Effects of Lactic Acid Bacteria Reducing the Content of Harmful Fungi and Mycotoxins on the Quality of Mixed Fermented Feed
Abstract
1. Introduction
2. Results
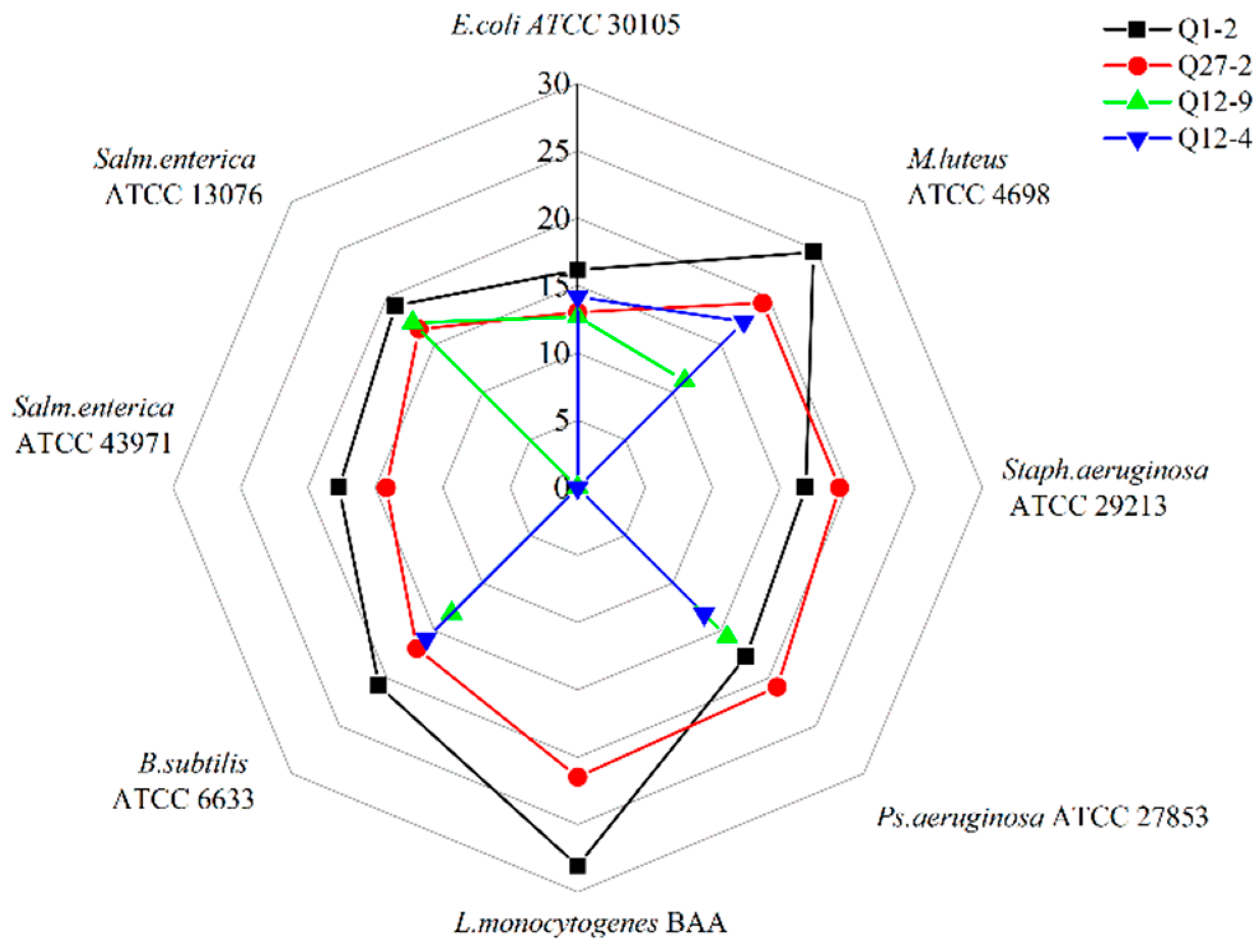
2.1. Screening of LAB with Antibacterial, Antifungal Activity, and Mycotoxin Adsorption Ability
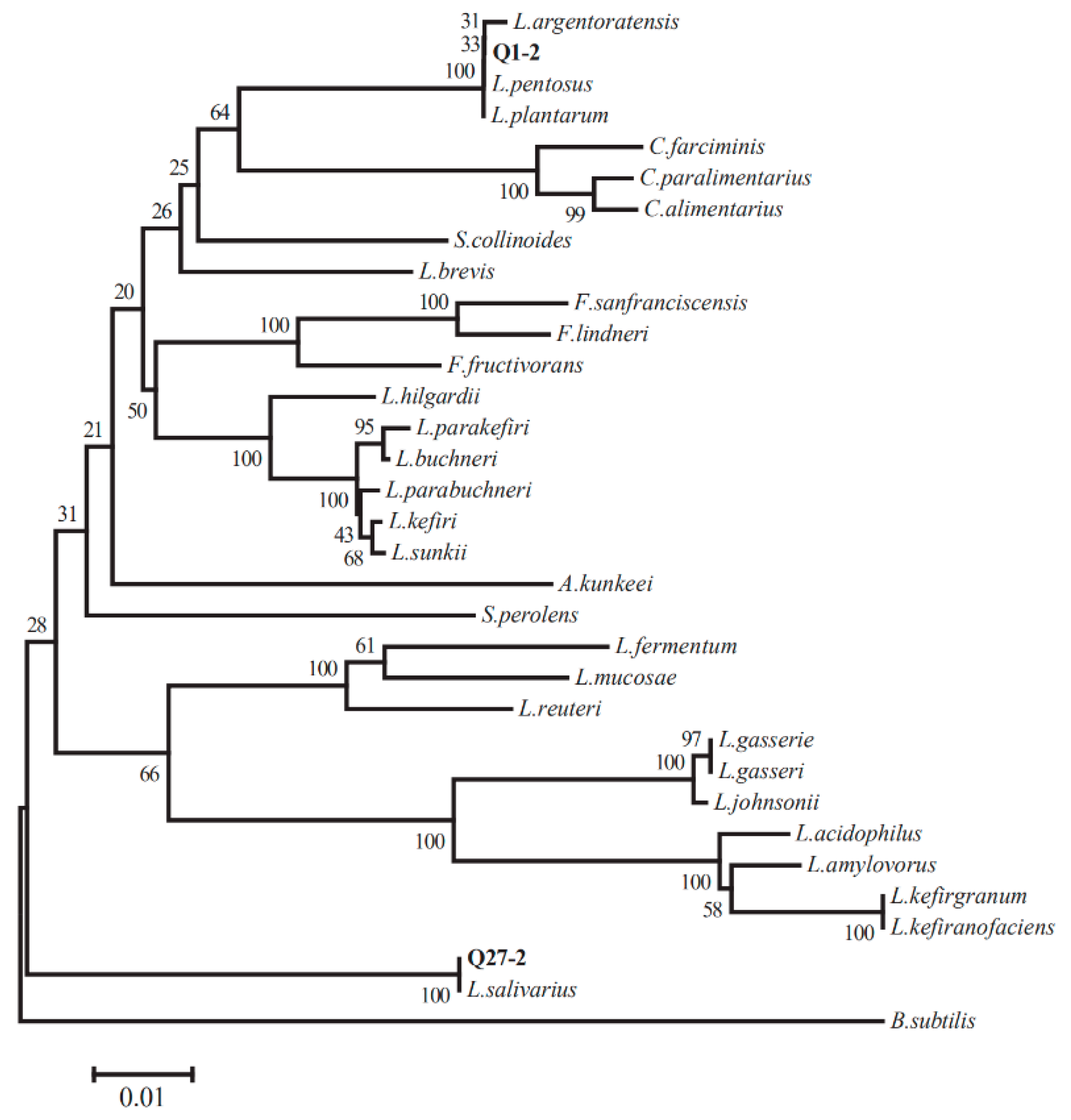
2.2. 16S rRNA Gene Sequence Analysis
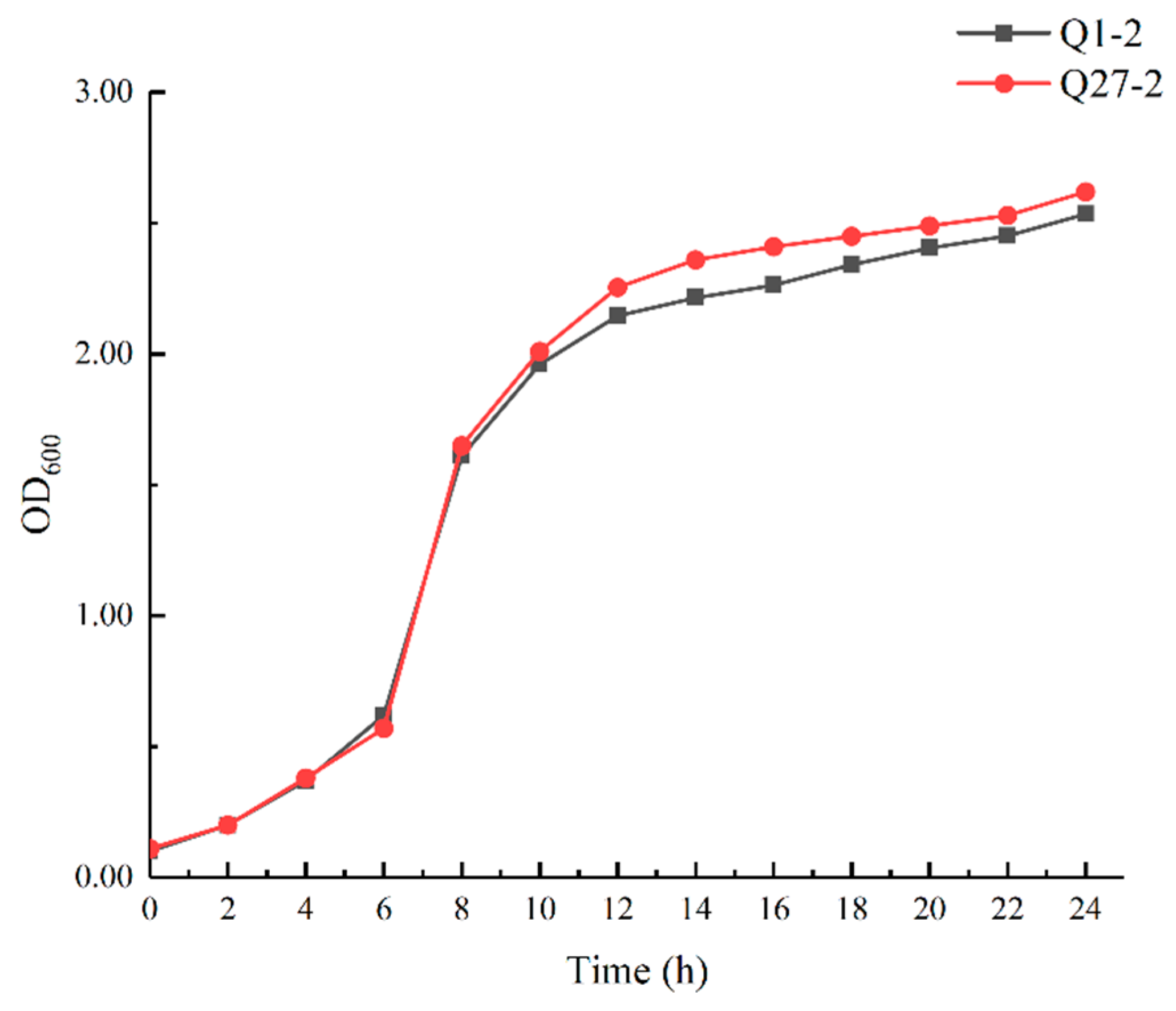
2.3. Growth Curve of Selected LAB Isolates
2.4. Physiological and Biochemical Characteristics
2.5. Fermentation Quality and Chemical Composition
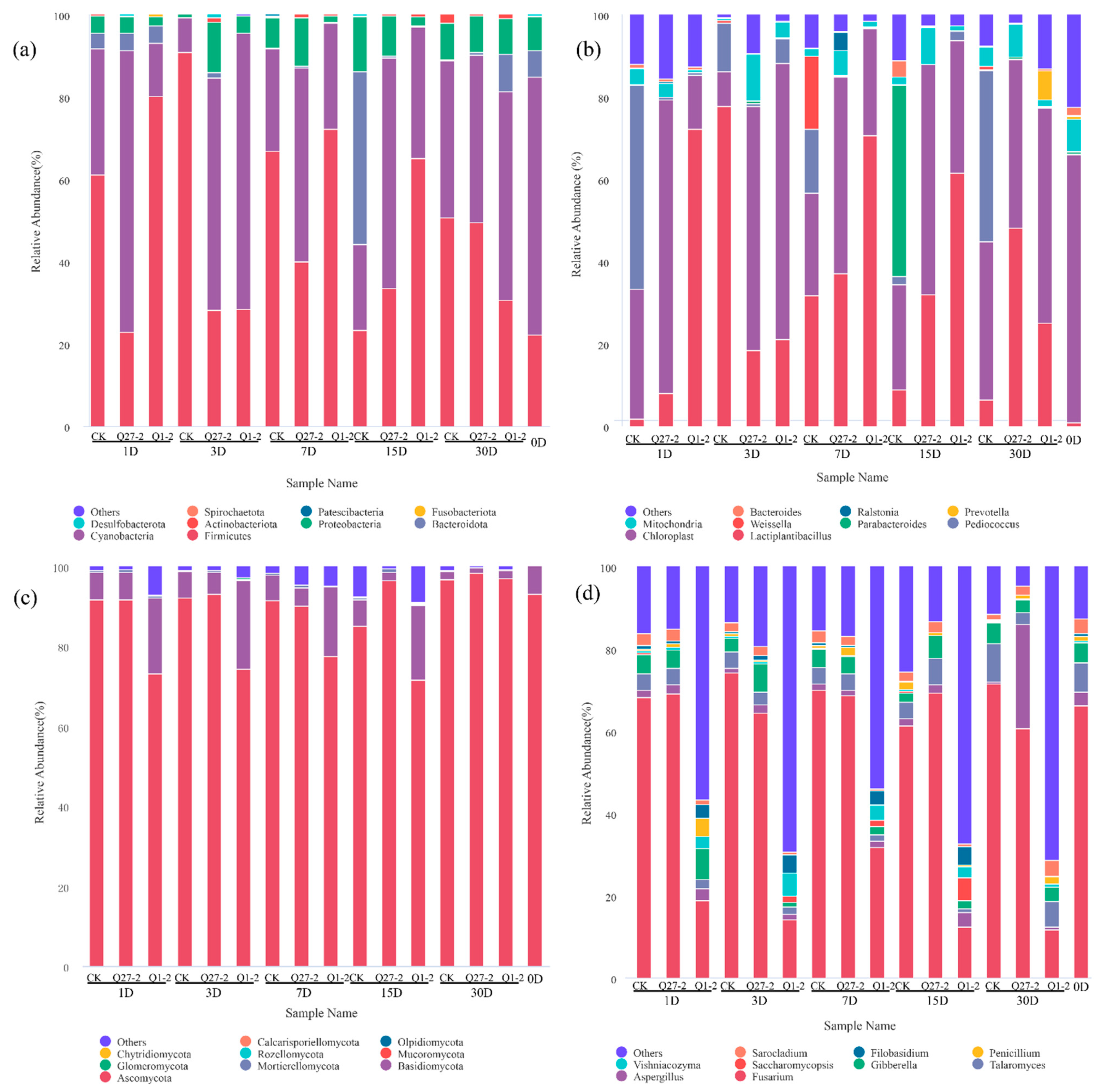
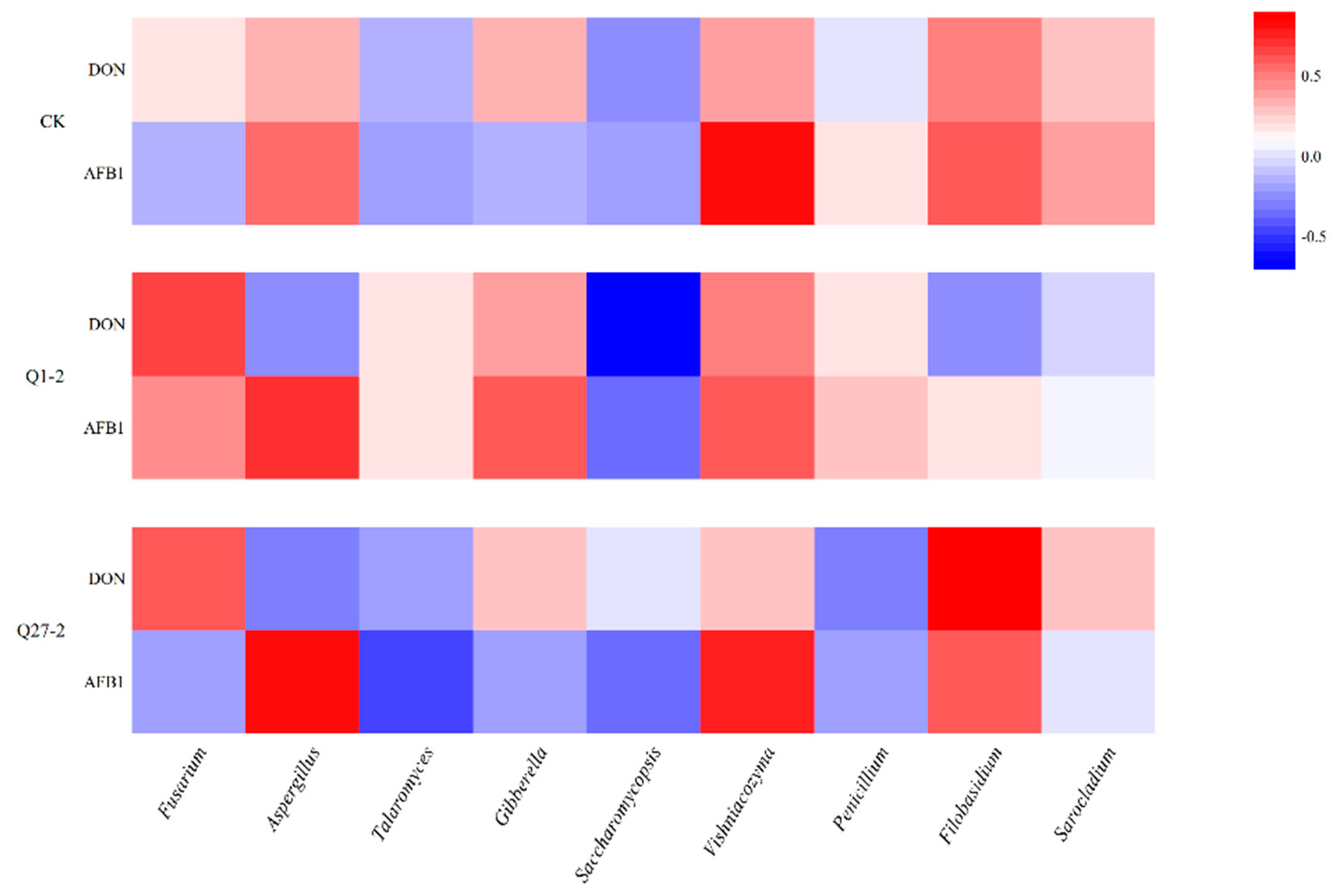
2.6. Microbial Diversity and Community Analysis during Fermentation
2.7. The Contents of Mycotoxins Aflatoxin B1 and Deoxynivalenol during Fermentation
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. LAB sample Collection and Isolation
5.2. Screening of LAB
5.2.1. Screening for Antibacterial LAB Strains
5.2.2. Screening for Antifungal LAB Strains
5.2.3. Screening LAB strains for Mycotoxin Removing Capacity
5.2.4. Adsorption Capacity of Lactic Acid Bacteria Inactivated by Heat
5.3. DNA Extraction and 16S rRNA Gene Amplification and Sequencing
5.4. Biological Characteristics of LAB Strains
5.5. Fermentation Feed Production
5.6. Chemical Composition Analysis
5.7. Bacterial and Fungal Community Analysis
5.8. Determination of AFB1 and DON Mycotoxins in Fermented Feed
5.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, R.; Kiarie, E.G.; Yiannikouris, A.; Sun, L.; Karrow, N.A. Nutritional impact of mycotoxins in food animal production and strategies for mitigation. J. Anim. Sci. Biotechnol. 2022, 13, 69. [Google Scholar] [CrossRef] [PubMed]
- Schatzmayr, G.; Zehner, F.; Täubel, M.; Schatzmayr, D.; Klimitsch, A.; Loibner, A.P.; Binder, E.M. Microbiologicals for deactivating mycotoxins. Mol. Nutr. Food Res. 2006, 50, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Amal, S.H.; Soher, E. Aly Biological detoxification of mycotoxins: A review. Ann. Microbiol. 2014, 64, 905–919. [Google Scholar]
- Loi, M.; Fanelli, F.; Liuzzi, V.C.; Logrieco, A.F.; Mulè, G. Mycotoxin Biotransformation by Native and Commercial Enzymes: Present and Future Perspectives. Toxins 2017, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, C.; Lyu, Y.; Cheng, W.; Guo, P. Advances in Biodegradation of Aflatoxins. Agric. Sci. Technol. 2016, 17, 951–955. [Google Scholar]
