A Computational Inter-Species Study on Safrole Phase I Metabolism-Dependent Bioactivation: A Mechanistic Insight into the Study of Possible Differences among Species
Abstract
:1. Introduction
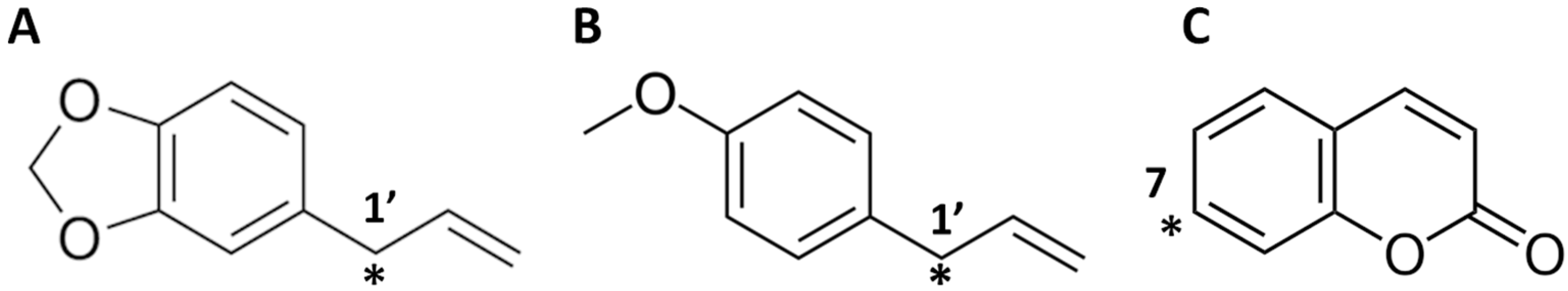
2. Results and Discussion
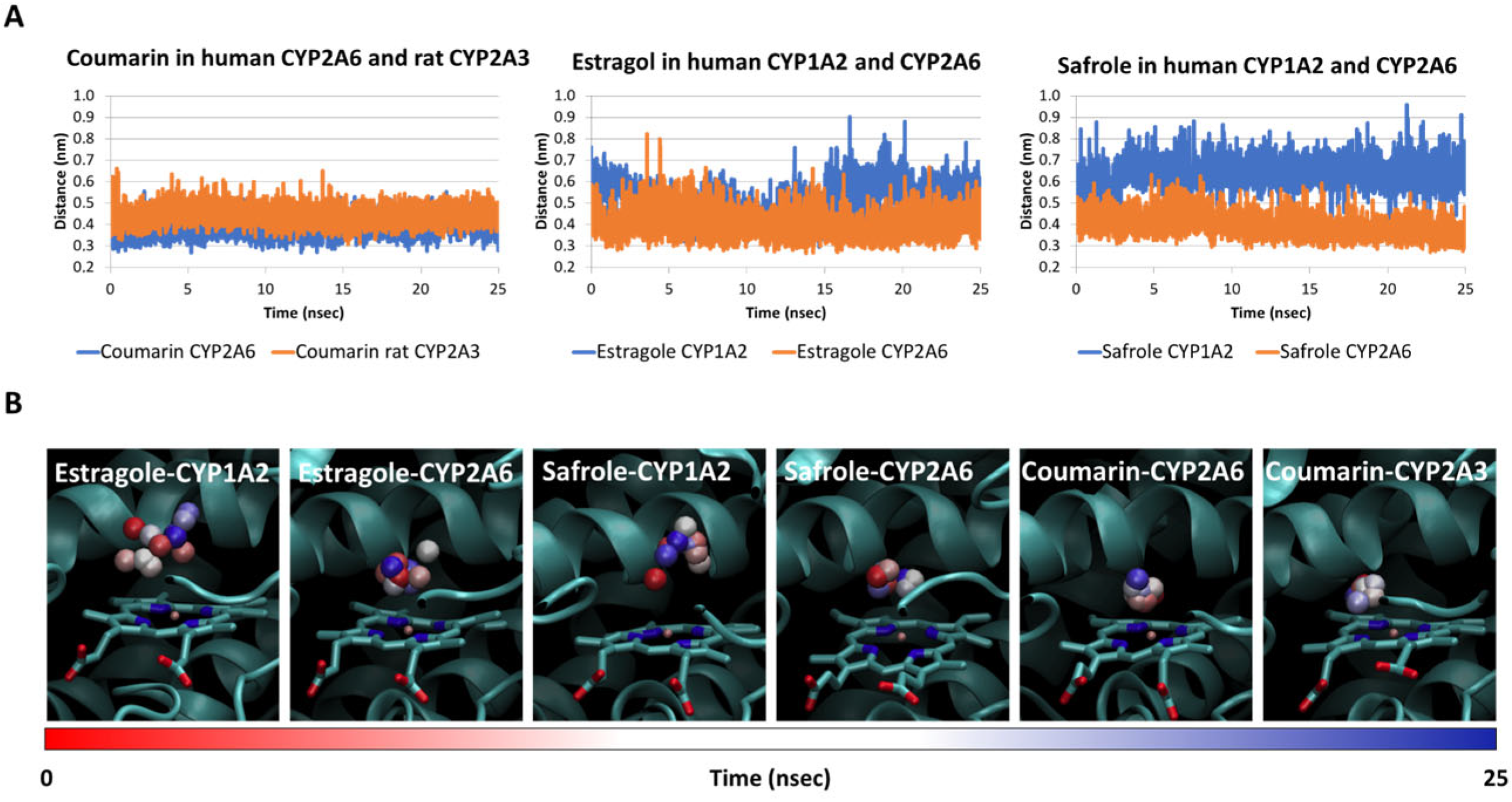
2.1. Fit-for-Purpose Validation
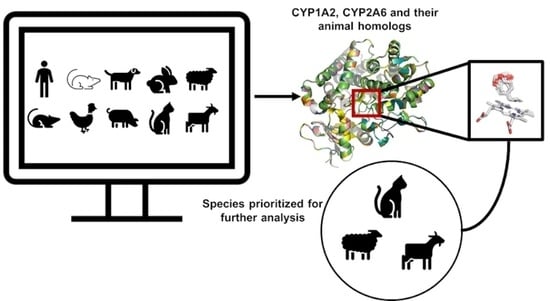
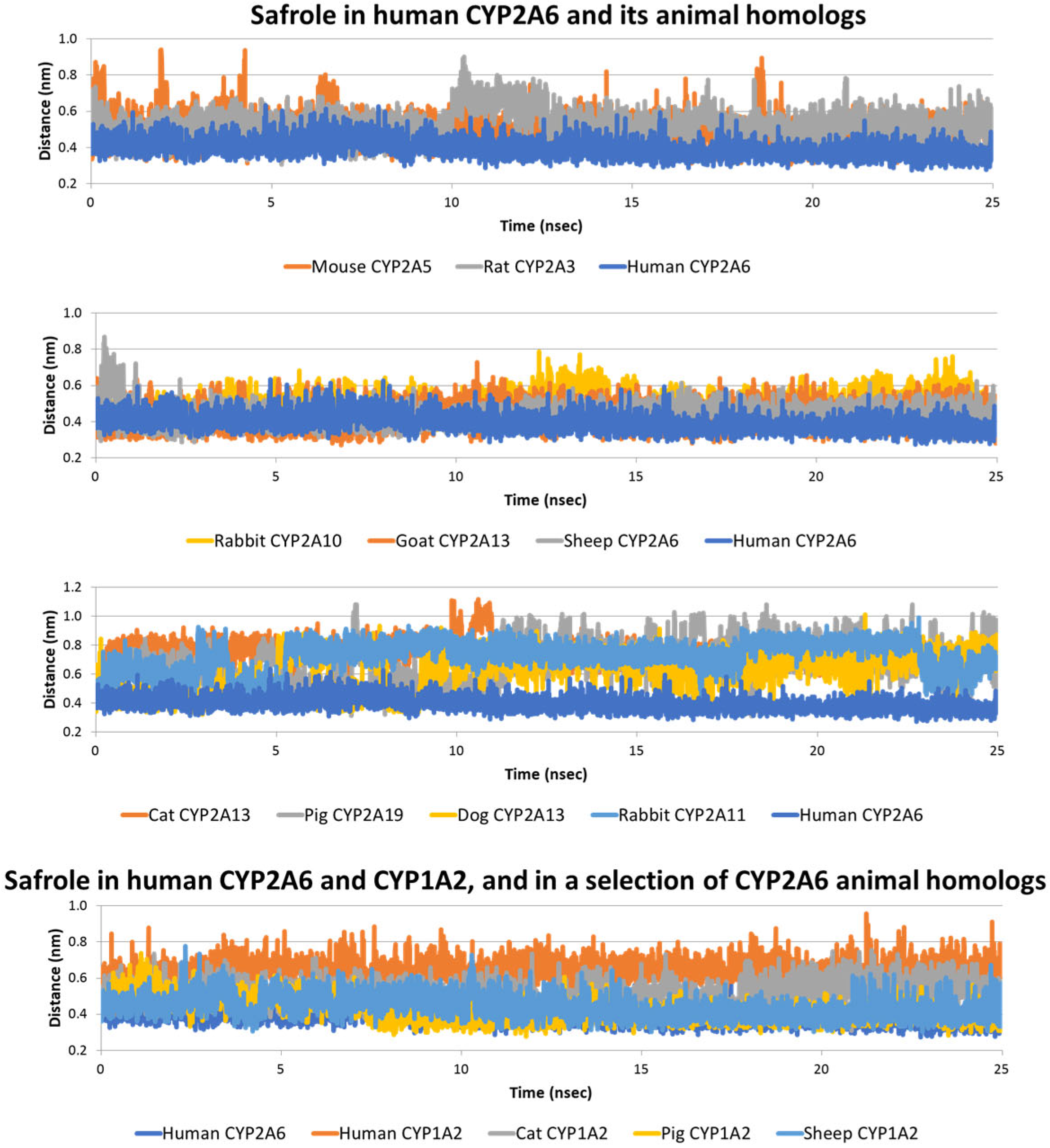
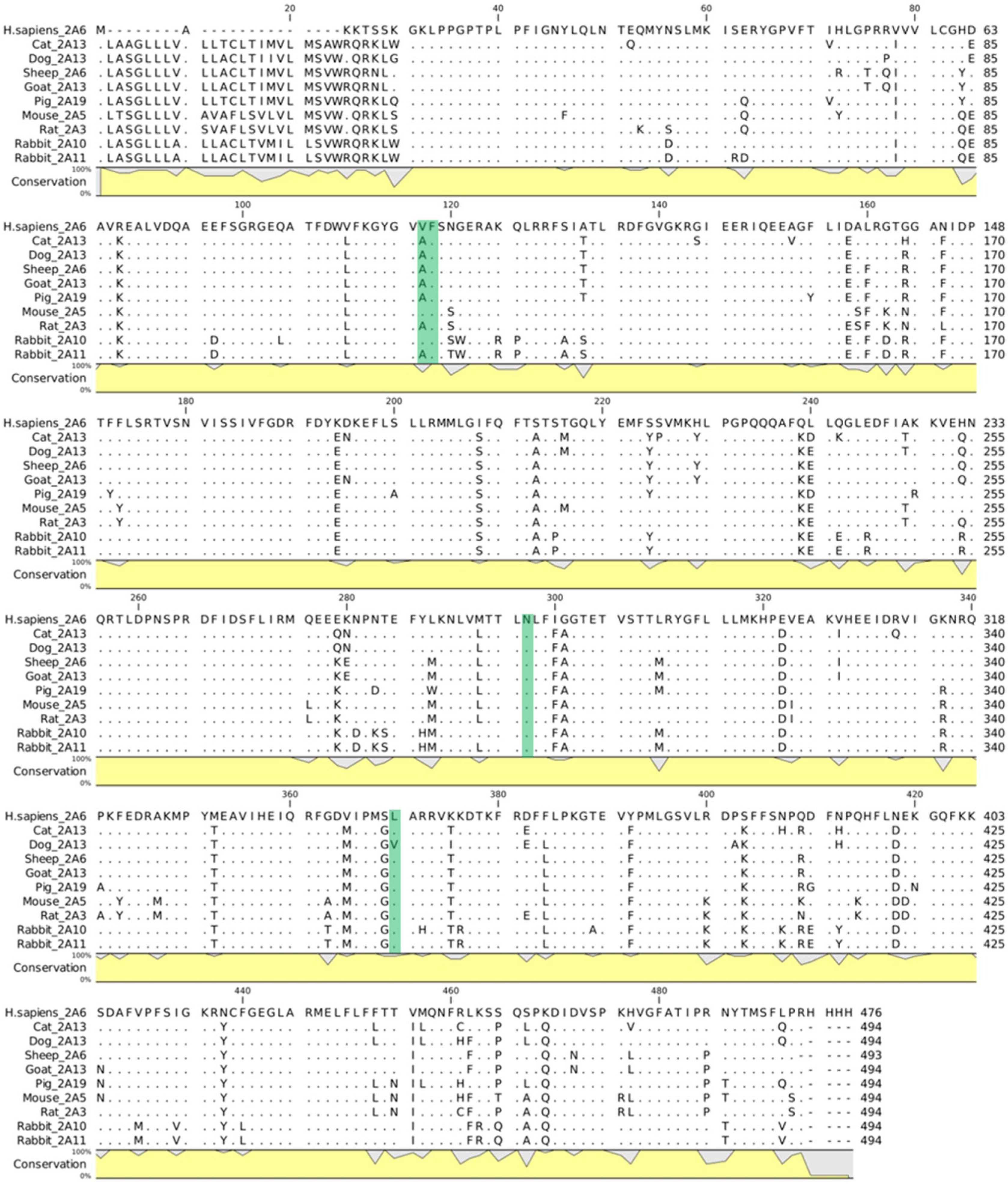
2.2. Interaction of Safrole with Animal Homologs of Human CYP1A2 and CYP2A6
2.3. Estimate of Binding Energy
3. Conclusions
4. Materials and Methods
4.1. Data Source
4.2. Homology Modelling
4.3. Docking Simulations
4.4. Molecular Dynamics
4.5. Statistical Analysis
4.6. Cluster and Binding Affinity Analysis of Protein-Ligand Complex Trajectories
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gotz, M.E.; Sachse, B.; Schafer, B.; Eisenreich, A. Myristicin and Elemicin: Potentially Toxic Alkenylbenzenes in Food. Foods 2022, 11, 1988. [Google Scholar] [CrossRef] [PubMed]
- Eisenreich, A.; Gotz, M.E.; Sachse, B.; Monien, B.H.; Herrmann, K.; Schafer, B. Alkenylbenzenes in Foods: Aspects Impeding the Evaluation of Adverse Health Effects. Foods 2021, 10, 2139. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, R.G. Phenylpropenes: Occurrence, Distribution, and Biosynthesis in Fruit. J. Agric. Food Chem. 2018, 66, 2259–2272. [Google Scholar] [CrossRef] [PubMed]
- Jeurissen, S.M.F.; Punt, A.; Boersma, M.G.; Bogaards, J.J.P.; Fiamegos, Y.C.; Schilter, B.; van Bladeren, P.J.; Cnubben, N.H.P.; Rietjens, I. Human cytochrome p450 enzyme specificity for the bioactivation of estragole and related alkenylbenzenes. Chem. Res. Toxicol. 2007, 20, 798–806. [Google Scholar] [CrossRef]
