Type II Toxin–Antitoxin Systems in Pseudomonas aeruginosa
Abstract
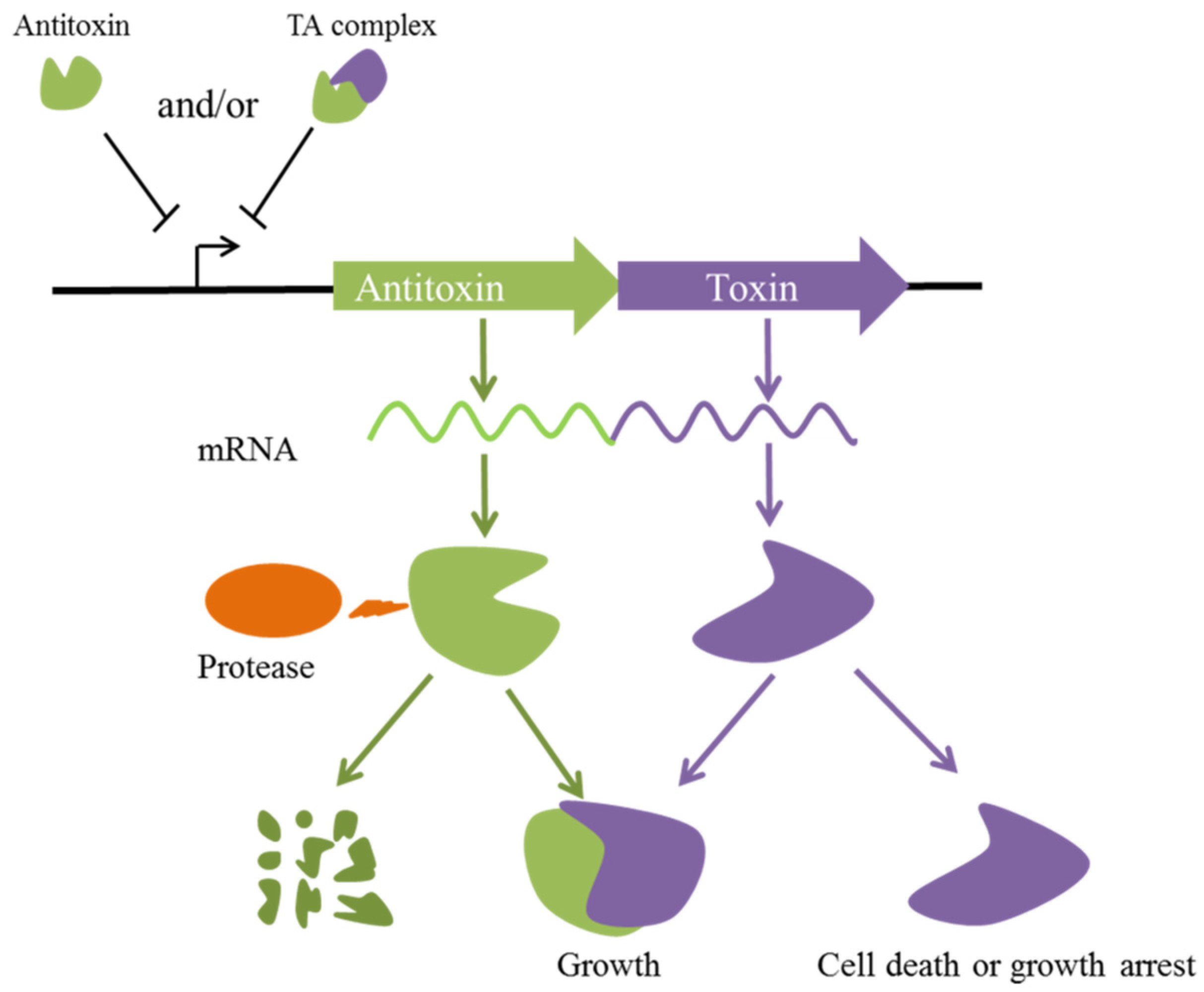
:1. Introduction
2. Transcriptional Regulation of Type II TA Systems in P. aeruginosa
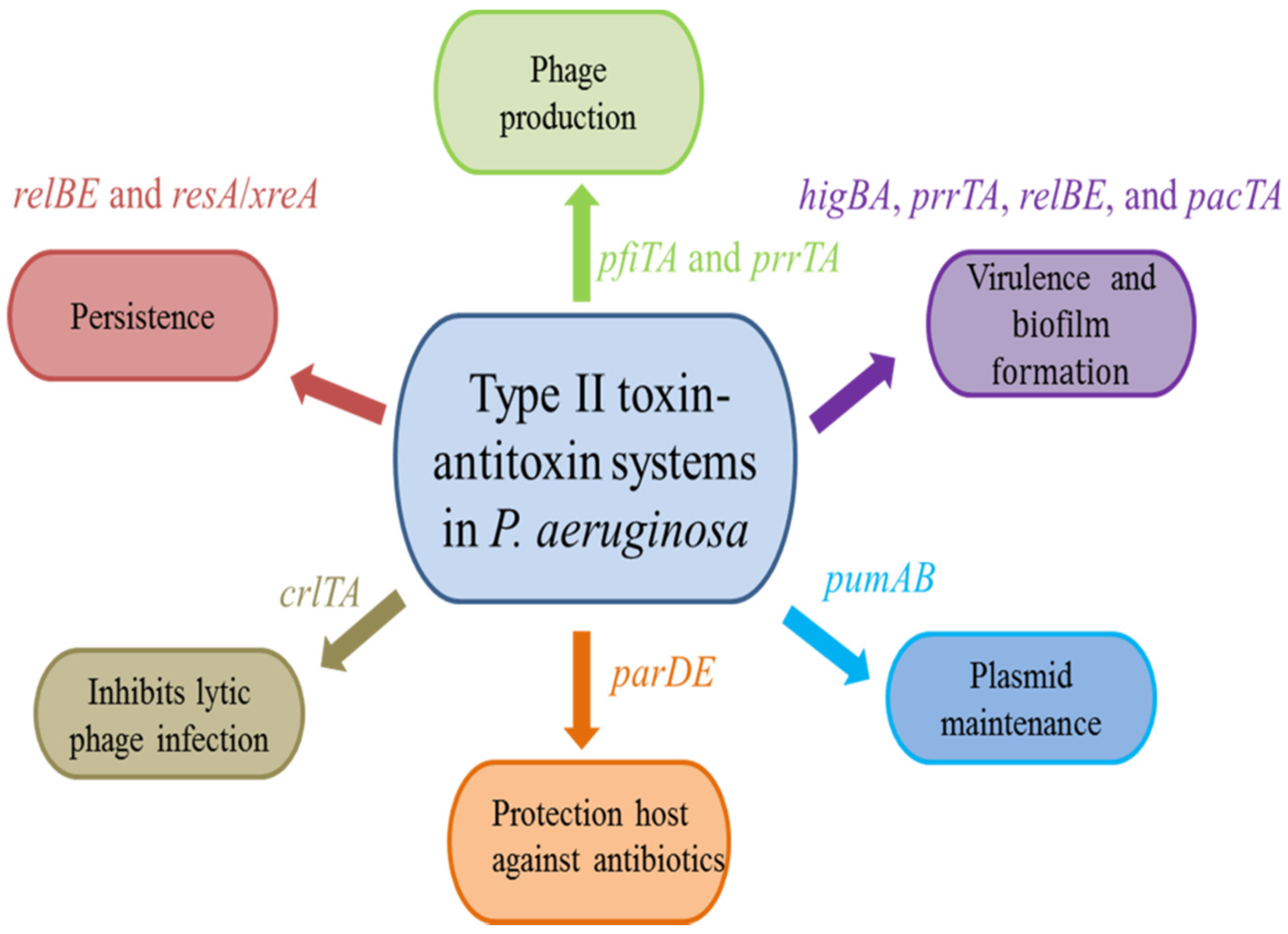
3. Biological Functions of Type II TA Systems in P. aeruginosa
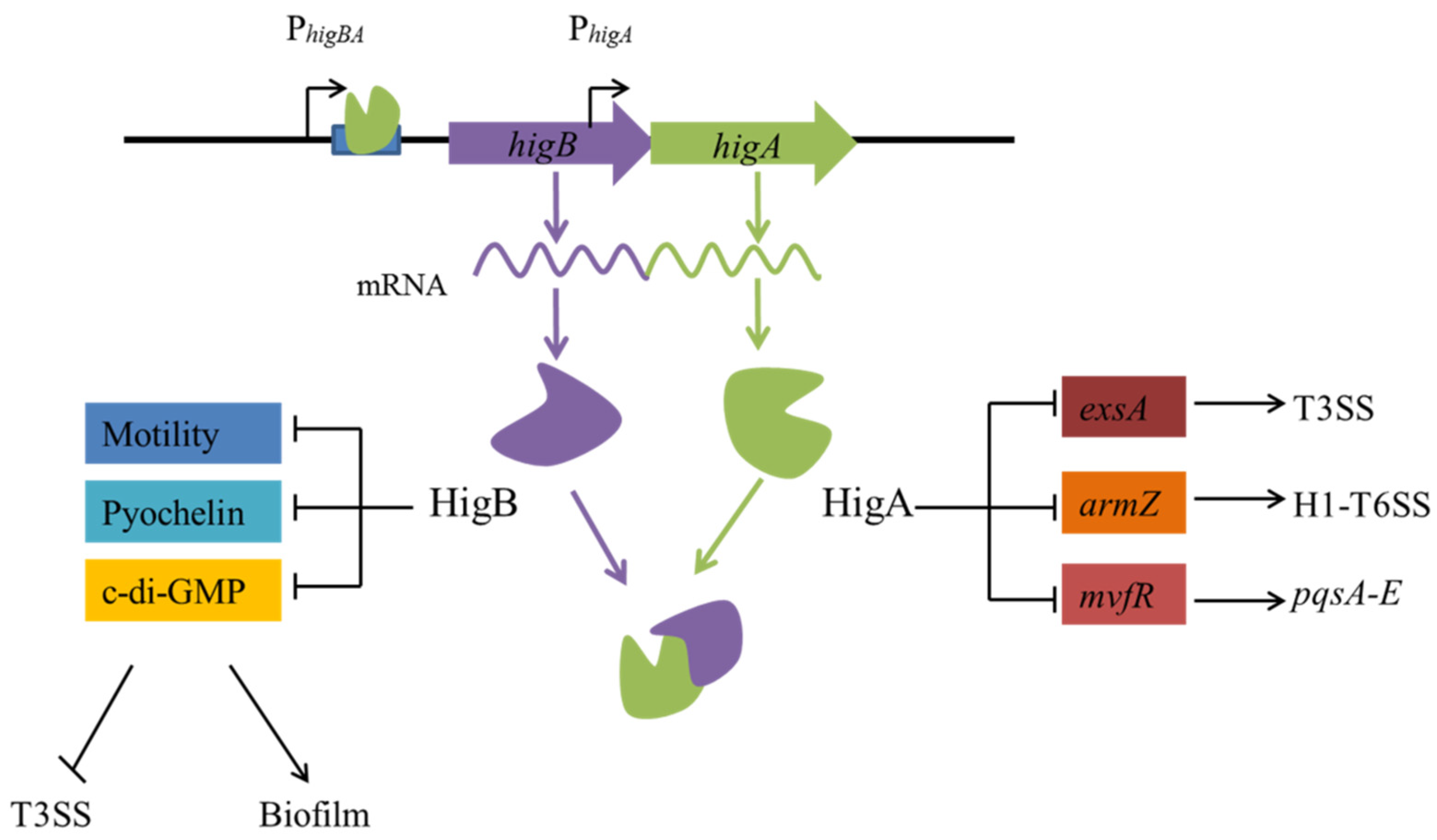
3.1. Virulence and Biofilm Formation
3.2. Protection Host against Antibiotics
3.3. Persistence
3.4. Plasmid Maintenance
3.5. Phage Production
4. Potential Application of Type II TA Systems in P. aeruginosa
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurenas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and evolution of bacterial toxin-antitoxin systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Gong, L.; Cheng, F.; Yu, H.; Zhao, D.; Wang, R.; Wang, T.; Zhang, S.; Zhou, J.; Shmakov, S.A.; et al. Toxin-antitoxin RNA pairs safeguard CRISPR-Cas systems. Science 2021, 372, eabe5601. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.E.; Kline, B.C. The F plasmid ccd autorepressor is a complex of CcdA and CcdB proteins. Mol. Genet. Genom. 1989, 219, 26–32. [Google Scholar] [CrossRef]
- Jurenas, D.; Van Melderen, L.; Garcia-Pino, A. Mechanism of regulation and neutralization of the AtaR-AtaT toxin-antitoxin system. Nat. Chem. Biol. 2019, 15, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Type II Toxin-Antitoxin Systems: Evolution and Revolutions. J. Bacteriol. 2020, 202, e00763-19. [Google Scholar] [CrossRef] [Green Version]
- Jimmy, S.; Saha, C.K.; Kurata, T.; Stavropoulos, C.; Oliveira, S.R.A.; Koh, A.; Cepauskas, A.; Takada, H.; Rejman, D.; Tenson, T.; et al. A widespread toxin-antitoxin system exploiting growth control via alarmone signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 10500–10510. [Google Scholar] [CrossRef]
- Jankevicius, G.; Ariza, A.; Ahel, M.; Ahel, I. The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA. Mol. Cell 2016, 64, 1109–1116. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Lord, D.M.; Cheng, H.Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef] [Green Version]
- Aakre, C.D.; Phung, T.N.; Huang, D.; Laub, M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell 2013, 52, 617–628. [Google Scholar] [CrossRef] [Green Version]