- Ma, R.; Zhang, L.; Liu, M.; Su, Y.-T.; Xie, W.-M.; Zhang, N.-Y.; Dai, J.-F.; Wang, Y.; Rajput, S.A.; Qi, D.-S.; et al. Individual and Combined Occurrence of Mycotoxins in Feed Ingredients and Complete Feeds in China. Toxins 2018, 10, 113. [Google Scholar] [CrossRef]
- Haque, A.M.; Wang, Y.; Shen, Z.; Li, X.; Saleemi, M.K.; He, C. Mycotoxin contamination and control strategy in human, domestic animal and poultry: A review. Microb. Pathog. 2020, 142, 104095. [Google Scholar] [CrossRef] [PubMed]
- Kosicki, R.; Błajet-Kosicka, A.; Grajewski, J.; Twarużek, M. Multiannual mycotoxin survey in feed materials and feedingstuffs. Anim. Feed Sci. Technol. 2016, 215, 165–180. [Google Scholar] [CrossRef]
- Huang, G.; Huang, J. Analysis of mycotoxin contamination in feed and feed raw materials in 21 provinces and cities of China in 2017. Swine Prod. 2017, 11, 46–50. [Google Scholar]
- Marroquín-Cardona, A.G.; Johnson, N.M.; Phillips, T.D.; Hayes, A.W. Mycotoxins in a changing global environment–A review. Food Chem. Toxicol. 2014, 69, 220–230. [Google Scholar] [CrossRef]
- Fan, X.; Zhao, S.; Yang, F.; Wang, Y.; Wang, Y. Effects of Lactic Acid Bacterial Inoculants on Fermentation Quality, Bacterial Community, and Mycotoxins of Alfalfa Silage under Vacuum or Nonvacuum Treatment. Microorganisms 2021, 9, 2614. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Bartkevics, V.; Mozuriene, E.; Lele, V.; Zadeike, D.; Juodeikiene, G. The safety, technological, nutritional, and sensory challenges associated with Lacto-Fermentation of meat and meat products by using pure lactic acid bacteria strains and Plant-Lactic acid bacteria bioproducts. Front. Microbiol. 2019, 10, 1036. [Google Scholar] [CrossRef] [PubMed]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Siedler, S.; Balti, R.; Neves, A.R. Bioprotective mechanisms of lactic acid bacteria against fungal spoilage of food. Curr. Opin. Biotechnol. 2019, 56, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Siedler, S.; Rau, M.H.; Bidstrup, S.; Vento, J.M.; Aunsbjerg, S.D.; Bosma, E.F.; Mcnair, L.M.; Beisel, C.L.; Neves, A.R. Competitive exclusion is a major bioprotective mechanism of Lactobacilli against fungal spoilage in fermented milk products. Appl. Environ. Microbiol. 2020, 86, e02312–e02319. [Google Scholar] [CrossRef]
