An Exploration of Novel Bioactives from the Venomous Marine Annelid Glycera alba
Abstract
:1. Introduction
2. Results
2.1. Comparative Proteomics between Proboscis and Skin
2.1.1. Gel Electrophoresis of Crude and Fractionated Protein Extracts
2.1.2. Proteomics
2.2. Toxicity Testing
2.2.1. Histopathological Analysis of Mussel Gills
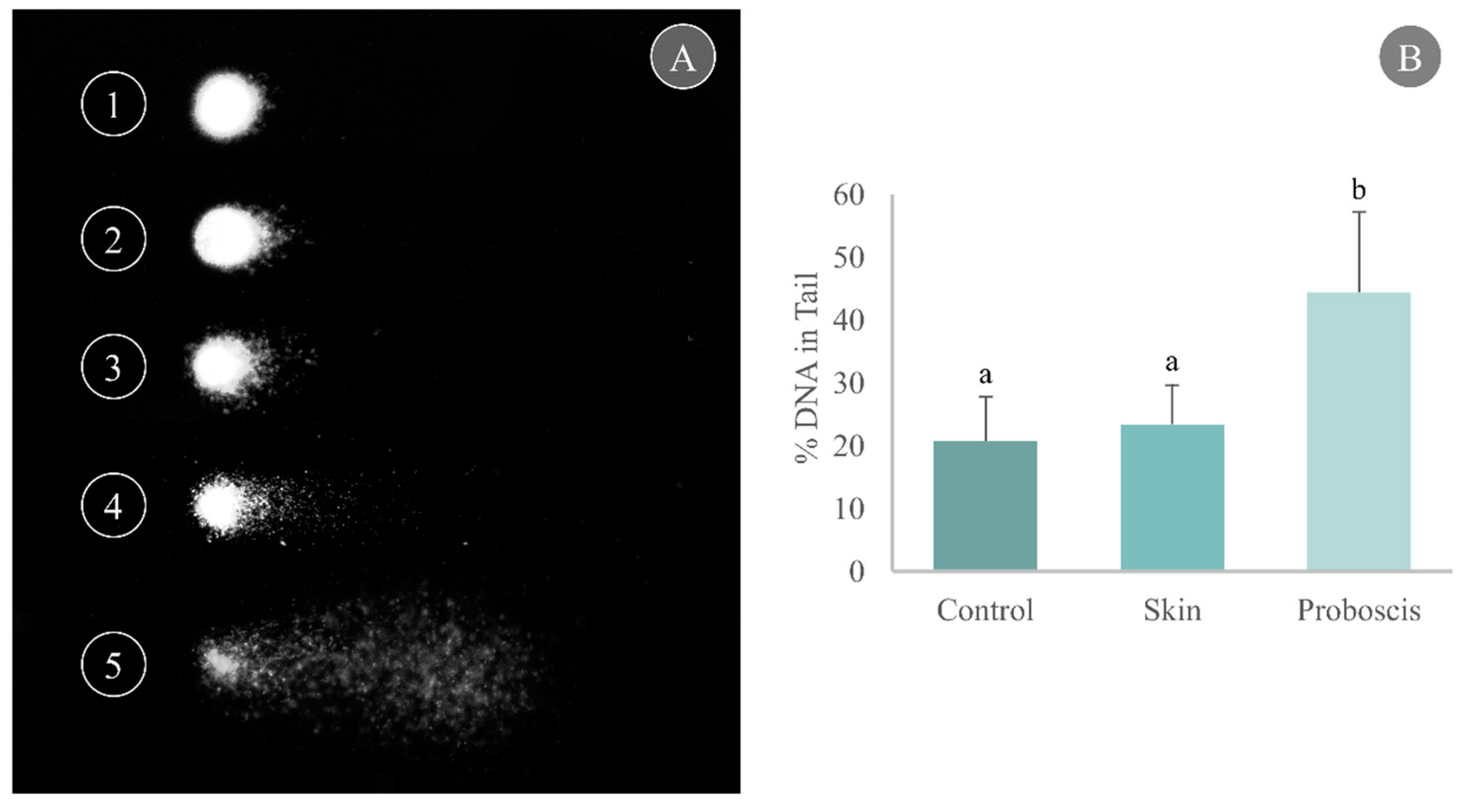
2.2.2. DNA Damage in Mussel Gills
2.2.3. Caspase-3 Activity as an Indicator of Apoptosis
3. Discussion
4. Materials and Methods
4.1. Animal Collection
4.2. Protein Extraction
4.2.1. Protein Electrophoresis
4.2.2. Proteomics
4.2.3. Protein Fractioning
4.3. Toxicity Testing
4.3.1. Histopathology
4.3.2. Comet Assay
4.3.3. Caspase-3 Activity
4.4. Statistical Analysis
4.4.1. Proteome Annotation and Search for Glycerotoxin (GLTx) Homologs
4.4.2. Toxicity Testing
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Why do we study animal toxins? Zool. Res. 2015, 36, 183–222. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Costa, P.M. The hidden biotechnological potential of marine invertebrates: The Polychaeta case study. Environ. Res. 2019, 173, 270–280. [Google Scholar] [CrossRef]
- Clark, G.C.; Casewell, N.R.; Elliott, C.T.; Harvey, A.L.; Jamieson, A.G.; Strong, P.N.; Turner, A.D. Friends or foes? Emerging impacts of biological toxins. Trends Biochem. Sci. 2019, 44, 365–379. [Google Scholar] [CrossRef]
- Burgess, J.G. New and emerging analytical techniques for marine biotechnology. Curr. Opin. Biotechnol. 2012, 23, 29–33. [Google Scholar] [CrossRef]
- Barbosa, A.J.M.; Roque, A.C.A. Free Marine Natural Products Databases for Biotechnology and Bioengineering. Biotechnol. J. 2019, 14, e1800607. [Google Scholar] [CrossRef]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef]
- Göransson, U.; Jacobsson, E.; Strand, M.; Andersson, H.S. The toxins of nemertean worms. Toxins 2019, 11, 120. [Google Scholar] [CrossRef]
- Kem, W.R. Alzheimer’s drug design based upon an invertebrate toxin (anabaseine) which is a potent nicotinic receptor agonist. Invertebr. Neurosci. 1997, 3, 251–259. [Google Scholar] [CrossRef]
- Iwanaga, S.; Lee, B.L. Recent advances in the innate immunity of invertebrate animals. BMB Rep. 2005, 38, 128–150. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, M.C.L.; Teixeira, V.L.; Santos, C.S.G. A Review of “Polychaeta” Chemicals and their Possible Ecological Role. J. Chem. Ecol. 2018, 44, 72–94. [Google Scholar] [CrossRef] [PubMed]
- Schmidtko, A.; Lötsch, J.; Freynhagen, R.; Geisslinger, G. Ziconotide for treatment of severe chronic pain. Lancet 2010, 375, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.C.; Madeira, C.; Brandão, C.A.; Puga, J.; Calado, R. Bioprospecting of marine invertebrates for new natural products—A chemical and zoogeographical perspective. Molecules 2012, 17, 9842–9854. [Google Scholar] [CrossRef] [PubMed]
