.S.; Huang, G.J.; Liu, C.H.; Hsieh, W.C.; Lin, C.C. Analysis of the efficacy of Taiwanese freeze-dried neurotoxic antivenom against Naja kaouthia, Naja siamensis and Ophiophagus hannah through proteomics and animal model approaches. PLoS Negl. Trop. Dis. 2017, 11, e0006138. [Google Scholar] [CrossRef]
- Casewell, N.R.; Jackson, T.N.W.; Laustsen, A.H.; Sunagar, K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef]
- Knudsen, C.; Laustsen, A.H. Recent Advances in Next Generation Snakebite Antivenoms. Trop. Med. Infect. Dis. 2018, 3, 42. [Google Scholar] [CrossRef]
- Utkin, Y.N. Modern trends in animal venom research-omics and nanomaterials. World J. Biol. Chem. 2017, 8, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Suryamohan, K.; Krishnankutty, S.P.; Guillory, J.; Jevit, M.; Schröder, M.S.; Wu, M.; Kuriakose, B.; Mathew, O.K.; Perumal, R.C.; Koludarov, I.; et al. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 2020, 52, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Chanda, A.; Mukherjee, A.K. Quantitative proteomics to reveal the composition of Southern India spectacled cobra (Naja naja) venom and its immunological cross-reactivity towards commercial antivenom. Int. J. Biol. Macromol. 2020, 160, 224–232. [Google Scholar] [CrossRef]
- Yap, M.K.K.; Fung, S.Y.; Tan, K.Y.; Tan, N.H. Proteomic characterization of venom of the medically important Southeast Asian Naja sumatrana (Equatorial spitting cobra). Acta Trop. 2014, 133, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-S.; Jiang, B.-R.; Hu, K.-C.; Liu, C.-H.; Hsieh, W.-C.; Lin, M.-H.; Sung, W.-C. Development of a Broad-Spectrum Antiserum against Cobra Venoms Using Recombinant Three-Finger Toxins. Toxins 2021, 13, 556. [Google Scholar] [CrossRef] [PubMed]
- Kornisiuk, E.; Jerusalinsky, D.; Cerveñansky, C.; Harvey, A.L. Binding of muscarinic toxins MT × 1 and MT × 2 from the venom of the green mamba Dendroaspis angusticeps to cloned human muscarinic cholinoceptors. Toxicon 1995, 33, 11–18. [Google Scholar] [CrossRef]
- Chien, K.Y.; Chiang, C.M.; Hseu, Y.C.; Vyas, A.A.; Rule, G.S.; Wu, W. Two distinct types of cardiotoxin as revealed by the structure and activity relationship of their interaction with zwitterionic phospholipid dispersions. J. Biol. Chem. 1994, 269, 14473–14483. [Google Scholar] [CrossRef]
- Rajagopalan, N.; Pung, Y.F.; Zhu, Y.Z.; Wong, P.T.H.; Kumar, P.P.; Kini, R.M. β-cardiotoxin: A new three-finger toxin from Ophiophagus hannah (king cobra) venom with beta-blocker activity. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2007, 21, 3685–3695. [Google Scholar] [CrossRef]
- Jang, J.-Y.; Kumar, T.K.S.; Jayaraman, G.; Yang, P.-W.; Yu, C. Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid. Biochemistry 1997, 36, 14635–14641. [Google Scholar] [CrossRef]
- Chiou, S.H.; Raynor, R.L.; Zheng, B.; Chambers, T.C.; Kuo, J.F. Cobra venom cardiotoxin (cytotoxin) isoforms and neurotoxin: Comparative potency of protein kinase C inhibition and cancer cell cytotoxicity and modes of enzyme inhibition. Biochemistry 1993, 32, 2062–2067. [Google Scholar] [CrossRef]
- Tans, G.; Rosing, J. Snake venom activators of factor X: An overview. Pathophysiol. Haemost. Thromb. 2001, 31, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.C.; Wu, W.B.; Huang, T.F. A snake venom metalloproteinase, kistomin, cleaves platelet glycoprotein VI and impairs platelet functions. J. Thromb. Haemost. 2008, 6, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Menezes, M.C.; Kitano, E.S.; Bauer, V.C.; Oliveira, A.K.; Cararo-Lopes, E.; Nishiyama, M.Y., Jr.; Zelanis, A.; Serrano, S.M.T. Early response of C2C12 myotubes to a sub-cytotoxic dose of hemorrhagic metalloproteinase HF3 from Bothrops jararaca venom. J. Proteom. 2019, 198, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Rau, J.C.; Beaulieu, L.M.; Huntington, J.A.; Church, F.C. Serpins in thrombosis, hemostasis and fibrinolysis. J. Thromb. Haemost. 2007, 5, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Sala, A.; Cabassi, C.S.; Santospirito, D.; Polverini, E.; Flisi, S.; Cavirani, S.; Taddei, S. Novel Naja atra cardiotoxin 1 (CTX-1) derived antimicrobial peptides with broad spectrum activity. PLoS ONE 2018, 13, e0190778. [Google Scholar] [CrossRef]
