Two-Step Epimerization of Deoxynivalenol by Quinone-Dependent Dehydrogenase and Candida parapsilosis ACCC 20221
Abstract
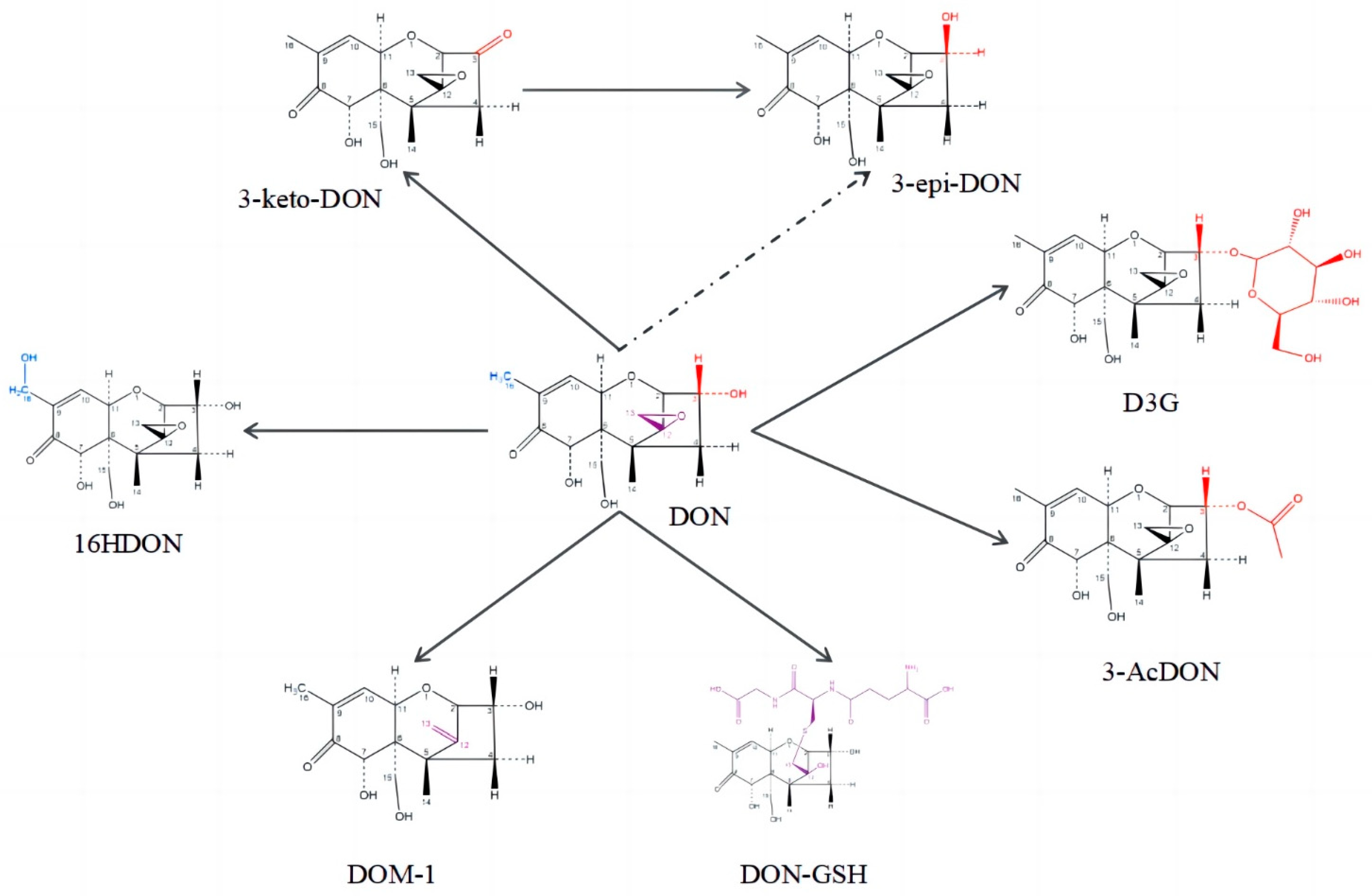
:1. Introduction
2. Results
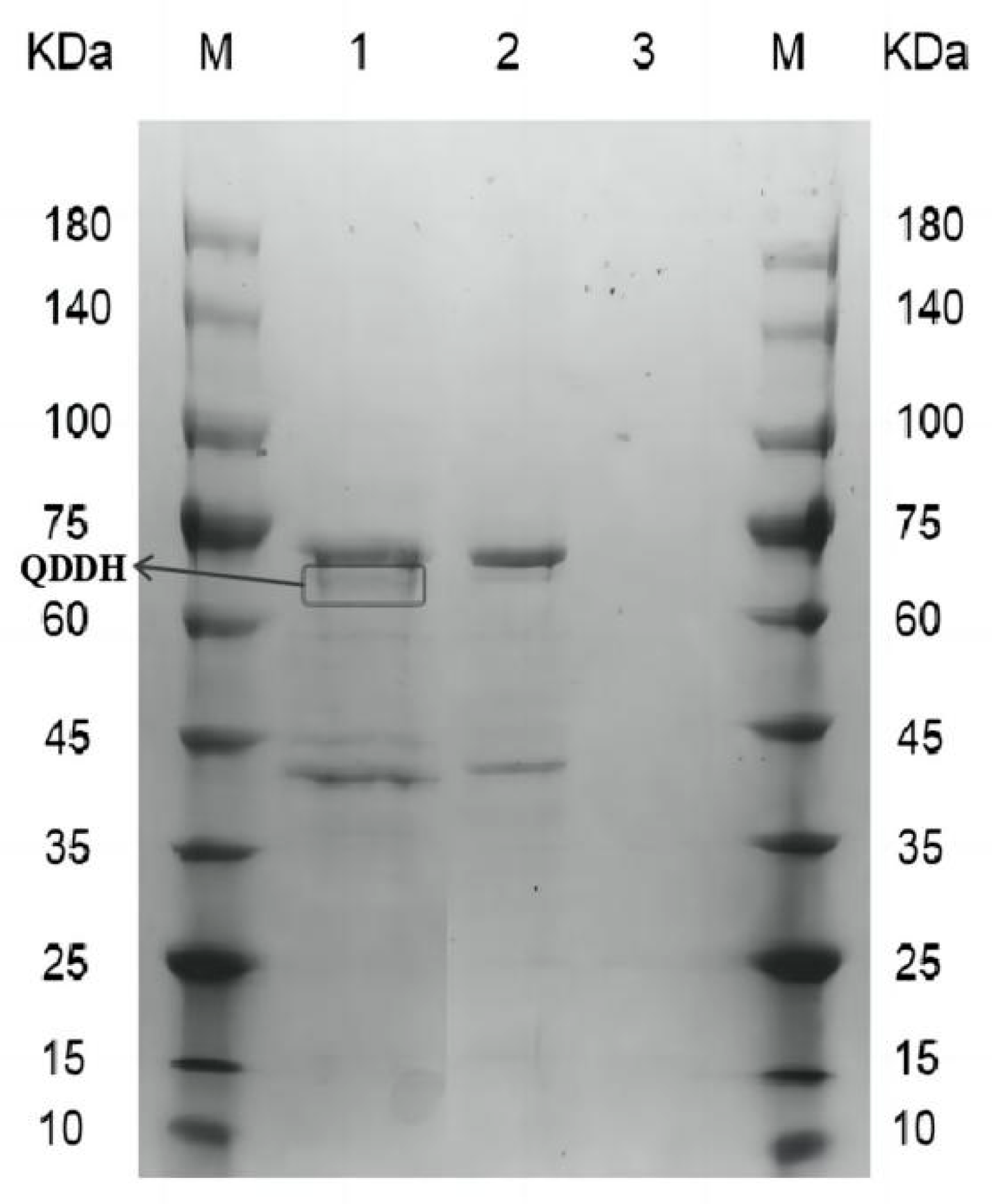
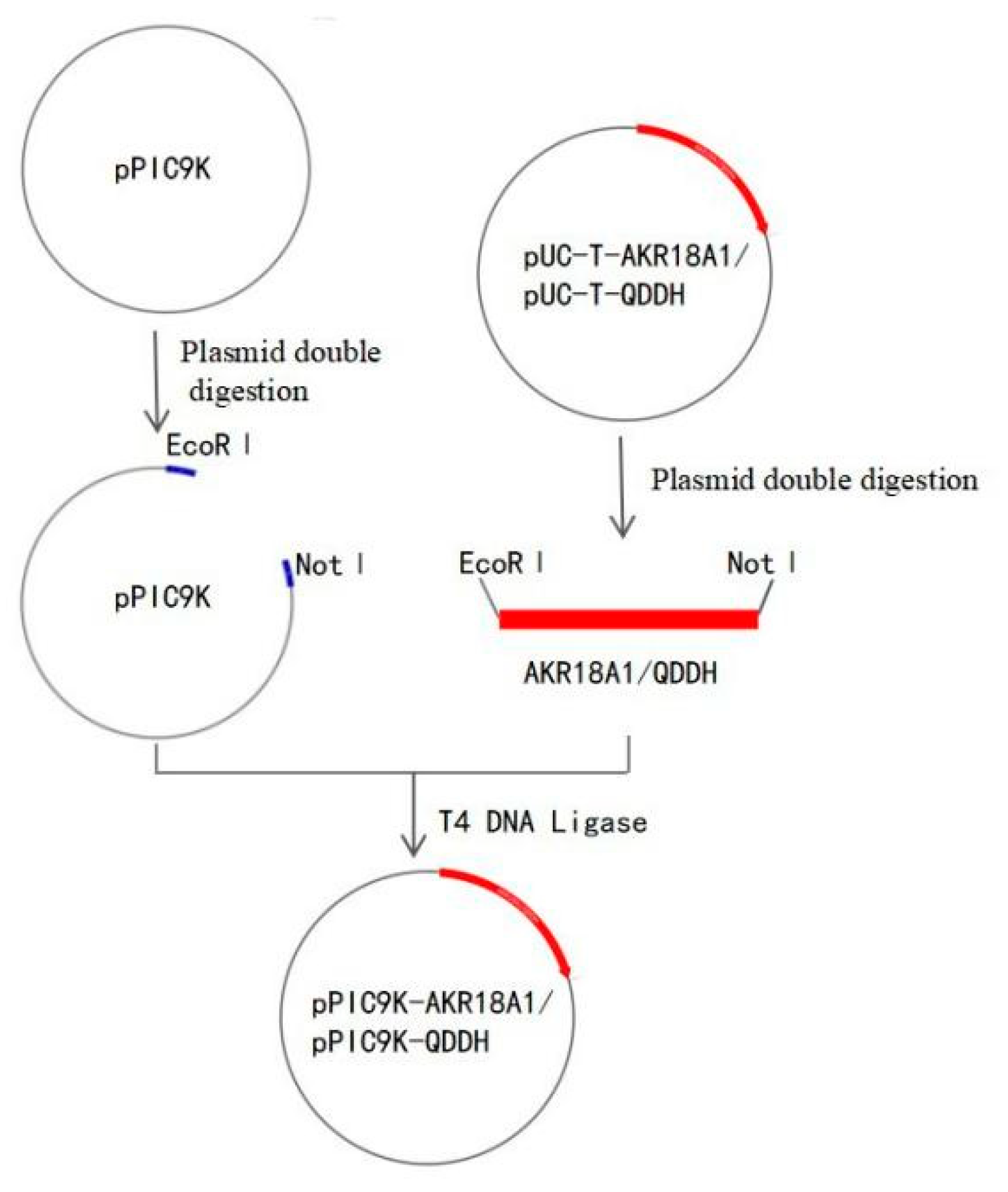
2.1. Recombinant Oxidase Expressed in Pichia pastoris
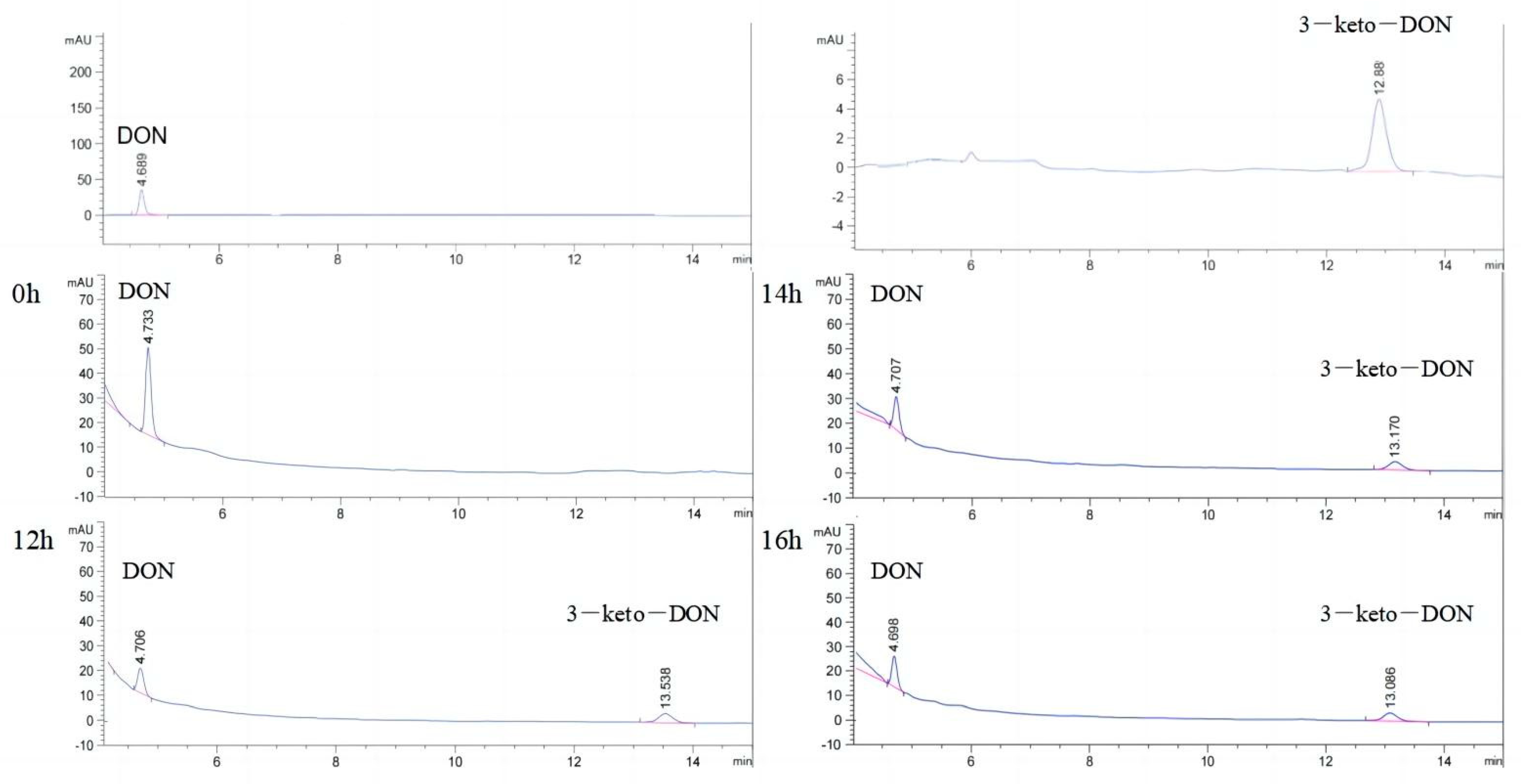
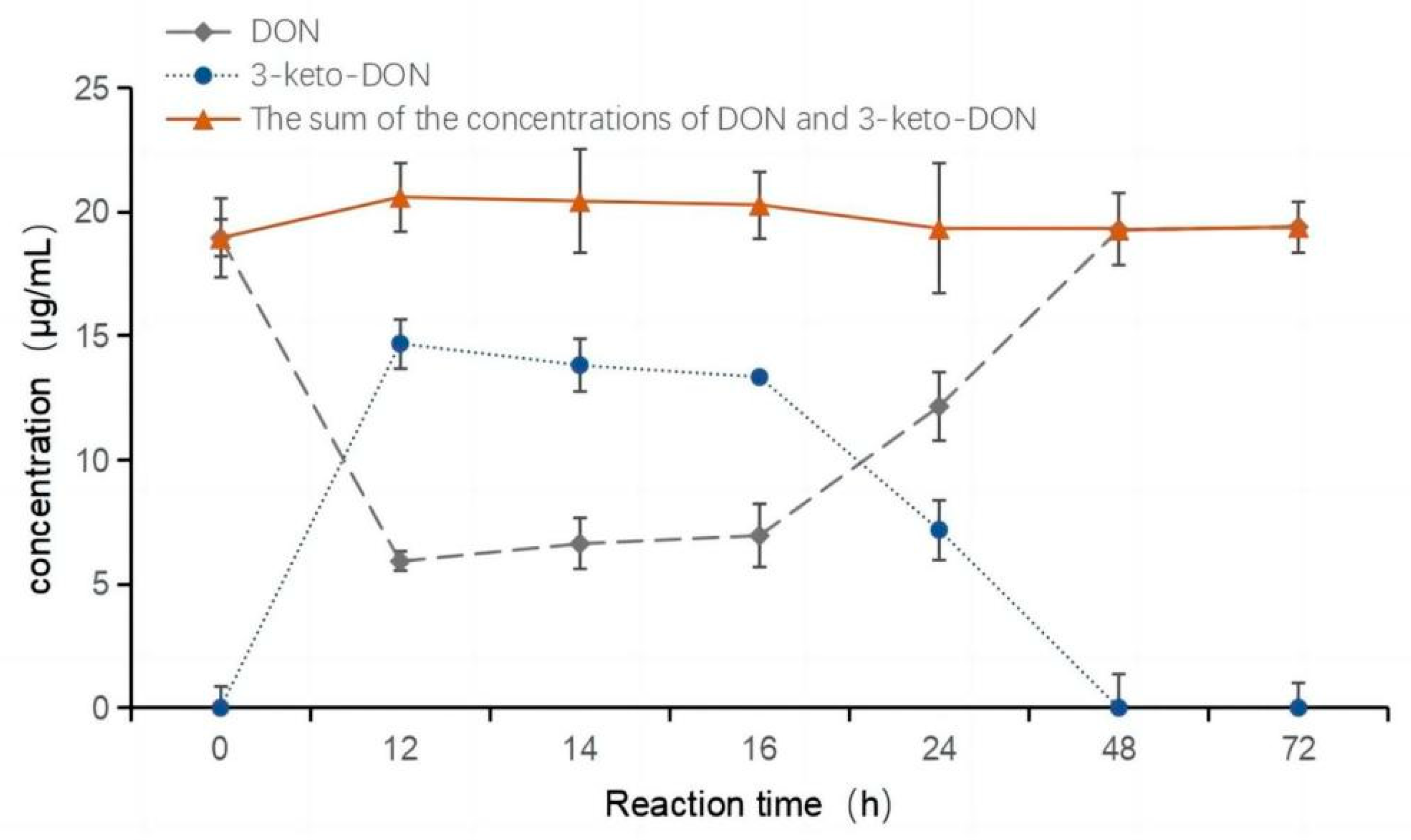
2.2. Oxidation of DON to 3-keto-DON
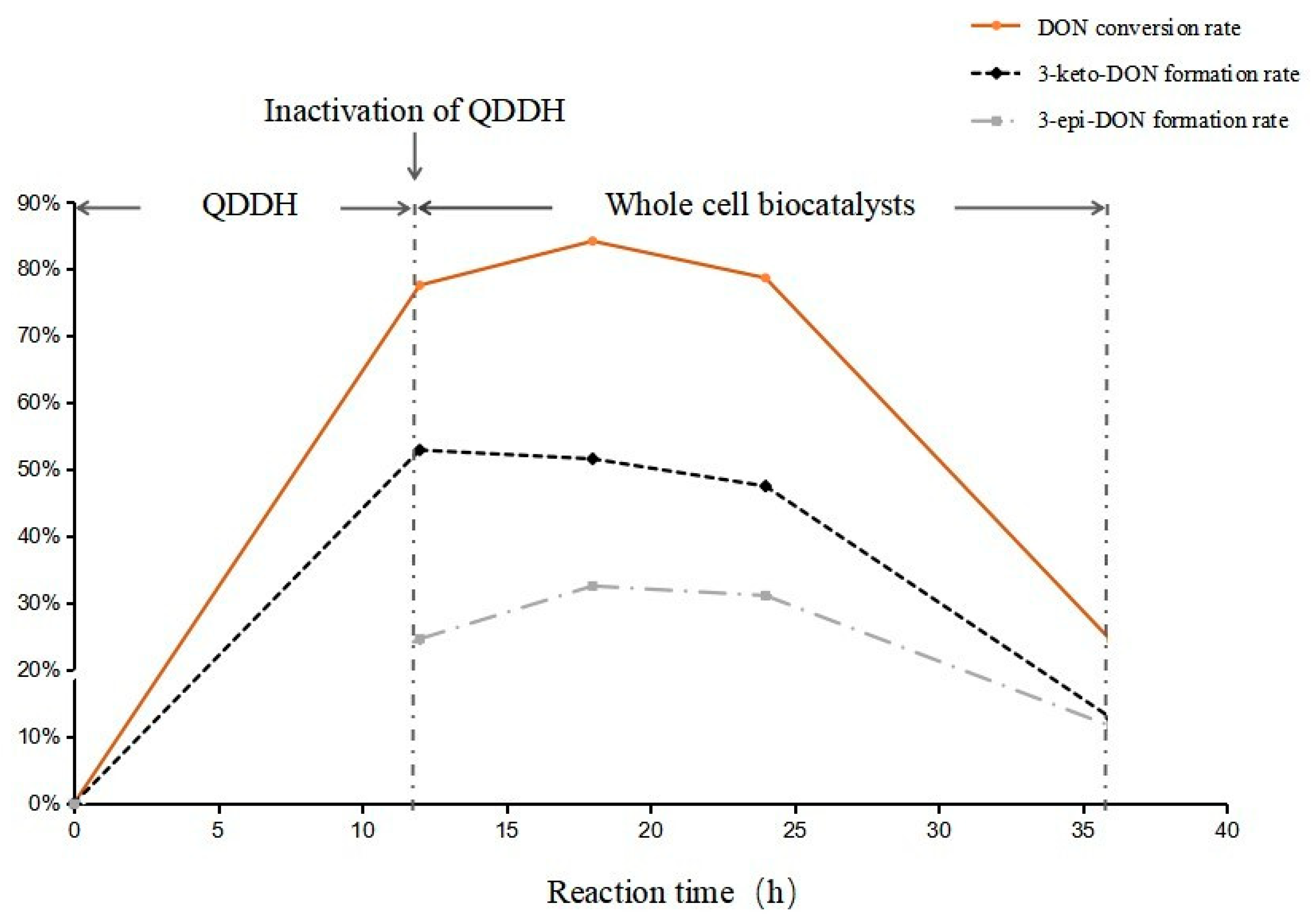
2.3. Highly Active Candida parapsilosis Strain Catabolizes 3-keto-DON into 3-epi-DON
| Strains | Enzyme | Substrates | Main Products | References |
|---|---|---|---|---|
| Candida parapsilosis | SCRII | 2-hydroxyacetophenone | (S)-phenyl ethanediol | [46,47] |
| CpCR | Benzaldehyde | (R)-Benzylalcohol | [49] | |
| CPR-C1 | ethyl ketopantoyl lactone | ethyl d-pantoyl lactone | [51] | |
| CPR-C2 | ketopantoyl lactone | d-pantoyl lactone | [48] | |
| Yarrowia lipolytica | YaCRI | 2-hydroxyacetophenone | (S)-1-phenyl-1,2-ethanediol | [53] |
| YaCRⅡ | ||||
| YlCR | 2-propanol | (R)-2-chloro-1-phenylethol | [54] | |
| YlCR2 | COBE * | (S)-CHBE * | [52] | |
| Candida magnoliae | CmCR | COBE * | (S)-CHBE * | [57] |
| S1 | COBE * | (S)-CHBE * | [58] | |
| Pichia pastoris | PsCRI | COBE * | (S)-CHBE * | [56] |
| PsCRII | COBE * | (S)-CHBE * | [55] | |