- IARC. IARC Monographs on the Evaluation of the Carcinogenic Risks to Humans. In OveraIl Evaluations of Carcinogenicity: An Updating of IARC Monographs; IARC: Lyon, France, 1987. [Google Scholar]
- Bampidis, V.; Azimonti, G.; Bastos, M.D.; Christensen, H.; Durjava, M.F.; Kouba, M.; Lopez-Alonso, M.; Puente, S.L.; Marcon, F.; Mayo, B.; et al. Safety and efficacy of a feed additive consisting of a tincture from the bark of Cinnamomum verum J. Presl (cinnamon tincture) for use in all animal species (FEFANA asbl). Efsa J. 2021, 19, e06986. [Google Scholar] [CrossRef]
- Hausner, E.; Poppenga, R.H. Chapter 26—Hazards Associated with the Use of Herbal and Other Natural Products. In Small Animal Toxicology, 3rd ed.; Peterson, M.E., Talcott, P.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 335–356. [Google Scholar]
- Prinsloo, G.; Nogemane, N.; Street, R. The use of plants containing genotoxic carcinogens as foods and medicine. Food Chem. Toxicol. 2018, 116, 27–39. [Google Scholar] [CrossRef]
- Siano, F.; Ghizzoni, C.; Gionfriddo, F.; Colombo, E.; Servillo, L.; Castaldo, D. Determination of estragole, safrole and eugenol methyl ether in food products. Food Chem. 2003, 81, 469–475. [Google Scholar] [CrossRef]
- Bampidis, V.; Azimonti, G.; Bastos, M.D.; Christensen, H.; Durjava, M.F.; Kouba, M.; Lopez-Alonso, M.; Puente, S.L.; Marcon, F.; Mayo, B.; et al. Safety and efficacy of a feed additive consisting of an essential oil from Cinnamomum camphora (L.) J. Presl (camphor white oil) for use in all animal species (FEFANA asbl). Efsa J. 2022, 20, e06985. [Google Scholar] [CrossRef]