- Axel, C.; Zannini, E.; Arendt, E.K. Mold spoilage of bread and its biopreservation: A review of current strategies for bread shelf life extension. Crit. Rev. Food Sci. Nutr. 2017, 57, 3528–3542. [Google Scholar] [CrossRef]
- Parappilly, S.J.; Idicula, D.V.; Chandran, A.; Radhakrishnan, K.M.; George, S.M. Antifungal activity of human gut lactic acid bacteria against aflatoxigenic Aspergillus flavus MTCC 2798 and their potential application as food biopreservative. J. Food Saf. 2021, 41, e12942. [Google Scholar] [CrossRef]
- Crowley, S.; Mahony, J.; Sinderen, D. Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends Food Sci. Technol. 2013, 33, 93–109. [Google Scholar] [CrossRef]
- Luz, C.; Ferrer, J.; Mañes, J.; Meca, G. Toxicity reduction of ochratoxin A by lactic acid bacteria. Food Chem. Toxicol. 2018, 112, 60–66. [Google Scholar] [CrossRef]
- Bangar, S.P.; Sharma, N.; Kumar, M.; Ozogul, F.; Purewal, S.S.; Trif, M. Recent developments in applications of lactic acid bacteria against mycotoxin production and fungal contamination. Food Biosci. 2021, 44, 101444. [Google Scholar] [CrossRef]
- Hernandez-Mendoza, A.; Garcia, H.S.; Steele, J.L. Screening of Lactobacillus casei strains for their ability to bind aflatoxin B1. Food Chem. Toxicol. 2009, 47, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Li, J.; Dong, Z.; Shao, T. The reconstitution mechanism of napier grass microiota during the ensiling of alfalfa and their contributions to fermentation quality of silage. Bioresour. Technol. 2020, 297, 122391. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Li, B.; Zheng, M.; Niu, D.; Xu, C. Effects of Pediococcus pentosaceus on fermentation, aerobic stability and microbial communities during ensiling and aerobic spoilage of total mixed ration silage containing alfalfa (Medicago sativa L.). Grassl. Sci. 2020, 66, 215–224. [Google Scholar] [CrossRef]
- Muck, R.E.; Nadeau, E.M.G.; McAllister, T.A.; Contreras-Govea, F.E.; Santos, M.C.; Kung, L. Silage review: Recent advances and future uses of silage additives. J. Dairy Sci. 2018, 101, 3980–4000. [Google Scholar] [CrossRef]