- Pla, D.; Petras, D.; Saviola, A.J.; Modahl, C.M.; Sanz, L.; Pérez, A.; Juárez, E.; Frietze, S.; Dorrestein, P.C.; Mackessy, S.P.; et al. Transcriptomics-guided bottom-up and top-down venomics of neonate and adult specimens of the arboreal rear-fanged Brown Treesnake, Boiga irregularis, from Guam. J. Proteom. 2018, 174, 71–84. [Google Scholar] [CrossRef]
- Xu, N.; Zhao, H.-Y.; Yin, Y.; Shen, S.-S.; Shan, L.-L.; Chen, C.-X.; Zhang, Y.-X.; Gao, J.-F.; Ji, X. Combined venomics, antivenomics and venom gland transcriptome analysis of the monocoled cobra (Naja kaouthia) from China. J. Proteom. 2017, 159, 19–31. [Google Scholar] [CrossRef]
- Durban, J.; Sanz, L.; Trevisan-Silva, D.; Neri-Castro, E.; Alagón, A.; Calvete, J.J. Integrated Venomics and Venom Gland Transcriptome Analysis of Juvenile and Adult Mexican Rattlesnakes Crotalus simus, C. tzabcan, and C. culminatus Revealed miRNA-modulated Ontogenetic Shifts. J. Proteome Res. 2017, 16, 3370–3390. [Google Scholar] [CrossRef]
- Zelanis, A.; Tashima, A.K.; Rocha, M.M.T.; Furtado, M.F.; Camargo, A.C.M.; Ho, P.L.; Serrano, S.M.T. Analysis of the Ontogenetic Variation in the Venom Proteome/Peptidome of Bothrops jararaca Reveals Different Strategies to Deal with Prey. J. Proteome Res. 2010, 9, 2278–2291. [Google Scholar] [CrossRef]
- Durban, J.; Pérez, A.; Sanz, L.; Gómez, A.; Bonilla, F.; Rodríguez, S.; Chacón, D.; Sasa, M.; Angulo, Y.; Gutiérrez, J.M.; et al. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genom. 2013, 14, 234. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Markland, F.S., Jr.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef]
- Nicholson, J.; Mirtschin, P.; Madaras, F.; Venning, M.; Kokkinn, M. Digestive properties of the venom of the Australian Coastal Taipan, Oxyuranus scutellatus (Peters, 1867). Toxicon 2006, 48, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, C.P.; Menaldo, D.L.; Zoccal, K.F.; Boldrini-França, J.; Peigneur, S.; Arantes, E.C.; Rosa, J.C.; Faccioli, L.H.; Tytgat, J.; Sampaio, S.V. First report on BaltCRP, a cysteine-rich secretory protein (CRISP) from Bothrops alternatus venom: Effects on potassium channels and inflammatory processes. Int. J. Biol. Macromol. 2019, 140, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Deka, A.; Sharma, M.; Mukhopadhyay, R.; Devi, A.; Doley, R. Naja kaouthia venom protein, Nk-CRISP, upregulates inflammatory gene expression in human macrophages. Int. J. Biol. Macromol. 2020, 160, 602–611. [Google Scholar] [CrossRef]
- Sun, Q.-Y.; Chen, G.; Guo, H.; Chen, S.; Wang, W.-Y.; Xiong, Y.-L. Prolonged cardiac xenograft survival in guinea pig-to-rat model by a highly active cobra venom factor. Toxicon 2003, 42, 257–262. [Google Scholar] [CrossRef]
- Ullah, A.; Ullah, K.; Ali, H.; Betzel, C.; ur Rehman, S. The Sequence and a Three-Dimensional Structural Analysis Reveal Substrate Specificity Among Snake Venom Phosphodiesterases. Toxins 2019, 11, 625. [Google Scholar] [CrossRef]
- Jiang, Y.; Yan, L.; Lee, W.; Xu, X.; Zhang, Y.; Zhao, R.; Zhang, Y.; Wang, W. Venom gland transcriptomes of two elapid snakes (Bungarus multicinctus and Naja atra) and evolution of toxin genes. BMC Genom. 2011, 12, 1. [Google Scholar] [CrossRef] [Green Version]
- Shan, L.-L.; Gao, J.-F.; Zhang, Y.-X.; Shen, S.-S.; He, Y.; Wang, J.; Ma, X.-M.; Ji, X. Proteomic characterization and comparison of venoms from two elapid snakes (Bungarus multicinctus and Naja atra) from China. J. Proteom. 2016, 138, 83–94. [Google Scholar] [CrossRef]
- El-Aziz, T.M.A.; Soares, A.G.; Stockand, J.D. Snake Venoms in Drug Discovery: Valuable Therapeutic Tools for Life Saving. Toxins 2019, 11, 564. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Fung, S.Y.; Tan, N.H. Venom-gland transcriptome and venom proteome of the Malaysian king cobra (Ophiophagus hannah). BMC Genom. 2015, 16, 687. [Google Scholar] [CrossRef] [PubMed]
- Koh, D.C.I.; Armugam, A.; Jeyaseelan, K. Snake venom components and their applications in biomedicine. Cell. Mol. Life. Sci. 2006, 63, 3030–3041. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Li, P. The IUCN Red List of Threatened Species 2014: E.T192109A2040894. Available online: https://doi.org/10.2305/IUCN.UK.2014-3.RLTS.T192109A2040894.en (accessed on 28 April 2019).