- Bode, A.M.; Dong, Z.G. Toxic Phytochemicals and Their Potential Risks for Human Cancer. Cancer Prev. Res. 2015, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Martati, E.; Boersma, M.G.; Spenkelink, A.; Khadka, D.B.; van Bladeren, P.J.; Rietjens, I.; Punt, A. Physiologically Based Biokinetic (PBBK) Modeling of Safrole Bioactivation and Detoxification in Humans as Compared with Rats. Toxicol. Sci. 2012, 128, 301–316. [Google Scholar] [CrossRef]
- Carmichael, P.L.; Baltazar, M.T.; Cable, S.; Cochrane, S.; Dent, M.; Li, H.Q.; Middleton, A.; Muller, I.; Reynolds, G.; Westmoreland, C.; et al. Ready for Regulatory Use: NAMs and NGRA for Chemical Safety Assurance. Altex Altern. Anim. Exp. 2022, 39, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Takemura, H.; Shimoi, K.; Yamamoto, K. A 3D Model of CYP1B1 Explains the Dominant 4-Hydroxylation of Estradiol. J. Chem. Inf. Model. 2010, 50, 1173–1178. [Google Scholar] [CrossRef]
- Dorne, J.; Cirlini, M.; Louisse, J.; Pedroni, L.; Galaverna, G.; Dellafiora, L. A Computational Understanding of Inter-Individual Variability in CYP2D6 Activity to Investigate the Impact of Missense Mutations on Ochratoxin A Metabolism. Toxins 2022, 14, 207. [Google Scholar] [CrossRef]
- Sridhar, J.; Goyal, N.; Liu, J.W.; Foroozesh, M. Review of Ligand Specificity Factors for CYP1A Subfamily Enzymes from Molecular Modeling Studies Reported to-Date. Molecules 2017, 22, 1143. [Google Scholar] [CrossRef] [Green Version]
- von Weymarn, L.B.; Murphy, S.E. Coumarin metabolism by rat esophageal microsomes and cytochrome P450 2A3. Chem. Res. Toxicol. 2001, 14, 1386–1392. [Google Scholar] [CrossRef] [PubMed]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.P.; George, N.; Fexova, S.; Fonseca, N.A.; Fullgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas update: From tissues to single cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastian, F.B.; Roux, J.; Niknejad, A.; Comte, A.; Costa, S.S.F.; de