- Songailiene, I.; Juozapaitis, J.; Tamulaitiene, G.; Ruksenaite, A.; Sulcius, S.; Sasnauskas, G.; Venclovas, C.; Siksnys, V. HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation. Mol. Cell 2020, 80, 955–970.e957. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Gao, X.; Zhu, K.; Yin, H.; Mao, X.; Wojdyla, J.A.; Qin, B.; Huang, H.; Wang, M.; Sun, Y.C.; et al. Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism. Commun. Biol. 2020, 3, 216. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, S.; Li, B.; Tang, K.; Yao, J.; Wood, T.K.; Wang, P.; Wang, X. Conjugative plasmid-encoded toxin-antitoxin system PrpT/PrpA directly controls plasmid copy number. Proc. Natl. Acad. Sci. USA 2021, 118, e2011577118. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; Wu, A.Y.; Iredell, J.R. Biological Functions of Type II Toxin-Antitoxin Systems in Bacteria. Microorganisms 2021, 9, 1276. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhai, Y.; Wei, M.; Zheng, C.; Jiao, X. Toxin-antitoxin systems: Classification, biological roles, and applications. Microbiol. Res. 2022, 264, 127159. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Dai, Z.; Wu, T.; Xu, S.; Zhou, L.; Tang, W.; Hu, E.; Zhan, L.; Chen, M.; Yu, G. Characterization of toxin-antitoxin systems from public sequencing data: A case study in Pseudomonas aeruginosa. Front. Microbiol. 2022, 13, 951774. [Google Scholar] [CrossRef]
- Wood, T.L.; Wood, T.K. The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiologyopen 2016, 5, 499–511. [Google Scholar] [CrossRef]
- Muthuramalingam, M.; White, J.C.; Murphy, T.; Ames, J.R.; Bourne, C.R. The toxin from a ParDE toxin-antitoxin system found in Pseudomonas aeruginosa offers protection to cells challenged with anti-gyrase antibiotics. Mol. Microbiol. 2019, 111, 441–454. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Shen, M.; Lu, S.; Le, S.; Tan, Y.; Wang, J.; Zhao, X.; Shen, W.; Guo, K.; Yang, Y.; et al. Identification and Characterization of the HicAB Toxin-Antitoxin System in the Opportunistic Pathogen Pseudomonas aeruginosa. Toxins 2016, 8, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savari, M.; Rostami, S.; Ekrami, A.; Bahador, A. Characterization of Toxin-Antitoxin (TA) Systems in Pseudomonas aeruginosa Clinical Isolates in Iran. Jundishapur J. Microbiol. 2016, 9, e26627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Li, S.; Li, H.; Jin, Y.; Bai, F.; Cheng, Z.; Wu, W. Identification of a Toxin-Antitoxin System That Contributes to Persister Formation by Reducing NAD in Pseudomonas aeruginosa. Microorganisms 2021, 9, 753. [Google Scholar] [CrossRef] [PubMed]
- Shmidov, E.; Lebenthal-Loinger, I.; Roth, S.; Karako-Lampert, S.; Zander, I.; Shoshani, S.; Danielli, A.; Banin, E. PrrT/A, a Pseudomonas aeruginosa Bacterial Encoded Toxin-Antitoxin System Involved in Prophage Regulation and Biofilm Formation. Microbiol. Spectr. 2022, 10, e01182-22. [Google Scholar] [CrossRef]