| Rhodococcus R6 | ReADH | 2-hydroxyacetophenone | (R)-1-phenyl-1,2-ethanediol | [60] |
| Rhodococcus pyridinivorans | - | para-acetylphenol | S-1-(para-hydroxyphenyl)ethanol | [61] |
| RpCR | COBE * | (S)-CHBE * | [62] | |
| Rhodococcus sp. ECU1014 | RhCR | epsilon-ketoester ethyl 8-chloro-6-oxooctanoate | ethyl (S)-8-chloro-6-hydroxyoctanoate | [63] |
2.4. Catabolite Identification of 3-keto-DON by LC–MS
2.5. Epimerization of DON
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Strains
5.2. Cloning, Expression, and Purification of AKR18A1 and QDDH in Pichia pastoris
5.3. Analysis of Recombinant Protein for DON Oxidation Activity
5.4. HPLC Analysis
5.5. Analysis of Candidate Strains for 3-keto-DON Reduction Activity
5.6. Liquid Chromatography–Mass Spectroscopy (LC–MS)
5.7. Catalytic Epimerization of Deoxynivalenol
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gruber-Dorninger, C.; Jenkins, T.; Schatzmayr, G. Global Mycotoxin Occurrence in Feed: A Ten-Year Survey. Toxins 2019, 11, 375. [Google Scholar] [CrossRef]
- Piacentini, K.C.; Rocha, L.O.; Savi, G.D.; Carnielli-Queiroz, L.; Almeida, F.G.; Minella, E.; Corrêa, B. Occurrence of deoxynivalenol and zearalenone in brewing barley grains from Brazil. Mycotoxin. Res. 2018, 34, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J.; Guan, X.L.; Zong, Y.; Hua, X.T.; Xing, F.G.; Wang, Y.; Wang, F.Z.; Liu, Y. Deoxynivalenol in wheat from the Northwestern region in China. Food Addit. Contam. B 2018, 11, 281–285. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, K.; Long, M.; Yang, S.; Zhang, Y. An update on immunotoxicity and mechanisms of action of six environmental mycotoxins. Food Chem. Toxicol. 2022, 163, 112895. [Google Scholar] [CrossRef]
- Guan, S.; He, J.W.; Young, J.C.; Zhu, H.H.; Li, X.Z.; Ji, C.; Zhou, T. Transformation of trichothecene mycotoxins by microorganisms from fish digesta. Aquaculture 2009, 290, 290–295. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Man, H.; Lee, Y.; Shi, J.; Xu, J. Metabolomics-guided analysis reveals a two-step epimerization of deoxynivalenol catalyzed by the bacterial consortium IFSN-C1. Appl. Microbiol. Biot. 2020, 104, 6045–6056. [Google Scholar] [CrossRef]
- Long, M.; Chen, X.L.; Zhang, Y.; Li, P.; He, J.B. Identification and characterization of deoxynivalenol-degrading bacterial strain Bacillus circulans C1-5-9. Fresen. Environ. Bull. 2016, 25, 2427–2435. [Google Scholar]
- He, W.J.; Shi, M.M.; Yang, P.; Huang, T.; Zhao, Y.; Wu, A.B.; Dong, W.B.; Li, H.P.; Zhang, J.B.; Liao, Y.C. A quinone-dependent dehydrogenase and two NADPH-dependent aldo/keto reductases detoxify deoxynivalenol in wheat via epimerization in a Devosia strain. Food Chem. 2020, 321, 126703. [Google Scholar] [CrossRef]