- Mackessy, S.P.; Williams, K.; Ashton, K.G.; Lannoo, M.J. Ontogenetic Variation in Venom Composition and Diet of Crotalus oreganus concolor: A Case of Venom Paedomorphosis? Copeia 2003, 2003, 769–782. [Google Scholar] [CrossRef]
- Modahl, C.M.; Doley, R.; Kini, R.M. Venom analysis of long-term captive Pakistan cobra (Naja naja) populations. Toxicon 2010, 55, 612–618. [Google Scholar] [CrossRef]
- Amazonas, D.R.; Freitas-De-Sousa, L.A.; Orefice, D.P.; de Sousa, L.F.; Martinez, M.G.; Mourão, R.H.V.; Chalkidis, H.M.; Camargo, P.B.; Moura-Da-Silva, A.M. Evidence for Snake Venom Plasticity in a Long-Term Study with Individual Captive Bothrops atrox. Toxins 2019, 11, 294. [Google Scholar] [CrossRef]
- De Farias, I.B.; de Morais-Zani, K.; Serino-Silva, C.; Sant’Anna, S.S.; da Rocha, M.M.; Grego, K.F.; Andrade-Silva, D.; Serrano, S.M.T.; Tanaka-Azevedo, A.M. Functional and proteomic comparison of Bothrops jararaca venom from captive specimens and the Brazilian Bothropic Reference Venom. J. Proteom. 2018, 174, 36–46. [Google Scholar] [CrossRef]
- Freitas-de-Sousa, L.A.; Amazonas, D.R.; Sousa, L.F.; Sant’Anna, S.S.; Nishiyama, M.Y., Jr.; Serrano, S.M.T.; Junqueira-de-Azevedo, I.L.M.; Chalkidis, H.M.; Moura-da-Silva, A.M.; Mourão, R.H.V. Comparison of venoms from wild and long-term captive Bothrops atrox snakes and characterization of Batroxrhagin, the predominant class PIII metalloproteinase from the venom of this species. Biochimie 2015, 118, 60–70. [Google Scholar] [CrossRef]
- Saad, E.; Curtolo Barros, L.; Biscola, N.; Pimenta, D.C.; Barraviera, S.R.C.S.; Barraviera, B.; Seabra Ferreira, R.S., Jr. Intraspecific variation of biological activities in venoms from wild and captive Bothrops jararaca. J. Toxicol. Environ. Health. Part A 2012, 75, 1081–1090. [Google Scholar] [CrossRef]
- Li, S.; Wang, J.; Zhang, X.; Ren, Y.; Wang, N.; Zhao, K.; Chen, X.; Zhao, C.; Li, X.; Shao, J.; et al. Proteomic characterization of two snake venoms: Naja naja atra and Agkistrodon halys. Biochem. J. 2004, 384, 119–127. [Google Scholar] [CrossRef]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.R.; Kerkkamp, H.M.E.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef] [PubMed]
- Kochva, E. The origin of snakes and evolution of the venom apparatus. Toxicon 1987, 25, 65–106. [Google Scholar] [CrossRef]
- Post, Y.; Puschhof, J.; Beumer, J.; Kerkkamp, H.M.; de Bakker, M.A.G.; Slagboom, J.; de Barbanson, B.; Wevers, N.R.; Spijkers, X.M.; Olivier, T.; et al. Snake Venom Gland Organoids. Cell 2020, 180, 233–247.e21. [Google Scholar] [CrossRef] [PubMed]
- Yamanouye, N.; Britto, L.R.; Carneiro, S.M.; Markus, R.P. Control of venom production and secretion by sympathetic outflow in the snake Bothrops jararaca. J. Exp. Biol. 1997, 200, 2547–2556. [Google Scholar] [CrossRef]
- Chey, W.Y. Regulation of pancreatic exocrine secretion. Int. J. Pancreatol. Off. J. Int. Assoc. Pancreatol. 1991, 9, 7–20. [Google Scholar] [CrossRef]
- Chandra, R.; Liddle, R.A. Modulation of pancreatic exocrine and endocrine secretion. Curr. Opin. Gastroenterol. 2013, 29, 517–522. [Google Scholar] [CrossRef]
- Beelman, C.A.; Parker, R. Degradation of mRNA in eukaryotes. Cell 1995, 81, 179–183. [Google Scholar] [CrossRef]
- Giorgianni, M.W.; Dowell, N.L.; Griffin, S.; Kassner, V.A.; Selegue, J.E.; Carroll, S.B. The origin and diversification of a novel protein family in venomous snakes. Proc. Natl. Acad. Sci. USA 2020, 117, 10911–10920. [Google Scholar] [CrossRef]
- Bayona-Serrano, J.D.; Viala, V.L.; Rautsaw, R.M.; Schramer, T.D.; Barros-Carvalho, G.A.; Nishiyama, M.Y.; Freitas-de-Sousa, L.A.; Moura-da-Silva, A.M.; Parkinson, C.L.; Grazziotin, F.G.; et al. Replacement and Parallel Simplification of Nonhomologous Proteinases Maintain Venom Phenotypes in Rear-Fanged Snakes. Mol. Biol. Evol. 2020, 37, 3563–3575. [Google Scholar] [CrossRef]
- Fry, B.G. From genome to “venome”: Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005, 15, 403–420. [Google Scholar] [CrossRef]