- Flint, R.; Rabalais, N. Polychaete Ecology and Niche Patterns: Texas Continental Shelf. Mar. Ecol. Prog. Ser. 1980, 3, 193–202. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Lopes, A.; Pereira, R.; Anjo, S.I.; Manadas, B.; Grosso, A.R.; Baptista, P.V.; Fernandes, A.R.; Costa, P.M. Endogenous Fluorescent Proteins in the Mucus of an Intertidal Polychaeta: Clues for Biotechnology. Mar. Drugs 2022, 20, 224. [Google Scholar] [CrossRef]
- Deheyn, D.D.; Latz, M.I. Internal and secreted bioluminescence of the marine polychaete Odontosyllis phosphorea (Syllidae). Invertebr. Biol. 2009, 128, 31–45. [Google Scholar] [CrossRef]
- von Reumont, B.M.; Campbell, L.I.; Richter, S.; Hering, L.; Sykes, D.; Hetmank, J.; Jenner, R.A.; Bleidorn, C. A polychaete’s powerful punch: Venom gland transcriptomics of Glycera reveals a complex cocktail of toxin homologs. Genome Biol. Evol. 2014, 6, 2406–2423. [Google Scholar] [CrossRef]
- Gibbs, P.E.; Bryan, G.W. Copper-the major metal component of glycerid polychaete jaws. J. Mar. Biol. Assoc. 1980, 60, 205–214. [Google Scholar] [CrossRef]
- Bon, C.; Saliou, B.; Thieffry, M.; Manaranche, R. Partial purification of α-glycerotoxin, a presynaptic neurotoxin from the venom glands of the polychaete annelid Glycera convoluta. Neurochem. Int. 1985, 7, 63–75. [Google Scholar] [CrossRef]
- Manaranche, R.; Thieffry, M.; Israel, M. Effect of the venom of Glycera convoluta on the spontaneous quantal release of transmitter. J. Cell Biol. 1980, 85, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.; Helm, C.; Meunier, F.A.; Hering, L.; Campbell, L.I.; Drukewitz, S.H.; Undheim, E.A.B.; Jenner, R.A.; Schiavo, G.; Bleidorn, C. Comparative analyses of glycerotoxin expression unveil a novel structural organization of the bloodworm venom system. BMC Evol. Biol. 2017, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Schenning, M.; Proctor, D.T.; Ragnarsson, L.; Barbier, J.; Lavidis, N.A.; Molgó, J.J.; Zamponi, G.W.; Schiavo, G.; Meunier, F.A. Glycerotoxin stimulates neurotransmitter release from N-type Ca2+ channel expressing neurons. J. Neurochem. 2006, 98, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, C.; Costa, P.M. Histochemical detection of free thiols in glandular cells and tissues of different marine Polychaeta. Histochem. Cell Biol. 2020, 154, 315–325. [Google Scholar] [CrossRef]
- D’Ambrosio, M.; Ramos, Í.; Martins, C.; Costa, P.M. An investigation into the toxicity of tissue extracts from two distinct marine Polychaeta. Toxicon X 2022, 14, 100116. [Google Scholar] [CrossRef]