- Carere, J.; Hassan, Y.I.; Lepp, D.; Zhou, T. The Identification of DepB: An Enzyme Responsible for the Final Detoxification Step in the Deoxynivalenol Epimerization Pathway in Devosia mutans 17-2-E-8. Front. Microbiol. 2018, 9, 1573. [Google Scholar] [CrossRef]
- He, J.W.; Bondy, G.S.; Zhou, T.; Caldwell, D.; Boland, G.J.; Scott, P.M. Toxicology of 3-epi-deoxynivalenol, a deoxynivalenol-transformation product by Devosia mutans 17-2-E-8. Food Chem. Toxicol. 2015, 84, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, Y.; Ji, F.; Xu, L.; Yu, M.; Shi, J.; Xu, J. Biodegradation of deoxynivalenol and its derivatives by Devosia insulae A16. Food Chem. 2019, 276, 436–442. [Google Scholar] [CrossRef]
- Ikunaga, Y.; Sato, I.; Grond, S.; Numaziri, N.; Yoshida, S.; Yamaya, H.; Hiradate, S.; Hasegawa, M.; Toshima, H.; Koitabashi, M.; et al. Nocardioides sp. strain WSN05-2, isolated from a wheat field, degrades deoxynivalenol, producing the novel intermediate 3-epi-deoxynivalenol. Appl. Microbiol. Biot. 2011, 89, 419–427. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.H.; Zhao, C.; Han, Y.T.; Liu, Y.C.; Zhang, X.L. Isolation and characterization of a novel deoxynivalenol-transforming strain Paradevosia shaoguanensis DDB001 from wheat field soil. Lett. Appl. Microbiol. 2017, 65, 414–422. [Google Scholar] [CrossRef] [PubMed]