- Fry, B.G.; Wüster, W. Assembling an Arsenal: Origin and Evolution of the Snake Venom Proteome Inferred from Phylogenetic Analysis of Toxin Sequences. Mol. Biol. Evol. 2004, 21, 870–883. [Google Scholar] [CrossRef] [PubMed]
- Zelanis, A.; Tashima, A.K. Unraveling snake venom complexity with ‘omics’ approaches: Challenges and perspectives. Toxicon 2014, 87, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H.; Engmark, M.; Milbo, C.; Johannesen, J.; Lomonte, B.; Gutiérrez, J.M.; Lohse, B. From Fangs to Pharmacology: The Future of Snakebite Envenoming Therapy. Curr. Pharm. Des. 2016, 22, 5270–5293. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.S.; Guimarães, J.A.; Prado, J.L. Purification and characterization of a sulfhydryl-dependent protease from Rhodnius prolixus midgut. Arch. Biochem. Biophys. 1978, 188, 315–322. [Google Scholar] [CrossRef]
- Habermann, E.; Hardt, K.L. A sensitive and specific plate test for the quantitation of phospholipases. Anal. Biochem. 1972, 50, 163–173. [Google Scholar] [CrossRef]
- Kishimoto, M.; Takahashi, T. A spectrophotometric microplate assay for L-amino acid oxidase. Anal. Biochem. 2001, 298, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, C.; Huang, T.-F. Inhibition of platelet aggregation by 5′-nucleotidase purified from Trimeresurus gramineus snake venom. Toxicon 1983, 21, 491–501. [Google Scholar] [CrossRef]
- Ellman, G.L.; Courtney, K.D.; Andres, V., Jr.; Feather-Stone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]









| Protein Accessions | Description |
|---|---|
| V8N8G0 | Alkaline phosphatase |
| ENSNNAP00000025215.1* | Acetylcholinesterase |
| Q9DF56 | Acidic phospholipase A2 |
| P00598 | Acidic phospholipase A2 1 |
| Q9DF33 | Acidic phospholipase A2 2 |
| ENSNNAP00000015782.1* | Acidic phospholipase A2 2 |
| P60045 | Acidic phospholipase A2 3 |
| Q6T179 | Acidic phospholipase A2 4 |
| Q9I900 | Acidic phospholipase A2 D |
| P25498 | Acidic phospholipase A2 E |
| A4FS04 | Acidic phospholipase A2 natratoxin |
| P86542 | Phospholipase A2 3 |
| P00599 | Basic phospholipase A2 1 |
| P60043 | Basic phospholipase A2 1 |
| V8ND68 | Phospholipase B-like |
| Q7LZI1 | Phospholipase A2 inhibitor 31 kDa subunit |
| ENSNNAP00000008460.1* | PLIalpha-like protein |
| V8NQ76 | Atriopeptidase |
| V8N495 | Carboxypeptidase |
| ENSNNAP00000003269.1* | Carboxypeptidase D |
| I2C090 | Ophiophagus venom factor |
| Q91132 | Cobra venom factor |
| Q91132 | Cobra venom factor |
| ENSNNAP00000021869.1* | Cobra venom factor |
| ENSNNAP00000023282.1* | Cobra venom factor |
| ENSNNAP00000010931.1* | Complement C3 |
| ENSNNAP00000013063.1* | Complement C3 |
| Q01833 | Complement C3 |
| Q90WI6 | C-type lectin BML-1 |
| E3P6P4 | Cystatin |
| P86543 | Cysteine-rich venom protein |
| P84807 | Cysteine-rich venom protein 25-A |
| P84808 | Cysteine-rich venom protein kaouthin-2 |
| Q7T1K6 | Cysteine-rich venom protein natrin-1 |
| Q53B46 | Beta-cardiotoxin CTX15 |
| P60305 | Cytotoxin 1 |
| P86541 | Cytotoxin 10 |
| Q9W6W6 | Cytotoxin 10 |
| ENSNNAP00000013731.1* | Cytotoxin 11 |
| Q98956 | Cytotoxin 1b |
| P85429 | Cytotoxin 1f |
| P01442 | Cytotoxin 2 |
| Q9DGH9 | Cytotoxin 2 |
| P01445 | Cytotoxin 2 |
| P01463 | Cytotoxin 2 |
| O93472 | Cytotoxin 2c |
| P01452 | Cytotoxin 4 |
| O93473 | Cytotoxin 4a |
| O73856 | Cytotoxin 4b |
| P07525 | Cytotoxin 5 |
| P01457 | Cytotoxin 5 |
| P24779 | Cytotoxin 5 |
| P25517 | Cytotoxin 5 |
| P24779 | Cytotoxin 5 |
| Q98965 | Cytotoxin 6 |
| P01465 | Cytotoxin 6 |
| P49122 | Cytotoxin 7 |
| O73859 | Cytotoxin 7 |
| P86540 | Cytotoxin 8 |
| P60311 | Cytotoxin KJC3 |
| P60308 | Cytotoxin SP15c |
| P62377 | Cytotoxin-like basic protein |
| P18328 | Muscarinic toxin 2 |
| P82462 | Muscarinic toxin-like protein 1 |
| P82463 | Muscarinic toxin-like protein 2 |
| P82464 | Muscarinic toxin-like protein 3 |
| Q9PSN6 | Neurotoxin 3 |
| Q9DEQ3 | Neurotoxin homolog NL1 |
| Q9W717 | Neurotoxin-like protein NTL2 |
| P60774 | Short neurotoxin 1 |
| P01427 | Short neurotoxin 1 |
| P14613 | Short neurotoxin 1 |
| P01431 | Short neurotoxin 1 |
| P68418 | Short neurotoxin 1 |
| P01424 | Short neurotoxin 1 |