- Moutinho Cabral, I.; Costa, P.M.; Madeira, C.; Grosso, A.R. A drug discovery approach based on comparative transcriptomics between two toxin-secreting marine annelids: Glycera alba and Hediste diversicolor. Mol. Omics 2022, 18, 731–744. [Google Scholar] [CrossRef]
- Dales, R.P. The polychaete stomodeum and the inter-relationships of the families of polychaeta. Proc. Zool. Soc. 2009, 139, 389–428. [Google Scholar] [CrossRef]
- Steele, J.A.; Uchytil, T.F.; Durbin, R.D. Chloroplast coupling factor 1: A species specific receptor for tentoxin. Proc. Natl. Acad. Sci. USA 1976, 73, 2245–2248. [Google Scholar] [CrossRef]
- Lax, A.; Shepherd, H.; Edwards, J. Tentoxin, a chlorosis-inducing toxin from Alternaria as a potential herbicide. Weed Technol. 1988, 2, 540–544. [Google Scholar] [CrossRef]
- Nobile, M.; Noceti, F.; Prestipino, G.; Possani, L.D. Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp. Brain Res. 1996, 110, 15–20. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Koike, H.; Sugiyama, Y.; Motoyoshi, K.; Wada, T.; Hishinuma, S.; Mita, M.; Morita, T. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 2002, 269, 2708–2715. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Connor, M.; Wilson, D.; Wilson, H.I.; Nicholson, G.M.; Smith, R.; Shaw, D.; Mackay, J.P.; Alewood, P.F.; Christie, M.J.; et al. Discovery and Structure of a Potent and Highly Specific Blocker of Insect Calcium Channels. J. Biol. Chem. 2001, 276, 40306–40312. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Smith, R.; Fletcher, J.I.; Wilson, H.; Wood, C.J.; Howden, M.E.; King, G.F. Structure-function studies of omega atracotoxin, a potent antagonist of insect voltage-gated calcium channels. Eur. J. Biochem. 1999, 264, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, A.P.; Grosso, A.R.; Baptista, P.V.; Fernandes, A.R.; Costa, P.M. A transcriptomic approach to the recruitment of venom proteins in a marine Polychaeta. Toxins 2021, 13, 97. [Google Scholar] [CrossRef]
- Verdes, A.; Simpson, D.; Holford, M. Are fireworms venomous? Evidence for the convergent evolution of toxin homologs in three species of fireworms (Annelida, Amphinomidae). Genome Biol. Evol. 2018, 10, 249–268. [Google Scholar] [CrossRef]
- Howes, J.M.; Wilkinson, M.C.; Theakston, R.D.G.; Laing, G.D. The purification and partial characterisation of two novel metalloproteinases from the venom of the West African carpet viper, Echis ocellatus. Toxicon 2003, 42, 21–27. [Google Scholar] [CrossRef]