Farias, T.M.; Moretti, S.; Parmentier, G.; de Laval, V.R.; Rosikiewicz, M.; et al. The Bgee suite: Integrated curated expression atlas and comparative transcriptomics in animals. Nucleic Acids Res. 2021, 49, D831–D847. [Google Scholar] [CrossRef]
- Righetti, L.; Rolli, E.; Dellafiora, L.; Galaverna, G.; Suman, M.; Bruni, R.; Dall’Asta, C. Thinking Out of the Box: On the Ability of Zea mays L. to Biotrasform Aflatoxin B1 Into Its Modified Forms. Front. Plant Sci. 2021, 11, 9158. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Jeurissen, S.M.F.; Bogaards, J.J.P.; Awad, H.M.; Boersma, M.G.; Brand, W.; Fiamegos, Y.C.; van Beek, T.A.; Alink, G.M.; Sudholter, E.J.R.; Cnubben, N.H.P.; et al. Human cytochrome P450 enzyme specificity for bioactivation of safrole to the proximate carcinogen 1′-hydroxysafrole. Chem. Res. Toxicol. 2004, 17, 1245–1250. [Google Scholar] [CrossRef] [PubMed]
- Martati, E.; Boersma, M.G.; Spenkelink, A.; Khadka, D.B.; Punt, A.; Vervoort, J.; van Bladeren, P.J.; Rietjens, I. Physiologically Based Biokinetic (PBBK) Model for Safrole Bioactivation and Detoxification in Rats. Chem. Res. Toxicol. 2011, 24, 818–834. [Google Scholar] [CrossRef] [PubMed]
- Kishida, T.; Muto, S.I.; Hayashi, M.; Tsutsui, M.; Tanaka, S.; Murakami, M.; Kuroda, J. Strain differences in hepatic cytochrome P450 1A and 3A expression between Sprague-Dawley and Wistar rats. J. Toxicol. Sci. 2008, 33, 447–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Vangone, A.; Schaarschmidt, J.; Koukos, P.; Geng, C.L.; Citro, N.; Trellet, M.E.; Xue, L.C.; Bonvin, A. Large-scale prediction of binding affinity in protein-small ligand complexes: The PRODIGY-LIG web server. Bioinformatics 2019, 35, 1585–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Chen, J.; Cheng, T.J.; Gindulyte, A.; He, J.; He, S.Q.; Li, Q.L.