- Ni, M.; Lin, J.; Gu, J.; Lin, S.; He, M.; Guo, Y. Antitoxin CrlA of CrlTA Toxin-Antitoxin System in a Clinical Isolate Pseudomonas aeruginosa Inhibits Lytic Phage Infection. Front. Microbiol. 2022, 13, 892021. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, S.; Ye, Z.; Song, Y.; Chen, L.; Tong, A.; He, Y.; Bao, R. The novel type II toxin-antitoxin PacTA modulates Pseudomonas aeruginosa iron homeostasis by obstructing the DNA-binding activity of Fur. Nucleic Acids Res. 2022, 50, 10586–10600. [Google Scholar] [CrossRef]
- Hernandez-Ramirez, K.C.; Chavez-Jacobo, V.M.; Valle-Maldonado, M.I.; Patino-Medina, J.A.; Diaz-Perez, S.P.; Jacome-Galarza, I.E.; Ortiz-Alvarado, R.; Meza-Carmen, V.; Ramirez-Diaz, M.I. Plasmid pUM505 encodes a Toxin-Antitoxin system conferring plasmid stability and increased Pseudomonas aeruginosa virulence. Microb. Pathog. 2017, 112, 259–268. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Tang, K.; Wang, W.; Guo, Y.; Wang, X. Prophage encoding toxin/antitoxin system PfiT/PfiA inhibits Pf4 production in Pseudomonas aeruginosa. Microb. Biotechnol. 2020, 13, 1132–1144. [Google Scholar] [CrossRef] [Green Version]
- Hayes, F.; Van Melderen, L. Toxins-antitoxins: Diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 386–408. [Google Scholar] [CrossRef]
- Moreno-Cordoba, I.; Chan, W.T.; Nieto, C.; Espinosa, M. Interactions of the Streptococcus pneumoniae Toxin-Antitoxin RelBE Proteins with Their Target DNA. Microorganisms 2021, 9, 851. [Google Scholar] [CrossRef]
- Moreno-Cordoba, I.; Diago-Navarro, E.; Barendregt, A.; Heck, A.J.; Alfonso, C.; Diaz-Orejas, R.; Nieto, C.; Espinosa, M. The toxin-antitoxin proteins relBE2Spn of Streptococcus pneumoniae: Characterization and association to their DNA target. Proteins 2012, 80, 1834–1846. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pino, A.; Balasubramanian, S.; Wyns, L.; Gazit, E.; De Greve, H.; Magnuson, R.D.; Charlier, D.; van Nuland, N.A.; Loris, R. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 2010, 142, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, M.; Borch, J.; Jorgensen, M.G.; Gerdes, K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 2008, 69, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, S.; Luo, G.; Shen, Y.; Li, C.; Zhu, Y.; Huang, Q.; Mou, X.; Tang, X.; Liu, T.; et al. Type II Antitoxin HigA Is a Key Virulence Regulator in Pseudomonas aeruginosa. ACS Infect. Dis. 2021, 7, 2930–2940. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, B.; Li, M.; Shi, J.; Long, Y.; Jin, Y.; Bai, F.; Cheng, Z.; Jin, S.; Wu, W. HigB Reciprocally Controls Biofilm Formation and the Expression of Type III Secretion System Genes through Influencing the Intracellular c-di-GMP Level in Pseudomonas aeruginosa. Toxins 2018, 10, 424. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, M.; Sadeghifard, N.; Maleki, A.; Yeo, C.C.; Ghafourian, S. relBE Toxin-antitoxin System as a Reliable Anti-biofilm Target in Pseudomonas aeruginosa. J. Appl. Microbiol. 2022, 133, 683–695. [Google Scholar] [CrossRef]
- Zander, I.; Shmidov, E.; Roth, S.; Ben-David, Y.; Shoval, I.; Shoshani, S.; Danielli, A.; Banin, E. Characterization of PfiT/PfiA toxin-antitoxin system of Pseudomonas aeruginosa that affects cell elongation and prophage induction. Environ. Microbiol. 2020, 22, 5048–5057. [Google Scholar] [CrossRef]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013, 121, 1–58. [Google Scholar] [CrossRef]
- Lee, K.; Yoon, S.S. Pseudomonas aeruginosa Biofilm, a Programmed Bacterial Life for Fitness. J. Microbiol. Biotechnol. 2017, 27, 1053–1064. [Google Scholar] [CrossRef] [Green Version]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Wang, X.; Wood, T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ. Microbiol. 2011, 77, 5577–5583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.; Mandell, J.B.; Donegan, N.P.; Cheung, A.L.; Ma, W.; Rothenberger, S.; Shanks, R.M.Q.; Richardson, A.R.; Urish, K.L. The Toxin-Antitoxin MazEF Drives Staphylococcus aureus Biofilm Formation, Antibiotic Tolerance, and Chronic Infection. mBio 2019, 10, e01658-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Long, Y.; Liu, Y.; Liu, Y.; Chen, R.; Shi, J.; Zhang, L.; Jin, Y.; Yang, L.; Bai, F.; et al. HigB of Pseudomonas aeruginosa Enhances Killing of Phagocytes by Up-Regulating the Type III Secretion System in Ciprofloxacin Induced Persister Cells. Front. Cell. Infect. Microbiol. 2016, 6, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Sun, C.; Li, Y.; Tang, K.; Ni, S.; Wang, X. Antitoxin HigA inhibits virulence gene mvfR expression in Pseudomonas aeruginosa. Environ. Microbiol. 2019, 21, 2707–2723. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Deziel, E.; He, J.; Lepine, F.; Lesic, B.; Castonguay, M.H.; Milot, S.; Tampakaki, A.P.; Stachel, S.E.; Rahme, L.G. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 2006, 62, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Brutinel, E.D.; Yahr, T.L. Control of gene expression by type III secretory activity. Curr. Opin. Microbiol. 2008, 11, 128–133. [Google Scholar] [CrossRef] [Green Version]