- Koua, D.; Kuhn-Nentwig, L. Spider neurotoxins, short linear cationic peptides and venom protein classification improved by an automated competition between exhaustive profile HMM classifiers. Toxins 2017, 9, 245. [Google Scholar] [CrossRef]
- Tambourgi, D.V.; Bio, F.; Magnoli, C.; Van Den Berg, C.W.; Morgan, B.P.; De Araujo, P.S.; Alves, E.W.; Dias, W.; Silva, D. Sphingomyelinases in the venom of the spider Loxosceles intermedia are responsible for both dermonecrosis and complement-dependent hemolysis. Biochem. Biophys. Res. Commun. 1998, 251, 366–373. [Google Scholar] [CrossRef]
- D’Ambrosio, M.; Gonçalves, C.; Calmão, M.; Rodrigues, M.; Costa, P.M. Localization and bioreactivity of cysteine-rich secretions in the marine gastropod Nucella lapillus. Mar. Drugs 2021, 19, 276. [Google Scholar] [CrossRef]
- Heyborne, W.H.; Mackessy, S.P.; Introduction, I. Cysteine-rich secretory proteins in reptile venoms. In Handbook of Venoms and Toxins of Reptiles; CRC Press: Boca Raton, FL, USA, 2010; pp. 321–332. [Google Scholar]
- Costa, P.M. The Handbook of Histopathological Practices in Aquatic Environments; Academic Press: Cambridge, UK, 2018; ISBN 9780128120323. [Google Scholar]
- Inoue, S.; Browne, G.; Melino, G.; Cohen, G.M. Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death Differ. 2009, 16, 1053–1061. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Mendes, V.M.; Manadas, B.; Grosso, A.R.; de Matos, A.P.A.; Baptista, P.V.; Costa, P.M.; Fernandes, A.R. Specific antiproliferative properties of proteinaceous toxin secretions from the marine annelid Eulalia sp. onto ovarian cancer cells. Mar. Drugs 2021, 19, 31. [Google Scholar] [CrossRef]
- Vogeler, S.; Carboni, S.; Li, X.; Joyce, A. Phylogenetic analysis of the caspase family in bivalves: Implications for programmed cell death, immune response and development. BMC Genom. 2021, 22, 80. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Hames, B.D. Gel Electrophoresis of Proteins: A Practical Approach, 3rd ed.; Oxford University Press: New York, NY, USA, 1998. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2008, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.; Costa, P.M. Technical Updates to the Comet Assay in vivo for Assessing DNA Damage in Zebrafish Embryos from Fresh and Frozen Cell Suspensions. Zebrafish 2020, 17, 220–228. [Google Scholar] [CrossRef]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef]