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Sansen, S.; Yano, J.K.; Reynald, R.L.; Schoch, G.A.; Griffin, K.J.; Stout, C.D.; Johnson, E.F. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J. Biol. Chem. 2007, 282, 14348–14355. [Google Scholar] [CrossRef] [Green Version]
- Sansen, S.; Hsu, M.H.; Stout, C.D.; Johnson, E.F. Structural insight into the altered substrate specificity of human cytochrome P450 2A6 mutants. Arch. Biochem. Biophys. 2007, 464, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Louisse, J.; Dorne, J.L.C.M.; Dellafiora, L. Investigating the interaction between organic anion transporter 1 and ochratoxin A: An in silico structural study to depict early molecular events of substrate recruitment and the impact of single point mutations. Toxicol. Lett. 2022, 355, 19–30. [Google Scholar] [CrossRef]
- Dellafiora, L.; Oswald, I.P.; Dorne, J.L.; Galaverna, G.; Battilani, P.; Dall’Asta, C. An in silico structural approach to characterize human and rainbow trout estrogenicity of mycotoxins: Proof of concept study using zearalenone and alternariol. Food Chem. 2020, 312, 126088. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado-Rojas, W.; Olivero-Verbel, J. Potential interaction of natural dietary bioactive compounds with COX-2. J. Mol. Graph. Model. 2011, 30, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone Φ, Ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Silva, D.A.; Yan, Y.J.; Huang, X.H. Force field development for cofactors in the photosystem II. J. Comput. Chem. 2012, 33, 1969–1980. [Google Scholar] [CrossRef]