| P01432 | Short neurotoxin 3 |
| E2IU01 | Long neurotoxin 7 |
| E2ITZ9 | Cobrotoxin-b |
| P85092 | Toxin AdTx1 |
| P29180 | Weak neurotoxin 6 |
| O42256 | Weak neurotoxin 6 |
| Q2VBN2 | Weak neurotoxin WNTX34 |
| P01401 | Weak toxin CM-11 |
| P25679 | Weak toxin CM-9a |
| P85520 | Oxiana weak toxin |
| Q9YGI4 | Probable weak neurotoxin NNAM2 |
| P25669 | Long neurotoxin 2 |
| Q2VBP3 | Long neurotoxin LNTX37 |
| P01391 | Alpha-cobratoxin |
| ENSNNAP00000012975.1* | Cobrotoxin |
| P82849 | Cobrotoxin II |
| P59275 | Cobrotoxin-b |
| D9IX97 | Natriuretic peptide Na-NP |
| V8NF35 | Dipeptidyl peptidase 2 |
| V8P9G9 | Venom dipeptidylpeptidase IV |
| ENSNNAP00000025792.1* | Dipeptidyl peptidase 7 |
| V8P395 | Glutathione peroxidase |
| ENSNNAP00000008637.1* | Hyaluronidase-2-like isoform X1 |
| ENSNNAP00000024830.1* | Insulin like growth factor 1 |
| ENSNNAP00000006918.1* | Insulin like growth factor binding protein 3 |
| V8N7H9 | BPTI/Kunitz domain-containing protein-like isoform X2 |
| P20229 | Kunitz-type serine protease inhibitor |
| ENSNNAP00000023266.1* | Kunitz-type serine protease inhibitor 4-like |
| A8QL51 | L-amino acid oxidase |
| ENSNNAP00000002373.1* | L-amino-acid oxidase |
| A8QL58 | L-amino-acid oxidase |
| V8P5W4 | Putative serine carboxypeptidase CPVL |
| ENSNNAP00000011792.1* | Serpin family E member 2 |
| A0A2I4HXH5 | 5′-nucleotidase |
| V8NYW9 | 5′-nucleotidase |
| A8QL53 | Snake venom serine protease NaSP |
| P86545 | Thrombin-like enzyme TLP |
| V8NCP7 | Vascular endothelial growth factor C |
| P61899 | Venom nerve growth factor |
| Q5YF90 | Venom nerve growth factor 1 |
| A0A2D0TC04 | Venom phosphodiesterase |
| P83234 | Vespryn-21 |
| D3TTC2 | Zinc metalloproteinase-disintegrin-like atragin |
| ENSNNAP00000015130.1* | Zinc metalloproteinase-disintegrin-like atragin |
| D5LMJ3 | Zinc metalloproteinase-disintegrin-like atrase-A |
| ENSNNAP00000015250.1* | Zinc metalloproteinase-disintegrin-like atrase-A |
| ENSNNAP00000015377.1* | Zinc metalloproteinase-disintegrin-like atrase-A |
| D6PXE8 | Zinc metalloproteinase-disintegrin-like atrase-B |
| Q9PVK7 | Zinc metalloproteinase-disintegrin-like cobrin |
| A8QL59 | Zinc metalloproteinase-disintegrin-like NaMP |
| Q10749 | Snake venom metalloproteinase-disintegrin-like mocarhagin |
| P82942 | Hemorrhagic metalloproteinase-disintegrin-like kaouthiagin |
| % Proteome | Neonate Males | Neonate Females | Adult Males | Adult Females |
|---|---|---|---|---|
| PLB | 0.07% | 0.06% | 0.10% | 0.10% |
| HAase | 0.09% | 0.10% | 0.11% | 0.11% |
| DPP-IV | 0.14% | 0.14% | 0.18% | 0.14% |
| VEGF | 0.22% | 0.22% | 0.23% | 0.21% |
| PLI | 0.19% | 0.28% | 0.26% | 0.30% |
| CTL | 0.35% | 0.37% | 0.47% | 0.51% |
| LAAO | 0.38% | 0.32% | 0.24% | 0.27% |
| 5′-NT | 0.43% | 0.39% | 0.40% | 0.45% |
| QC | 0.49% | 0.50% | 0.40% | 0.39% |
| SVSP | 0.50% | 0.50% | 0.26% | 0.27% |
| Acidic PLA2 | 0.53% | 0.52% | 0.94% | 1.3% |
| Basic PLA2 | 0.02% | 0.02% | 0.07% | 0.1% |
| KUN | 0.95% | 0.70% | 0.71% | 0.73% |
| NP | 1.05% | 1.83% | 1.03% | 1.08% |
| VESP | 1.12% | 1.18% | 0.72% | 0.86% |
| AChE | 1.44% | 1.23% | 1.80% | 1.41% |
| NGF | 2.52% | 2.70% | 2.82% | 2.94% |
| PDE | 3.49% | 3.30% | 4.35% | 4.51% |
| CVF | 3.53% | 3.92% | 4.70% | 4.78% |
| SVMP(P-III) | 10.23% | 10.13% | 5.02% | 4.93% |
| CRISP | 11.39% | 9.37% | 7.08% | 6.65% |
| LNX(3-FTx) | 0.10% | 0.09% | 0.13% | 0.14% |
| SNX(3-FTx) | 15.67% | 15.73% | 11.00% | 13.65% |
| WNX(3-FTx) | 5.08% | 6.18% | 5.09% | 5.79% |
| CTX(3-FTx) | 40.00% | 40.22% | 51.77% | 48.63% |
| ID | Distribution of Glycosylation Sites (N-linked Asparagine) | ||
|---|---|---|---|
| Metalloprotease Structural Domains | Disintegrin Structural Domain | Cysteine-Rich Structural Domains | |
| P82942 | 304 | - | - |
| D6PXE8 | 320 | - | 509 |
| D3TTC2 | - | 438 | - |
| Q9PVK7 | - | 438 | - |
| Q10749 | 303 | - | 509 |
| D5LMJ3 | 221, 273, 304 | 438 | 527 |
| A8QL59 | 225, 268, 319 | - | 551 |
| Gene ID | Gene Chromosome | Gene Coding Protein | Abbreviation |
|---|---|---|---|
| ENSNNAG00000009518 | 3 | Alpha-elapitoxin-Nk2a | α-CbTx(3-FTx) |