- Lee, R.F.; Steinert, S. Use of the single cell gel electrophoresis/comet assay for detecting DNA damage in aquatic (marine and freshwater) animals. Mutat. Res.-Rev. Mutat. Res. 2003, 544, 43–64. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Soudy, M.; Anwar, A.M.; Ahmed, E.A.; Osama, A.; Ezzeldin, S.; Mahgoub, S.; Magdeldin, S. UniprotR: Retrieving and visualizing protein sequence and functional information from Universal Protein Resource (UniProt knowledgebase). J. Proteom. 2020, 213, 103613. [Google Scholar] [CrossRef] [PubMed]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- Nagai, H. Marine protein toxins. In Handbook of Marine Natural Products; Fattorusso, E., Gerwick, W., Taglialatela-Scafati, O., Eds.; Springer: Dordrecht, Switzerland, 2012; pp. 1389–1420. [Google Scholar] [CrossRef]
- Liscovitch, M. Receptor-regulated phospholipases. Encycl. Endocr. Dis. 2004, 4, 167–173. [Google Scholar]
- Rangaraju, S.; Khoo, K.K.; Feng, Z.P.; Crossley, G.; Nugent, D.; Khaytin, I.; Chi, V.; Pham, C.; Calabresi, P.; Pennington, M.W.; et al. Potassium channel modulation by a toxin domain in matrix metalloprotease 23. J. Biol. Chem. 2010, 285, 9124–9136. [Google Scholar] [CrossRef]
- Chew, J.S.C.; Saleem, M.; Florkowski, C.M.; George, P.M. Cystatin C–A Paradigm of evidence based laboratory medicine. Clin. Biochem. Rev. 2008, 29, 47–62. [Google Scholar]
- Durani, V.; Magliery, T.J. Protein engineering and stabilization from sequence statistics: Variation and covariation analysis. Meth. Enzymol. 2013, 523, 237–256. [Google Scholar] [CrossRef]
- Singer, E.; Markó, L.; Paragas, N.; Barasch, J.; Dragun, D.; Müller, D.N.; Budde, K.; Schmidt-Ott, K.M. Neutrophil gelatinase-associated lipocalin: Pathophysiology and clinical applications. Acta Physiol. 2013, 207, 663–672. [Google Scholar] [CrossRef]
- Jiang, R.; Zhang, B.; Kurokawa, K.; So, Y.; Kim, E.; Hwang, H.O.; Lee, J.; Shiratsuchi, A.; Zhang, J.; Nakanishi, Y.; et al. 93-kDa Twin-domain serine protease inhibitor (serpin) has a regulatory function on the beetle toll proteolytic signaling cascade. J. Biol. Chem. 2011, 286, 35087–35095. [Google Scholar] [CrossRef]
- Darwiche, R.; Kelleher, A.; Hudspeth, E.M.; Schneiter, R.; Asojo, O.A. Structural and functional characterization of the CAP domain of pathogen-related yeast 1 (Pry1) protein. Sci. Rep. 2006, 6, 28838. [Google Scholar] [CrossRef]
- Lopes-Ferreira, M.; Magalhães, G.S.; Fernandez, J.H.; Junqueira-de-Azevedo, I.D.L.; Le Ho, P.; Lima, C.; Valente, R.H.; Moura-da-Silva, A.M. Structural and biological characterization of Nattectin, a new C-type lectin from the venomous fish Thalassophryne nattereri. Biochimie 2011, 93, 971–980. [Google Scholar] [CrossRef]