- Panneerselvam, S.; Yesudhas, D.; Durai, P.; Anwar, M.A.; Gosu, V.; Choi, S. A Combined Molecular Docking/Dynamics Approach to Probe the Binding Mode of Cancer Drugs with Cytochrome P450 3A4. Molecules 2015, 20, 14915–14935. [Google Scholar] [CrossRef]
- Del Favero, G.; Mayer, R.M.; Dellafiora, L.; Janker, L.; Niederstaetter, L.; Dall’Asta, C.; Gerner, C.; Marko, D. Structural Similarity with Cholesterol Reveals Crucial Insights into Mechanisms Sustaining the Immunomodulatory Activity of the Mycotoxin Alternariol. Cells 2020, 9, 847. [Google Scholar] [CrossRef] [PubMed]
| Molecule | Species | CYP2A6 and its Homologues | CYP1A2 |
|---|---|---|---|
| Estragole | Human | 162 | 154 |
| Coumarin | Rat | 144 | n.p. |
| Human | 146 | n.p. | |
| Safrole | Human | 162 | 159 |
| Cat | 174 | 153 | |
| Chicken | n.p. | 150 | |
| Dog | 164 | 152 | |
| Goat | 173 | 156 | |
| Mouse | 157 | 171 | |
| Pig | 172 | 155 | |
| Rat | 167 | 155 | |
| Sheep | 153 | 159 | |
| Rabbit | 174 | 161 | |
| 169 |
| Molecule | Species | CYP2A6 and its Homologues | CYP1A2 | ||
|---|---|---|---|---|---|
| Distance (nm) | Biotransformation 1 | Distance (nm) | Biotransformation 1 | ||
| Estragole | Human | 0.40 ± 0.06 | Yes (experimental) 2 | 0.51 ± 0.09 | Yes (experimental) 2 |
| Coumarin | Rat | 0.45 ± 0.04 | Yes (experimental) 3 | n.p. | - |
| Human | 0.39 ± 0.04 | Yes (experimental) 3 | n.p. | - | |
| Safrole | Human | 0.39 ± 0.04 | Yes (experimental) 2 | 0.64 ± 0.09 | No (experimental) 2, 4 |
| Cat | 0.74 ± 0.08 | No (calculated) | 0.52 ± 0.07 | Yes (calculated) | |
| Dog | 0.63 ± 0.14 | No (calculated) | 0.47 ± 0.06 | Yes (calculated) | |
| Pig | 0.65 ± 0.15 | No (calculated) | 0.43 ± 0.07 | Yes (calculated) | |
| Goat | 0.43 ± 0.07 | Yes (calculated) | 0.47 ± 0.06 | Yes (calculated) | |