- McGarvey, J.A.; Franco, R.B.; Palumbo, J.D.; Hnasko, R.; Stanker, L.; Mitloehner, F.M. Bacterial population dynamics during the ensiling of Medicago sativa (alfalfa) and subsequent exposure to air. J. Appl. Microbiol. 2013, 114, 1661–1670. [Google Scholar] [CrossRef]
- Wambacq, E.; Vanhoutte, I.; Audenaert, K.; Gelder, L.D.; Haesaert, G. Occurrence, prevention and remediation of toxigenic fungi and mycotoxins in silage: A review. J. Sci. Food Agric. 2016, 96, 2284–2302. [Google Scholar] [CrossRef] [PubMed]
- Ahlberg, S.; Joutsjoki, V.; Laurikkala, S.; Varmanen, P.; Korhonen, H. Aspergillus flavus growth inhibition by Lactobacillus strains isolated from traditional fermented Kenyan milk and maize products. Arch. Microbiol. 2016, 199, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.Z.; Abdelall, M.F.; El-Mahdy, O.M. Detoxification of Aflatoxin B1 by Antifungal Compounds from Lactobacillus brevis and Lactobacillus paracasei, Isolated from Dairy Products. Probiotics Antimicrob. Proteins 2018, 10, 201–209. [Google Scholar] [CrossRef] [PubMed]
- El-Nezami, H.; Kankaanpaa, P.; Salminen, S.; Ahokas, J. Ability of dairy strains of lactic acid bacteria to bind a common food carcinogen, aflatoxin B1. Food Chem. Toxicol. 1998, 36, 321–326. [Google Scholar] [CrossRef]
- Silva, V.P.; Pereira, O.G.; Leandro, E.S.; Da Silva, T.C.; Ribeiro, K.G.; Mantovani, H.C.; Santos, S.A. Effects of lactic acid bacteria with bacteriocinogenic potential on the fermentation profile and chemical composition of alfalfa silage in tropical conditions. J. Dairy Sci. 2016, 99, 1895–1902. [Google Scholar] [CrossRef]
- Ni, K.; Wang, Y.; Li, D.; Cai, Y.; Pang, H. Characterization, identification and application of lactic acid bacteria isolated from forage paddy rice silage. PLoS ONE 2015, 10, e0121967. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.N.; Ferreira, G.; Teets, C.L.; Thomason, W.E.; Teutsch, C.D. Nutritional composition and in vitro digestibility of grass and legume winter (cover) crops. J. Dairy Sci. 2018, 101, 2037–2047. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; An, Y.; Wang, G.; Bian, S. Effect of silaging temperature on the pH and nutrition of Medicago sativa silage. Acta Prataculturae Sinica 2005, 14, 82–86. [Google Scholar]