| ENSNNAG00000009534 | 3 | Muscarinic toxin-like protein 2 | MTLP-2(3-FTx) |
| ENSNNAG00000011050 | 3 | Muscarinic toxin-like protein 3 homolog | MTLP-3(3-FTx) |
| ENSNNAG00000009404 | 3 | Cytotoxin 3a | CTX(3-FTx) |
| ENSNNAG00000009217 | 3 | Cytotoxin 4N | CTX(3-FTx) |
| ENSNNAG00000011032 | 3 | Cytotoxin 2 | CTX(3-FTx) |
| ENSNNAG00000011010 | 3 | Cytotoxin 5 | CTX(3-FTx) |
| ENSNNAG00000008699 | 3 | Probable weak neurotoxin NNAM1 | WNX(3-FTx) |
| ENSNNAG00000008719 | 3 | Tryptophan-containing weak neurotoxin | WNX(3-FTx) |
| ENSNNAG00000009048 | 3 | Neurotoxin-like protein NTL2 | NTL2(3-FTx) |
| ENSNNAG00000011613 | SOZL01001066.1 | Neurotoxin homolog NL1 | NL1(3-FTx) |
| ENSNNAG00000011709 | SOZL01001066.1 | Cardiotoxin 7 | CTX(3-FTx) |
| ENSNNAG00000008731 | 3 | Cobrotoxin | CBT(3-FTx) |
| ENSNNAG00000007474 | 1 | Snake venom 5′-nucleotidase | 5′ NT |
| ENSNNAG00000007520 | 1 | Snake venom 5′-nucleotidase | 5′ NT |
| novel.2091 | SOZL01000663.1 | 5′-nucleotidase | 5′ NT |
| ENSNNAG00000018750 | 2 | 5′-nucleotidase | 5′ NT |
| ENSNNAG00000016482 | SOZL01001525.1 | Acetylcholinesterase | AchE |
| ENSNNAG00000016482 | SOZL01001525.1 | Acetylcholinesterase | AchE |
| ENSNNAG00000015104 | MIC_6 | Aminopeptidase | AP |
| ENSNNAG00000008828 | 2 | Complement C3 | C3 |
| ENSNNAG00000008657 | 2 | Complement C3 | C3 |
| ENSNNAG00000012844 | SOZL01001814.1 | Cathelicidin-related peptide | CATH |
| ENSNNAG00000014282 | 1 | Cysteine-rich venom protein ophanin | CRISP |
| ENSNNAG00000014151 | 1 | Cysteine-rich venom protein ophanin | CRISP |
| ENSNNAG00000018595 | MIC_4 | Cysteine-rich secretory protein | CRISP |
| ENSNNAG00000000526 | 2 | C-type lectin | CTL |
| ENSNNAG00000018045 | 7 | C-type lectin lectoxin-Thr1 | CTL |
| ENSNNAG00000000430 | 2 | C-type lectin | CTL |
| ENSNNAG00000009410 | 1 | C-type lectin | CTL |
| ENSNNAG00000018053 | 7 | C-type lectin | CTL |
| ENSNNAG00000016651 | MIC_7 | C-type lectin | CTL |
| novel.1968 | MIC_9 | Cystatin (cysteine proteinase inhibitor) | CYS |
| ENSNNAG00000010061 | 2 | Cystatin-B | CYS |
| ENSNNAG00000005504 | MIC_9 | Cystatin | CYS |
| ENSNNAG00000012223 | 3 | Cystatin-2 | CYS |
| novel.1967 | MIC_9 | Cystatin | CYS |
| ENSNNAG00000006541 | 1 | Dipeptidyl peptidase IV | DPP-IV |
| ENSNNAG00000013472 | 2 | Glutathione peroxidase 3 | GPX |
| ENSNNAG00000013545 | 1 | Glutathione peroxidase 2 | GPX |
| ENSNNAG00000013509 | 2 | Glutathione peroxidase 6 | GPX |
| ENSNNAG00000008046 | 2 | Glutathione peroxidase 1 | GPX |
| ENSNNAG00000002583 | 2 | Hyaluronidase | HAase |
| ENSNNAG00000005815 | MIC_2 | Hyaluronidase | HAase |
| ENSNNAG00000018577 | MIC_3 | Kunitz-type serine protease inhibitor | KSPI |
| ENSNNAG00000018577 | MIC_3 | Kunitz-type protease inhibitor | KSPI |
| ENSNNAG00000015199 | 3 | Kunitz-type protease inhibitor | KSPI |
| ENSNNAG00000001233 | 1 | Kunitz-type protease inhibitor | KSPI |
| ENSNNAG00000001705 | 2 | L-amino-acid oxidase | LAAO |
| ENSNNAG00000001808 | 2 | L-amino-acid oxidase | LAAO |
| ENSNNAG00000001642 | 2 | L-amino-acid oxidase | LAAO |
| ENSNNAG00000001705 | 2 | L-amino-acid oxidase | LAAO |
| ENSNNAG00000013519 | MIC_2 | Nerve growth factor | NGF |
| ENSNNAG00000004808 | 3 | Venom nerve growth factor | NGF |
| ENSNNAG00000004800 | 3 | Venom nerve growth factor1 | NGF |
| ENSNNAG00000004808 | 3 | Venom nerve growth factor | NGF |
| ENSNNAG00000006727 | 4 | Natriuretic peptide Na-NP | NP |
| ENSNNAG00000006779 | 4 | Natriuretic peptide Na-NP | NP |
| ENSNNAG00000006809 | 4 | Natriuretic peptide Na-NP | NP |
| ENSNNAG00000006727 | 4 | Natriuretic peptide Na-NP | NP |
| ENSNNAG00000000097 | 1 | Venom phosphodiesterase 1 | PDE |
| ENSNNAG00000013762 | MIC_11 | Group IIE secretory phospholipase A2 | PLA2 |
| ENSNNAG00000003407 | 1 | Phospholipase A2 crotoxin basic subunit | PLA2 |
| ENSNNAG00000010611 | SOZL01000342.1 | Acidic phospholipase A2 | PLA2 |
| ENSNNAG00000003618 | 4 | Phospholipase A2 | PLA2 |
| ENSNNAG00000015696 | 7 | Phospholipase B | PLB |
| novel.1890 | MIC_8 | Phospholipase B | PLB |