- Allsopp, L.P.; Wood, T.E.; Howard, S.A.; Maggiorelli, F.; Nolan, L.M.; Wettstadt, S.; Filloux, A. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 7707–7712. [Google Scholar] [CrossRef] [Green Version]
- Gotfredsen, M.; Gerdes, K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 1998, 29, 1065–1076. [Google Scholar] [CrossRef]
- Pandey, D.P.; Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef]
- Coskun, U.S.S.; Cicek, A.C.; Kilinc, C.; Guckan, R.; Dagcioglu, Y.; Demir, O.; Sandalli, C. Effect of mazEF, higBA and relBE toxin-antitoxin systems on antibiotic resistance in Pseudomonas aeruginosa and Staphylococcus isolates. Malawi Med. J. 2018, 30, 67–72. [Google Scholar] [CrossRef] [Green Version]
- Chan, W.T.; Domenech, M.; Moreno-Cordoba, I.; Navarro-Martinez, V.; Nieto, C.; Moscoso, M.; Garcia, E.; Espinosa, M. The Streptococcus pneumoniae yefM-yoeB and relBE Toxin-Antitoxin Operons Participate in Oxidative Stress and Biofilm Formation. Toxins 2018, 10, 378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golmoradi Zadeh, R.; Mirshekar, M.; Sadeghi Kalani, B.; Pourghader, J.; Barati, M.; Masjedian Jazi, F. The expression of type II TA system genes following persister cell formation in Pseudomonas aeruginosa isolates in the exponential and stationary phases. Arch. Microbiol. 2022, 204, 451. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.J.; Newsom, D.; Kelly, B.; Irie, Y.; Jennings, L.K.; Xu, B.; Limoli, D.H.; Harrison, J.J.; Parsek, M.R.; White, P.; et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1003984. [Google Scholar] [CrossRef] [PubMed]
- Waligora, E.A.; Ramsey, D.M.; Pryor, E.E., Jr.; Lu, H.; Hollis, T.; Sloan, G.P.; Deora, R.; Wozniak, D.J. AmrZ beta-sheet residues are essential for DNA binding and transcriptional control of Pseudomonas aeruginosa virulence genes. J. Bacteriol. 2010, 192, 5390–5401. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.J.; Ryder, C.R.; Mann, E.E.; Wozniak, D.J. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J. Bacteriol. 2013, 195, 1637–1644. [Google Scholar] [CrossRef] [Green Version]
- Pasqua, M.; Visaggio, D.; Lo Sciuto, A.; Genah, S.; Banin, E.; Visca, P.; Imperi, F. Ferric Uptake Regulator Fur Is Conditionally Essential in Pseudomonas aeruginosa. J. Bacteriol. 2017, 199, e00472-17. [Google Scholar] [CrossRef] [Green Version]