| Rabbit | 0.72 ± 0.09 | No (calculated) | 0.47 ± 0.06 | Yes (calculated) | |
| 0.47 ± 0.08 | Yes (calculated) | ||||
| Chicken | n.p. | n.p. | 0.47 ± 0.09 | Yes (calculated) | |
| Sheep | 0.43 ± 0.06 | Yes (calculated) | 0.44 ± 0.06 | Yes (calculated) | |
| Mouse | 0.50 ± 0.09 | Yes (calculated) | 0.48 ± 0.06 | Yes (calculated) | |
| Rat | 0.52 ± 0.08 | Yes (calculated) | 0.47 ± 0.06 | Yes (calculated) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedroni, L.; Louisse, J.; Punt, A.; Dorne, J.L.C.M.; Dall’Asta, C.; Dellafiora, L. A Computational Inter-Species Study on Safrole Phase I Metabolism-Dependent Bioactivation: A Mechanistic Insight into the Study of Possible Differences among Species. Toxins 2023, 15, 94. https://doi.org/10.3390/toxins15020094
Pedroni L, Louisse J, Punt A, Dorne JLCM, Dall’Asta C, Dellafiora L. A Computational Inter-Species Study on Safrole Phase I Metabolism-Dependent Bioactivation: A Mechanistic Insight into the Study of Possible Differences among Species. Toxins. 2023; 15(2):94. https://doi.org/10.3390/toxins15020094
Chicago/Turabian StylePedroni, Lorenzo, Jochem Louisse, Ans Punt, Jean Lou C. M. Dorne, Chiara Dall’Asta, and Luca Dellafiora. 2023. "A Computational Inter-Species Study on Safrole Phase I Metabolism-Dependent Bioactivation: A Mechanistic Insight into the Study of Possible Differences among Species" Toxins 15, no. 2: 94. https://doi.org/10.3390/toxins15020094
APA StylePedroni, L., Louisse, J., Punt, A., Dorne, J. L. C. M., Dall’Asta, C., & Dellafiora, L. (2023). A Computational Inter-Species Study on Safrole Phase I Metabolism-Dependent Bioactivation: A Mechanistic Insight into the Study of Possible Differences among Species. Toxins, 15(2), 94. https://doi.org/10.3390/toxins15020094