| novel.1889 | MIC_8 | Phospholipase B | PLB |
| ENSNNAG00000005694 | 4 | PLIalpha-like protein | PLI |
| ENSNNAG00000003592 | MIC_3 | Phospholipase A2 inhibitor | PLI |
| ENSNNAG00000003672 | MIC_3 | Phospholipase A2 inhibitor | PLI |
| ENSNNAG00000013551 | 2 | Snake venom serine protease NaSP | SVSP |
| ENSNNAG00000015301 | MIC_3 | Snake venom serine protease | SVSP |
| ENSNNAG00000015396 | MIC_3 | Snake venom serine protease | SVSP |
| ENSNNAG00000017712 | 6 | Venom prothrombin activator | VPA |
| ENSNNAG00000017242 | 1 | Vascular endothelial growth factor A | VEGF |
| ENSNNAG00000011309 | MIC_5 | Vascular endothelial growth factor | VEGF |
| ENSNNAG00000008620 | MIC_9 | Vascular endothelial growth factor A | VEGF |
| ENSNNAG00000004103 | MIC_2 | Cysteine-rich with EGF-like domain | CREGF |
| ENSNNAG00000001343 | 2 | Cysteine-rich with EGF-like domain protein | CREGF |
| ENSNNAG00000007968 | 4 | Vascular endothelial growth factor C | VEGF |
| ENSNNAG00000017242 | 1 | Vascular endothelial growth factor A | VEGF |
| ENSNNAG00000013375 | SOZL01001403.1 | Cobra venom factor | CVF |
| ENSNNAG00000008178 | 2 | Cobra venom factor | CVF |
| ENSNNAG00000013375 | SOZL01001403.1 | Cobra venom factor | CVF |
| ENSNNAG00000008178 | 2 | Cobra venom factor | CVF |
| novel.1103 | 3 | Snake toxin and toxin-like protein | STLK |
| novel.1007 | 3 | Snake toxin and toxin-like protein | STLK |
| novel.1104 | 3 | Snake toxin and toxin-like protein | STLK |
| novel.2094 | SOZL01000688.1 | Snake toxin and toxin-like protein | STLK |
| ENSNNAG00000009894 | MIC_1 | Zinc metalloproteinase-disintegrin-like NaMP | SVMP |
| ENSNNAG00000017863 | MIC_8 | Disintegrin and metalloproteinase domain-containing protein 9 | SVMP |
| ENSNNAG00000013917 | 2 | A disintegrin and metalloproteinase with thrombospondin motifs 12 | SVMP |
| ENSNNAG00000011023 | Z | Disintegrin and metalloproteinase domain-containing protein 11 | SVMP |
| ENSNNAG00000010003 | MIC_1 | Zinc metalloproteinase-disintegrin-like atragin | SVMP |
| ENSNNAG00000008597 | MIC_6 | A disintegrin and metalloproteinase with thrombospondin motifs 17 | SVMP |
| ENSNNAG00000005308 | MIC_1 | Disintegrin and metalloproteinase domain-containing protein 9 | SVMP |
| novel.1209 | 4 | Zinc metalloprotease | SVMP |
| FPKM% | Neonate Males | Neonate Females | Adult Males | Adult Females |
|---|---|---|---|---|
| PLI | 0.001% | 0.001% | 0.001% | 0.004% |
| C3 | 0.005% | 0.002% | 0.007% | 0.008% |
| CREGF | 0.015% | 0.012% | 0.006% | 0.053% |
| AP | 0.015% | 0.013% | 0.007% | 0.062% |
| DPP-IV | 0.017% | 0.007% | 0.014% | 0.022% |
| VEGF | 0.027% | 0.015% | 0.013% | 0.059% |
| PLB | 0.039% | 0.037% | 0.053% | 0.083% |
| HAase | 0.057% | 0.031% | 0.067% | 0.067% |
| VPA | 0.069% | 0.025% | 0.010% | 0.063% |
| CATH | 0.072% | 0.006% | 0.011% | 0.022% |
| AChE | 0.091% | 0.072% | 0.049% | 0.167% |
| CYS | 0.107% | 0.081% | 0.064% | 0.172% |
| PDE | 0.112% | 0.059% | 0.134% | 0.136% |
| SVSP | 0.168% | 0.124% | 0.145% | 0.210% |
| LAAO | 0.213% | 0.121% | 0.055% | 0.207% |
| KUN | 0.221% | 0.151% | 0.203% | 0.221% |
| CVF | 0.223% | 0.096% | 0.213% | 0.179% |
| CTL | 0.253% | 0.212% | 0.177% | 0.456% |
| 5′-NT | 0.347% | 0.211% | 0.154% | 0.400% |
| CRISP | 0.615% | 0.346% | 0.135% | 0.497% |
| NGF | 0.972% | 0.887% | 1.386% | 1.450% |
| SVMP | 0.985% | 0.515% | 0.078% | 0.653% |
| Basic PLA2 | 1.305% | 1.244% | 3.405% | 1.829% |
| Acidic PLA2 | 0.001% | 0.001% | 0.001% | 0.001% |
| STLK | 1.673% | 2.182% | 1.853% | 3.056% |
| GPX | 2.352% | 1.521% | 1.386% | 2.593% |
| NP | 4.738% | 3.806% | 2.556% | 6.932% |
| LNX(3-FTx) | 0.751% | 2.630% | 1.918% | 7.968% |
| CTX(3-FTx) | 37.825% | 38.359% | 26.955% | 29.696% |
| WNX(3-FTx) | 12.815% | 9.078% | 3.417% | 3.429% |
| SNX(3-FTx) | 33.919% | 38.158% | 55.530% | 39.307% |
| Protein Accessions | Gene | Protein Name |
|---|---|---|
| A0A2D0TC04 | ENSNNAG00000000097 | Venom phosphodiesterase |
| A8QL53 | ENSNNAG 00000013551 | Snake venom serine protease NaSP |
| ENSNNAG00000015396 | Snake venom serine protease NaSP | |
| A8QL58 | ENSNNAG00000001808 | L-amino-acid oxidase |