- Sun, F.; Gao, H.; Zhang, Y.; Wang, L.; Fang, N.; Tan, Y.; Guo, Z.; Xia, P.; Zhou, D.; Yang, R. Fur is a repressor of biofilm formation in Yersinia pestis. PLoS ONE 2012, 7, e52392. [Google Scholar] [CrossRef] [Green Version]
- Garcia, C.A.; Alcaraz, E.S.; Franco, M.A.; De Rossi, B.N.P. Iron is a signal for Stenotrophomonas maltophilia biofilm formation, oxidative stress response, OMPs expression, and virulence. Front. Microbiol. 2015, 6, 926. [Google Scholar] [CrossRef] [Green Version]
- Roberts, R.C.; Strom, A.R.; Helinski, D.R. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J. Mol. Biol. 1994, 237, 35–51. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Iredell, J. A ParDE-family toxin antitoxin system in major resistance plasmids of Enterobacteriaceae confers antibiotic and heat tolerance. Sci. Rep. 2019, 9, 9872. [Google Scholar] [CrossRef] [Green Version]
- Ames, J.R.; Muthuramalingam, M.; Murphy, T.; Najar, F.Z.; Bourne, C.R. Expression of different ParE toxins results in conserved phenotypes with distinguishable classes of toxicity. Microbiologyopen 2019, 8, e902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez-Diaz, M.I.; Diaz-Magana, A.; Meza-Carmen, V.; Johnstone, L.; Cervantes, C.; Rensing, C. Nucleotide sequence of Pseudomonas aeruginosa conjugative plasmid pUM505 containing virulence and heavy-metal resistance genes. Plasmid 2011, 66, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Ramirez, K.C.; Valerio-Arellano, B.; Valle-Maldonado, M.I.; Ruiz-Herrera, L.F.; Meza-Carmen, V.; Ramirez-Diaz, M.I. Virulence Conferred by PumA Toxin from the Plasmid-Encoded PumAB Toxin-Antitoxin System is Regulated by Quorum System. Curr. Microbiol. 2020, 77, 2535–2543. [Google Scholar] [CrossRef] [PubMed]
- Sweere, J.M.; Van Belleghem, J.D.; Ishak, H.; Bach, M.S.; Popescu, M.; Sunkari, V.; Kaber, G.; Manasherob, R.; Suh, G.A.; Cao, X.; et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019, 363, eaat9691. [Google Scholar] [CrossRef]
- Wang, X.; Wood, T.K. Cryptic prophages as targets for drug development. Drug Resist. Updates 2016, 27, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Quiroga, C.; Chen, Q.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K.; Wang, X. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014, 42, 6448–6462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, J.; Guo, Y.; Wang, P.; Zeng, Z.; Li, B.; Tang, K.; Liu, X.; Wang, X. Type II toxin/antitoxin system ParESO /CopASO stabilizes prophage CP4So in Shewanella oneidensis. Environ. Microbiol. 