- Jung, J.S.; Ravindran, B.; Soundharrajan, I.; Awasthi, M.K.; Choi, K.C. Improved performance and microbial community dynamics in anaerobic fermentation of triticale silages at different stages. Bioresour. Technol. 2022, 345, 126485. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Risu, N.; Ge, G.; Jia, Y.; Cai, Y. Dynamic changes and characterization of the protein and carbohydrate fractions of native grass grown in Inner Mongolia during ensiling and the aerobic stage. Asian Australas. J. Anim. Sci. 2020, 33, 556–567. [Google Scholar] [CrossRef]
- Moore, J.E.; Kunkle, W.E. Evaluation of equations for estimating voluntary intake of forages and forage-based diets. J. Anim. Sci. 1999, 1, 204. [Google Scholar]
- Ni, K.; Wang, X.; Lu, Y.; Guo, L.; Li, X.; Yang, F. Exploring the silage quality of alfalfa ensiled with the residues of astragalus and hawthorn. Bioresour. Technol. 2020, 297, 122249. [Google Scholar] [CrossRef] [PubMed]
- Ezeji, J.C.; Sarikonda, D.K.; Hopperton, A.; Erkkila, H.L.; Cohen, D.E.; Martinez, S.P.; Rodriguez-Palacios, A. Parabacteroides distasonis: Intriguing aerotolerant gut anaerobe with emerging antimicrobial resistance and pathogenic and probiotic roles in human health. Gut Microbes 2021, 13, 1922241. [Google Scholar] [CrossRef]
- Ogunade, I.M.; Martinez-Tuppia, C.; Queiroz, O.C.M.; Jiang, Y.; Drouin, Y.; Wu, F.; Vyas, D.; Adesogan, A.T. Silage review: Mycotoxins in silage: Occurrence, effects, prevention, and mitigation. J. Dairy Sci. 2018, 101, 4034–4059. [Google Scholar] [CrossRef] [PubMed]
- Zychowski, K.E.; Pohlenz, C.; Mays, T.; Romoser, A.; Hume, M.; Buentello, A.; Gatlin, D.M.; Phillips, T.D. The effect of NovaSil dietary supplementation on the growth and health performance of Nile tilapia (Oreochromis niloticus) fed aflatoxin-B1 contaminated feed. Aquaculture 2013, 376–379, 117–123. [Google Scholar] [CrossRef]
- Wang, M.; Xu, S.; Wang, T.; Jia, T.; Xu, Z.; Wang, X.; Yu, Z. Effect of inoculants and storage temperature on the microbial, chemical and mycotoxin composition of corn silage. Asian Australas. J. Anim. Sci. 2018, 31, 1903–1912. [Google Scholar] [CrossRef]