- He, W.J.; Zhang, L.; Yi, S.Y.; Tang, X.L.; Yuan, Q.S.; Guo, M.W.; Wu, A.B.; Qu, B.; Li, H.P.; Liao, Y.C. An aldo-keto reductase is responsible for Fusarium toxin-degrading activity in a soil Sphingomonas strain. Sci. Rep. 2017, 7, 9549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qin, X.; Guo, Y.; Zhang, Q.; Ma, Q.; Ji, C.; Zhao, L. Enzymatic degradation of deoxynivalenol by a novel bacterium, Pelagibacterium halotolerans ANSP101. Food Chem. Toxicol 2020, 140, 126701–126703. [Google Scholar] [CrossRef]
- He, C.H.; Fan, Y.H.; Liu, G.F.; Zhang, H.B. Isolation and Identification of a Strain of Aspergillus tubingensis with Deoxynivalenol Biotransformation Capability. Int. J. Mol. Sci. 2008, 9, 2366–2375. [Google Scholar] [CrossRef]
- Yang, S.H.; Wu, Y.; Yang, J.; Yan, R.; Bao, Y.H.; Wang, K.P.; Liu, G.Q.; Wang, W. Isolation and identification of an extracellular enzyme from Aspergillus niger with Deoxynivalenol biotransformation capability. Emir. J. Food Agr. 2017, 29, 742–750. [Google Scholar] [CrossRef]
- Garda-Buffon, J.; Kupski, L.; Badiale-Furlong, E. Deoxynivalenol (DON) degradation and peroxidase enzyme activity in submerged fermentation. Cienc. E Tecnol. Aliment. 2011, 31, 198–203. [Google Scholar] [CrossRef]
- Cheng, B.C.; Wan, C.X.; Yang, S.L.; Xu, H.Y.; Wei, H.; Liu, J.S.; Tian, W.H.; Zeng, M. Detoxification of Deoxynivalenol by Bacillus Strains. J. Food Saf. 2010, 30, 599–614. [Google Scholar] [CrossRef]