- Hamid, R.; Khan, M.A.; Ahmad, M.; Ahmad, M.M.; Abdin, M.Z.; Musarrat, J.; Javed, S. Chitinases: An update. J. Pharm. Bioallied Sci. 2013, 5, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Barth, T.; Coelho Mandacaru, S.; Charneau, S.; Valle de Souza, M.; Ornelas Ricart, C.A.; Ferreira Noronha, E.; Araújo Souza, A.; de Freitas, S.M.; Roespstorff, P.; Fontes, W.; et al. Biochemical and structural characterization of a protein complex containing a hyaluronidase and a CRISP-like protein isolated from the venom of the spider Acanthoscurria natalensis. J. Proteom. 2019, 192, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.R.; Ding, M.H.; Zhang, L.W.; Zhang, W.G.; Liu, L.; Li, D. Expression of a bee venom phospholipase A2 from Apis cerana cerana in the baculovirus-insect cell. J. Zhejiang Univ. Sci. B 2010, 11, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Murayama, N.; Saguchi, K.; Mentele, R.; Assakura, M.T.; Ohi, H.; Fujita, Y.; Camargo, A.C.; Higuchi, S.; Serrano, S.M. The unusual high molecular mass of Bothrops protease A, a trypsin-like serine peptidase from the venom of Bothrops jararaca, is due to its high carbohydrate content. Biochim. Biophys. Acta Proteins Proteom. 2003, 1652, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.R.; Takeuchi, M.; Kobayashi, Y. Purification and characterization of serine carboxypeptidases from Absidia zychae. Biosci. Biotech. Biochem. 1993, 57, 618–622. [Google Scholar] [CrossRef]
- Price, D.R.G.; Bell, H.A.; Hinchliffe, G.; Fitches, E.; Weaver, R.; Gatehouse, J.A. A venom metalloproteinase from the parasitic wasp Eulophus pennicornis is toxic towards its host, tomato moth (Lacanobia oleracae). Insect. Mol. Biol. 2009, 18, 195–202. [Google Scholar] [CrossRef]

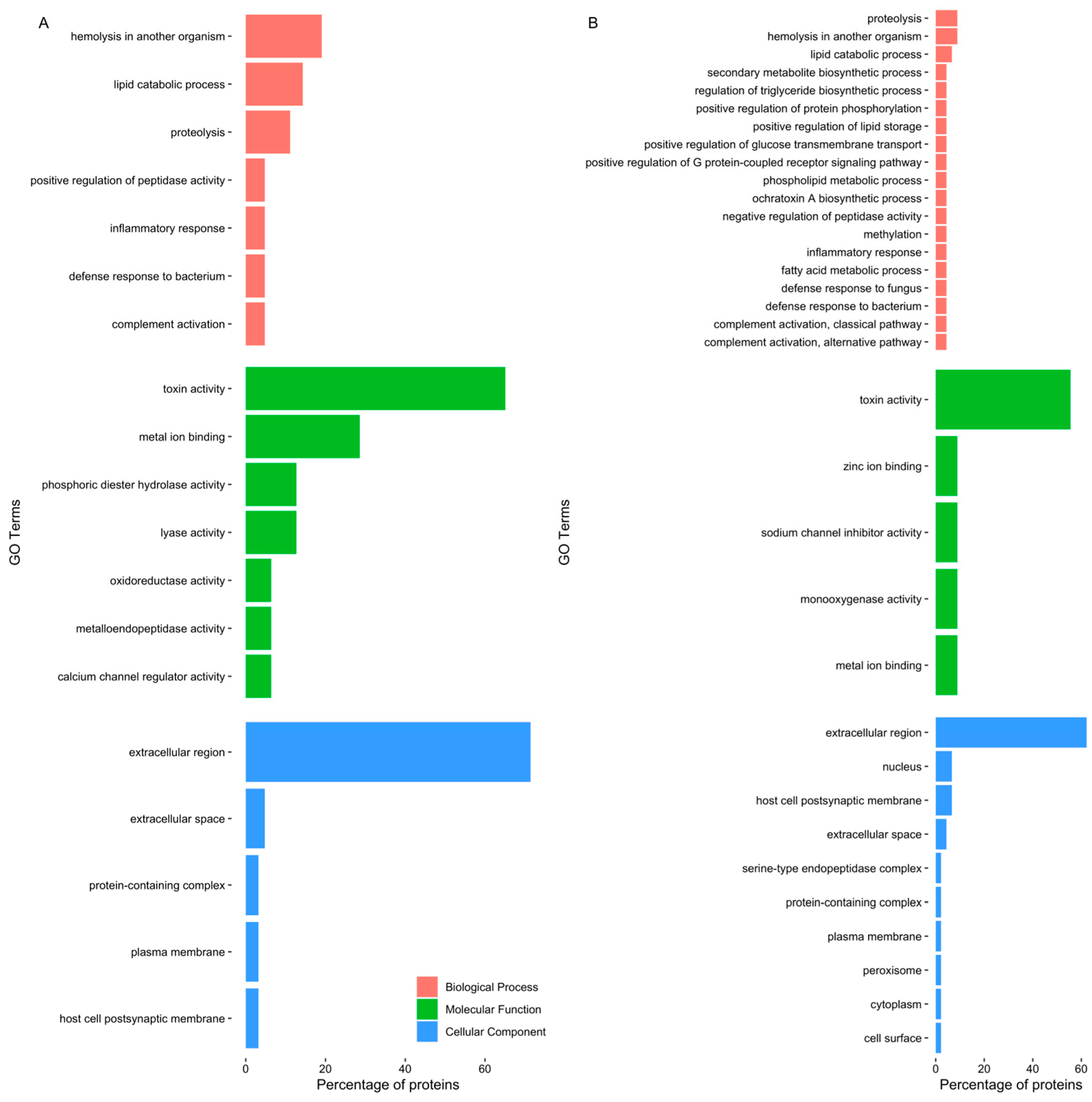


| Protein Match | Accession | % Cov (95) | Number of Matched Peptides | Organ |
|---|---|---|---|---|
| Glycerotoxin paralog 1 (G. tridactyla) | A0A1U9VX98 | 28.87–29.37 | 29 | Proboscis |
| Glycerotoxin paralog 1 (G. tridactyla) | A0A1U9VX95 | 27.02–28.86 | 2–3 | Proboscis |
| Glycerotoxin paralog 1 (G. tridactyla) | A0A1U9VX95 | 0.92 | 1 | Skin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos, S.; Rodrigo, A.P.; Moutinho Cabral, I.; Mendes, V.M.; Manadas, B.; D’Ambrosio, M.; Costa, P.M. An Exploration of Novel Bioactives from the Venomous Marine Annelid Glycera alba. Toxins 2023, 15, 655. https://doi.org/10.3390/toxins15110655
Campos S, Rodrigo AP, Moutinho Cabral I, Mendes VM, Manadas B, D’Ambrosio M, Costa PM. An Exploration of Novel Bioactives from the Venomous Marine Annelid Glycera alba. Toxins. 2023; 15(11):655. https://doi.org/10.3390/toxins15110655
Chicago/Turabian StyleCampos, Sónia, Ana P. Rodrigo, Inês Moutinho Cabral, Vera M. Mendes, Bruno Manadas, Mariaelena D’Ambrosio, and Pedro M. Costa. 2023. "An Exploration of Novel Bioactives from the Venomous Marine Annelid Glycera alba" Toxins 15, no. 11: 655. https://doi.org/10.3390/toxins15110655