- Ferrero, F.; Prencipe, S.; Spadaro, D.; Gullino, M.L.; Cavallarin, L.; Piano, S.; Tabacco, E.; Borreani, G. Increase in aflatoxins due to Aspergillus section Flavi multiplication during the aerobic deterioration of corn silage treated with different bacteria inocula. J. Dairy Sci. 2019, 102, 1176–1193. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tan, Z.; Wu, G.; Wang, L.; Qin, G.; Wang, Y.; Pang, H. Microbial community and fermentation characteristic of whole-crop wheat silage treated by lactic acid bacteria and Artemisia argyi during ensiling and aerobic exposure. Front. Microbiol. 2022, 13, 1004495. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shen, Q.; Zhou, W.; Mo, H.; Pan, D.; Hu, L. Nanocapsular Dispersion of Cinnamaldehyde for Enhanced Inhibitory Activity against Aflatoxin Production by Aspergillus flavus. Molecules 2015, 20, 6022–6032. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, A.; Ebrahimi, M.; Mortazavi, S.A.; Abedfar, A. Application of the selected antifungal LAB isolate as a protective starter culture in pan whole-wheat sourdough bread. Food Control. 2019, 95, 298–307. [Google Scholar] [CrossRef]
- Zuo, R.; Chang, J.; Yin, Q.; Wang, P.; Yang, Y.; Wang, X.; Wang, G.; Zheng, Q. Effect of the combined probiotics with aflatoxin B1-degrading enzyme on aflatoxin detoxification, broiler production performance and hepatic enzyme gene expression. Food Chem. Toxicol. 2013, 59, 470–475. [Google Scholar] [CrossRef]
- Chen, J.; Pang, H.; Wang, L.; Ma, C.; Wu, G.; Liu, Y.; Guan, Y.; Zhang, M.; Qin, G.; Tan, Z. Bacteriocin-Producing Lactic Acid Bacteria Strains with Antimicrobial Activity Screened from Bamei Pig Feces. Foods 2022, 11, 709. [Google Scholar] [CrossRef]
- Magnusson, J. Antifungal Activity of Lactic Acid Bacteria. Ph.D. Thesis, Swedish University of Agricultural Sciences, Ultuna, Sweden, 2003. [Google Scholar]
- Wang, W.; Ma, H.; Yu, H.; Qin, G.; Tan, Z.; Wang, Y.; Pang, H. Screening of Lactobacillus plantarum Subsp. plantarum with Potential Probiotic Activities for Inhibiting ETEC K88 in Weaned Piglets. Molecules 2020, 25, 4481. [Google Scholar] [CrossRef]
- Li, Y.; Du, S.; Sun, L.; Cheng, Q.; Hao, J.; Lu, Q.; Ge, G.; Wang, Z.; Jia, Y. Effects of Lactic Acid Bacteria and Molasses Additives on Dynamic Fermentation Quality and Microbial Community of Native Grass Silage. Front. Microbiol. 2022, 13, 830121. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Pian, R.; Yang, F.; Chen, X.; Zhang, Q. The sustainable mitigation of ruminal methane and carbon dioxide emissions by co-ensiling corn stalk with Neolamarckia cadamba leaves for cleaner livestock production. J. Clean. Prod. 2021, 311, 127680. [Google Scholar] [CrossRef]