- Jia, R.; Cao, L.R.; Liu, W.B.; Shen, Z.Y. Detoxification of deoxynivalenol by Bacillus subtilis ASAG 216 and characterization the degradation process. Eur. Food Res. Technol. 2021, 247, 67–76. [Google Scholar] [CrossRef]
- Zhang, C.; Yu, X.; Xu, H.; Cui, G.; Chen, L. Action of Bacillus natto 16 on deoxynivalenol (DON) from wheat flour. J. Appl. Microbiol. 2021, 131, 2317–2324. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Niu, J.; Yang, H.; Lu, Z.; Zhou, L.; Meng, F.; Lu, F.; Chen, M. Epimerization of deoxynivalenol by the Devosia strain A6-243 assisted by pyrroloquinoline quinone. Toxins 2022, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Y.I.; He, J.W.; Perilla, N.; Tang, K.J.; Karlovsky, P.; Zhou, T. The enzymatic epimerization of deoxynivalenol by Devosia mutans proceeds through the formation of 3-keto-DON intermediate. Sci. Rep. 2017, 7, 6929. [Google Scholar] [CrossRef]
- He, J.W.; Yang, R.; Zhou, T.; Boland, G.J.; Scott, P.M.; Bondy, G.S. An epimer of deoxynivalenol: Purification and structure identification of 3-epi-deoxynivalenol. Food Addit. Contam. A 2015, 32, 1523–1530. [Google Scholar] [CrossRef]
- Zhang, H.H.; Zhang, H.; Qin, X.; Wang, X.L.; Wang, Y.; Bin, Y.; Xie, X.M.; Zheng, F.; Luo, H.Y. Biodegradation of Deoxynivalenol by Nocardioides sp. ZHH-013: 3-keto-Deoxynivalenol and 3-epi-Deoxynivalenol as Intermediate Products. Front. Microbiol. 2021, 12, 658421. [Google Scholar] [CrossRef]