| A8QL59 | ENSNNAG00000009894 | Zinc metalloproteinase-disintegrin -like NaMP |
| ENSNNAG00000008597 | Zinc metalloproteinase-disintegrin -like NaMP | |
| D5LMJ3 | ENSNNAG00000005308 | Zinc metalloproteinase-disintegrin -like atrase-A |
| ENSNNAG00000013917 | Zinc metalloproteinase-disintegrin -like atrase-A | |
| D9IX97 | ENSNNAG00000006809 | Natriuretic peptide Na-NP |
| ENSNNAG00000006727 | Natriuretic peptide Na-NP | |
| ENSNNAG00000006779 | Natriuretic peptide Na-NP | |
| E3P6P4 | ENSNNAG00000005504 | Cystatin |
| ENSNNAP00000002373.1 | ENSNNAG00000001642 | L-amino-acid oxidase (Fragment chain B) |
| ENSNNAG00000001705 | L-amino-acid oxidase | |
| ENSNNAP00000008460.1 | ENSNNAG00000005694 | PLI alpha-like protein |
| ENSNNAP00000008637.1 | ENSNNAG00000005815 | Hyaluronidase-2-like isoform X1 |
| ENSNNAG00000002583 | Hyaluronidase-2-like isoform X1 | |
| ENSNNAP00000009791.1 | ENSNNAG00000006541 | Venom dipeptidylpeptidase IV |
| ENSNNAP00000011851.1 | ENSNNAG00000008620 | Vascular endothelial growth factor C |
| ENSNNAG00000007968 | Vascular endothelial growth factor C | |
| ENSNNAG00000017242 | Vascular endothelial growth factor C | |
| ENSNNAP00000012520.1 | ENSNNAG00000008178 | Cobra venom factor |
| ENSNNAP00000012975.1 | ENSNNAG00000008731 | Cobrotoxin |
| ENSNNAP00000013063.1 | ENSNNAG00000008657 | Complement C3 |
| ENSNNAP00000014120.1 | ENSNNAG00000009518 | Alpha-cobratoxin |
| ENSNNAP00000015130.1 | ENSNNAG00000010003 | Zinc metalloproteinase-disintegrin -like atragin |
| ENSNNAP00000021869.1 | ENSNNAG00000013375 | Cobra venom factor |
| ENSNNAP00000023044.1 | ENSNNAG00000001233 | BPTI/Kunitz domain-containing protein-like isoform X2 |
| ENSNNAG00000015199 | BPTI/Kunitz domain-containing protein-like isoform X2 | |
| ENSNNAP00000025215.1 | ENSNNAG00000016482 | Acetylcholinesterase |
| P00598 | ENSNNAG00000010611 | Acidic phospholipase A2 1 |
| P01442 | ENSNNAG00000009217 | Cytotoxin 2 |
| ENSNNAG00000011010 | Cytotoxin 2 | |
| P01445 | ENSNNAG00000011032 | Cytotoxin 2 |
| P29180 | ENSNNAG00000008699 | Weak neurotoxin 6 |
| P61899 | ENSNNAG00000004808 | Venom nerve growth factor |
| P62377 | ENSNNAG00000011709 | Cytotoxin-like basic protein |
| P86540 | ENSNNAG00000009404 | Cytotoxin 8 |
| Q01833 | ENSNNAG00000008828 | Complement C3 |
| Q10749 | ENSNNAG00000017863 | Snake venom metalloproteinase -disintegrin-like mocarhagin |
| ENSNNAG00000011023 | Snake venom metalloproteinase -disintegrin-like mocarhagin | |
| Q5YF90 | ENSNNAG00000004800 | Venom nerve growth factor 1 |
| ENSNNAG00000013519 | Venom nerve growth factor 1 | |
| Q6T179 | ENSNNAG00000013762 | Acidic phospholipase A2 4 |
| ENSNNAG00000003407 | Acidic phospholipase A2 4 | |
| Q7LZI1 | ENSNNAG00000003592 | Phospholipase A2 inhibitor 31 kDa subunit |
| Q7T1K6 | ENSNNAG00000014151 | Cysteine-rich venom protein natrin-1 |
| Q9DEQ3 | ENSNNAG00000011613 | Neurotoxin homolog NL1 |
| ENSNNAG00000011050 | Neurotoxin homolog NL1 | |
| Q9W717 | ENSNNAG00000009048 | Neurotoxin-like protein NTL2 |
| Q9YGI4 | ENSNNAG00000008719 | Probable weak neurotoxin NNAM2 |
| V8P395 | ENSNNAG00000013472 | Glutathione peroxidase |
| ENSNNAG00000008046 | Glutathione peroxidase | |
| ENSNNAG00000013545 | Glutathione peroxidase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, X.; Chen, Q.; Wang, C.; Huang, W.; Lai, R.; Lu, Q.; He, Q.; Yu, X. Venom Variation of Neonate and Adult Chinese Cobras in Captivity Concerning Their Foraging Strategies. Toxins 2022, 14, 598. https://doi.org/10.3390/toxins14090598
Nie X, Chen Q, Wang C, Huang W, Lai R, Lu Q, He Q, Yu X. Venom Variation of Neonate and Adult Chinese Cobras in Captivity Concerning Their Foraging Strategies. Toxins. 2022; 14(9):598. https://doi.org/10.3390/toxins14090598
Chicago/Turabian StyleNie, Xuekui, Qianzi Chen, Chen Wang, Wangxiang Huang, Ren Lai, Qiumin Lu, Qiyi He, and Xiaodong Yu. 2022. "Venom Variation of Neonate and Adult Chinese Cobras in Captivity Concerning Their Foraging Strategies" Toxins 14, no. 9: 598. https://doi.org/10.3390/toxins14090598