| Indicators | Q1-2 | Q27-2 | Q12-9 | Q12-4 |
|---|---|---|---|---|
| Aspergillus niger | + | ++ | ++ | ++ |
| Aspergillus flavus | ++ | + | − | − |
| Aspergillus oryzae | + | + | ++ | − |
| Penicillium citrus | + | ++ | + | ++ |
| Trichoderma | ++ | + | − | + |
| Strains | AFB1 Binding (% of Total) | DON Binding (% of Total) | ||
|---|---|---|---|---|
| Viable | Heat Inactivation | Viable | Heat Inactivation | |
| Q1-2 | 48.21 ± 2.83c | 56.33 ± 1.60b | 63.40 ± 1.83a | 64.27 ± 1.22a |
| Q27-2 | 52.82 ± 3.24c | 53.21 ± 1.57c | 61.52 ± 2.59b | 69.36 ± 0.95a |
| Strains | Concentration of NaCl (%) | Temperature (°C) | pH | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 7 | 5 | 10 | 30 | 45 | 50 | 3 | 3.5 | 4 | 5 | 6 | 8 | 9 | 10 | |
| Q1-2 | + | + | + | + | + | w | − | w | + | + | + | + | + | + | + |
| Q27-2 | w | + | + | + | + | + | w | − | w | + | + | + | + | + | + |
| Items | Treatment | Fermenting Days | SEM | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 d | 3 d | 7 d | 15 d | 30 d | T | D | T × D | |||
| LA (%DM) | CK | 0.26 Ac | 0.23 Bc | 0.64 Cc | 1.33 Cb | 2.56 Ba | 0.149 | <0.05 | NS | NS |
| Q1-2 | 0.25 Ae | 1.29 Ad | 1.92 Ac | 2.55 Ab | 4.06 Aa | |||||
| Q27-2 | 0.26 Ad | 1.17 Acd | 1.51 Bc | 2.09 Bb | 4.26 Aa | |||||
| AA (%DM) | CK | ND | 0.01 | 0.31 | 0.66 | 1.49 A | 0.049 | <0.05 | <0.05 | <0.05 |
| Q1-2 | ND | 0.06 | 0.25 | 0.37 | 0.74 B | |||||
| Q27-2 | ND | 0.07 | 0.14 | 0.27 | 0.38 C | |||||
| PA (%DM) | CK | ND | 0.31 | 0.27 | 0.16 | 0.55 | - | - | - | - |
| Q1-2 | ND | 0.09 | ND | ND | ND | |||||
| Q27-2 | ND | 0.17 | ND | ND | ND | |||||
| BA (%DM) | CK | 0.15 | 0.24 | 0.34 | 0.55 | 0.71 | - | - | - | - |
| Q1-2 | ND | 0.12 | 0.07 | ND | ND | |||||
| Q27-2 | ND | 0.09 | ND | ND | ND | |||||
| NH3–N (g/kg DM) | CK | 5.81 Ab | 6.55 Aa | 5.16 Ac | 5.34 Ac | 5.15 Ac | 0.081 | <0.05 | <0.05 | NS |
| Q1-2 | 4.75 Bb | 5.39 Ba | 4.74 Bb | 4.27 Bc | 3.83 Bc | |||||
| Q27-2 | 4.37 Cb | 4.64 Ca | 3.84 Cc | 3.55 Cd | 3.31 Be | |||||
| Items | Treatment | Fermenting Days | SEM | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 d | 3 d | 7 d | 15 d | 30 d | T | D | T × D | |||