- Yu, H.; Zhou, T.; Gong, J.; Young, C.; Su, X.; Li, X.Z.; Zhu, H.; Tsao, R.; Yang, R. Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection. BMC Microbiol. 2010, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.L.; Ma, J.J.; Cheng, Y.Q.; Wang, H.A.; Sun, J.H.; Yan, Y.X. The toxicity mechanisms of DON to humans and animals and potential biological treatment strategies. Crit. Rev. Food Sci. 2021, 63, 790–812. [Google Scholar] [CrossRef]
- Hassan, Y.I.; Watts, C.; Li, X.Z.; Zhou, T. A Novel Peptide-Binding Motifs Inference Approach to Understand Deoxynivalenol Molecular Toxicity. Toxins 2015, 7, 1989–2005. [Google Scholar] [CrossRef]
- Pinto, A.; De Pierri, C.R.; Evangelista, A.G.; Gomes, A.; Luciano, F.B. Deoxynivalenol: Toxicology, Degradation by Bacteria, and Phylogenetic Analysis. Toxins 2022, 14, 90. [Google Scholar] [CrossRef]
- Sumarah, M.W. The Deoxynivalenol Challenge. J. Agr. Food Chem. 2022, 70, 9619–9624. [Google Scholar] [CrossRef] [PubMed]
- Freire, L.; Sant’Ana, A.S. Modified mycotoxins: An updated review on their formation, detection, occurrence, and toxic effects. Food Chem. Toxicol. 2018, 111, 189–205. [Google Scholar] [CrossRef]