2018, 20, 1224–1239. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Tang, K.; Wang, P.; Zeng, Z.; Guo, Y.; Wang, X. Excisionase in Pf filamentous prophage controls lysis-lysogeny decision-making in Pseudomonas aeruginosa. Mol. Microbiol. 2019, 111, 495–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wettstadt, S. Protect thy host: Pf4 phages shield Pseudomonas aeruginosa from antibiotics. Environ. Microbiol. 2020, 22, 2461–2462. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Pati, S.; Kaushik, H.; Singh, S.; Garg, L.C. Toxin-antitoxin systems and their medical applications: Current status and future perspective. Appl. Microbiol. Biotechnol. 2021, 105, 1803–1821. [Google Scholar] [CrossRef] [PubMed]
- Rownicki, M.; Lasek, R.; Trylska, J.; Bartosik, D. Targeting Type II Toxin-Antitoxin Systems as Antibacterial Strategies. Toxins 2020, 12, 568. [Google Scholar] [CrossRef]
- Culviner, P.H.; Laub, M.T. Global Analysis of the E. coli Toxin MazF Reveals Widespread Cleavage of mRNA and the Inhibition of rRNA Maturation and Ribosome Biogenesis. Mol. Cell 2018, 70, 868–880 e810. [Google Scholar] [CrossRef] [Green Version]
- Barth, V.C.; Woychik, N.A. The Sole Mycobacterium smegmatis MazF Toxin Targets tRNA(Lys) to Impart Highly Selective, Codon-Dependent Proteome Reprogramming. Front. Genet. 2019, 10, 1356. [Google Scholar] [CrossRef]
- Critchlow, S.E.; O’Dea, M.H.; Howells, A.J.; Couturier, M.; Gellert, M.; Maxwell, A. The interaction of the F plasmid killer protein, CcdB, with DNA gyrase: Induction of DNA cleavage and blocking of transcription. J. Mol. Biol. 1997, 273, 826–839. [Google Scholar] [CrossRef]
- Freire, D.M.; Gutierrez, C.; Garza-Garcia, A.; Grabowska, A.D.; Sala, A.J.; Ariyachaokun, K.; Panikova, T.; Beckham, K.S.H.; Colom, A.; Pogenberg, V.; et al. An NAD(+) Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death. Mol. Cell 2019, 73, 1282–1291 e1288. [Google Scholar] [CrossRef] [Green Version]
- Pecota, D.C.; Kim, C.S.; Wu, K.; Gerdes, K.; Wood, T.K. Combining the hok/sok, parDE, and pnd postsegregational killer loci to enhance plasmid stability. Appl. Environ. Microbiol. 1997, 63, 1917–1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daimon, Y.; Narita, S.; Akiyama, Y. Activation of Toxin-Antitoxin System Toxins Suppresses Lethality Caused by the Loss of sigmaE in Escherichia coli. J. Bacteriol. 2015, 197, 2316–2324. [Google Scholar] [CrossRef] [Green Version]