| DM (%FM) | CK | 42.58 | 42.46 | 43.07 | 43.10 | 42.80 | 0.7110 | NS | NS | NS |
| Q1-2 | 43.98 | 43.01 | 43.51 | 42.58 | 43.92 | |||||
| Q27-2 | 42.28 | 42.95 | 43.11 | 42.04 | 42.09 | |||||
| CP (%DM) | CK | 19.47 | 19.03 | 18.82 | 18.27 | 17.92 | 0.0670 | <0.05 | <0.05 | <0.05 |
| Q1-2 | 19.27 | 18.80 | 18.36 | 17.71 | 17.19 | |||||
| Q27-2 | 19.31 | 18.78 | 18.21 | 17.39 | 16.86 | |||||
| NDF (%DM) | CK | 50.71 | 45.12 | 44.30 | 44.00 | 41.63 | 0.6400 | NS | <0.05 | NS |
| Q1-2 | 50.65 | 46.68 | 44.62 | 44.60 | 41.88 | |||||
| Q27-2 | 50.52 | 48.80 | 44.55 | 44.19 | 42.07 | |||||
| ADF (%DM) | CK | 35.69 | 31.50 | 28.82 | 27.58 | 26.76 | 0.3180 | NS | <0.05 | NS |
| Q1-2 | 36.23 | 31.06 | 29.03 | 27.50 | 26.47 | |||||
| Q27-2 | 36.16 | 31.90 | 28.99 | 27.55 | 26.63 | |||||
| Items | Ingredients Composition, % of DM |
|---|---|
| Alfalfa | 46.5 |
| Corn | 30 |
| Wheat bran | 10 |
| Soybean meal | 10 |
| Soybean oil | 2 |
| Premixture | 1 |
| Sodium chloride | 0.5 |
| Total | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, Y.; Lv, H.; Wu, G.; Chen, J.; Wang, M.; Zhang, M.; Pang, H.; Duan, Y.; Wang, L.; Tan, Z. Effects of Lactic Acid Bacteria Reducing the Content of Harmful Fungi and Mycotoxins on the Quality of Mixed Fermented Feed. Toxins 2023, 15, 226. https://doi.org/10.3390/toxins15030226
Guan Y, Lv H, Wu G, Chen J, Wang M, Zhang M, Pang H, Duan Y, Wang L, Tan Z. Effects of Lactic Acid Bacteria Reducing the Content of Harmful Fungi and Mycotoxins on the Quality of Mixed Fermented Feed. Toxins. 2023; 15(3):226. https://doi.org/10.3390/toxins15030226
Chicago/Turabian StyleGuan, Yifei, Haoxin Lv, Guofang Wu, Jun Chen, Min Wang, Miao Zhang, Huili Pang, Yaoke Duan, Lei Wang, and Zhongfang Tan. 2023. "Effects of Lactic Acid Bacteria Reducing the Content of Harmful Fungi and Mycotoxins on the Quality of Mixed Fermented Feed" Toxins 15, no. 3: 226. https://doi.org/10.3390/toxins15030226
APA StyleGuan, Y., Lv, H., Wu, G., Chen, J., Wang, M., Zhang, M., Pang, H., Duan, Y., Wang, L., & Tan, Z. (2023). Effects of Lactic Acid Bacteria Reducing the Content of Harmful Fungi and Mycotoxins on the Quality of Mixed Fermented Feed. Toxins, 15(3), 226. https://doi.org/10.3390/toxins15030226