- Butt, A.; Higman, V.A.; Williams, C.; Crump, M.P.; Hemsley, C.M.; Harmer, N.; Titball, R.W. The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochem. J. 2014, 459, 333–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Kim, Y.; Hong, S.H.; Ma, Q.; Brown, B.L.; Pu, M.; Tarone, A.M.; Benedik, M.J.; Peti, W.; Page, R.; et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 2011, 7, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Soo, V.W.; Wood, T.K. Antitoxin MqsA represses curli formation through the master biofilm regulator CsgD. Sci. Rep. 2013, 3, 3186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Benedik, M.J.; Wood, T.K. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environ. Microbiol. 2012, 14, 669–679. [Google Scholar] [CrossRef]
| TA System | Toxin | Antitoxin | Localisation | Targeted Cellular Process | Function |
|---|---|---|---|---|---|
| HigB/HigA | HigB | HigA | chromosome | Translation | Virulence and biofilm formation |
| PumA/PumB | PumA | PumB | plasmid | Unknown | Virulence and Plasmid maintenance |
| PfiA/PfiT | PfiT | PfiA | prophage | Unknown | Phage production and immunity |
| ParD/ParE | ParE | ParD | chromosome | Replication | Protection host against antibiotics |
| HicA/HicB | HicA | HicB | chromosome | Translation | Unknown |
| RelB/RelE | RelE | RelB | chromosome | Translation | Persistence and biofilm formation |
| ResA/XreA | ResA | XreA | chromosome | Metabolic stress | Persistence |
| PrrA/PrrT | PrrT | PrrA | chromosome | Unknown | Phage production and biofilm formation |
| CrlA/CrlT | CrlT | CrlA | chromosome | Translation | Inhibits lytic phage infection |
| PacA/PacT | PacT | PacA | chromosome | Translation | Virulence and biofilm formation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Guo, N.; Song, G.; Huang, Y.; Wang, L.; Zhang, Y.; Wang, T. Type II Toxin–Antitoxin Systems in Pseudomonas aeruginosa. Toxins 2023, 15, 164. https://doi.org/10.3390/toxins15020164
Li M, Guo N, Song G, Huang Y, Wang L, Zhang Y, Wang T. Type II Toxin–Antitoxin Systems in Pseudomonas aeruginosa. Toxins. 2023; 15(2):164. https://doi.org/10.3390/toxins15020164
Chicago/Turabian StyleLi, Meng, Nannan Guo, Gaoyu Song, Yi Huang, Lecheng Wang, Yani Zhang, and Tietao Wang. 2023. "Type II Toxin–Antitoxin Systems in Pseudomonas aeruginosa" Toxins 15, no. 2: 164. https://doi.org/10.3390/toxins15020164
APA StyleLi, M., Guo, N., Song, G., Huang, Y., Wang, L., Zhang, Y., & Wang, T. (2023). Type II Toxin–Antitoxin Systems in Pseudomonas aeruginosa. Toxins, 15(2), 164. https://doi.org/10.3390/toxins15020164