- Sato, I.; Ito, M.; Ishizaka, M.; Ikunaga, Y.; Sato, Y.; Yoshida, S.; Koitabashi, M.; Tsushima, S. Thirteen novel deoxynivalenol-degrading bacteria are classified within two genera with distinct degradation mechanisms. Fems. Microbiol. Lett. 2012, 327, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Maresca, M. From the Gut to the Brain: Journey and Pathophysiological Effects of the Food-Associated Trichothecene Mycotoxin Deoxynivalenol. Toxins 2013, 5, 784–820. [Google Scholar] [CrossRef]
- Awad, W.A.; Ghareeb, K.; Bohm, J.; Zentek, J. Decontamination and detoxification strategies for the Fusarium mycotoxin deoxynivalenol in animal feed and the effectiveness of microbial biodegradation. Food Addit. Contam. A 2010, 27, 510–520. [Google Scholar] [CrossRef]
- Pierron, A.; Mimoun, S.; Murate, L.S.; Loiseau, N.; Lippi, Y.; Bracarense, A.; Liaubet, L.; Schatzmayr, G.; Berthiller, F.; Moll, W.D.; et al. Intestinal toxicity of the masked mycotoxin deoxynivalenol-3-beta-d-glucoside. Arch. Toxicol. 2016, 90, 2037–2046. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, N.; Devreese, M.; De Baere, S.; De Backer, P.; Croubels, S. Modified Fusarium mycotoxins unmasked: From occurrence in cereals to animal and human excretion. Food Chem. Toxicol. 2015, 80, 17–31. [Google Scholar] [CrossRef]
- Carere, J.; Hassan, Y.I.; Lepp, D.; Zhou, T. The enzymatic detoxification of the mycotoxin deoxynivalenol: Identification of DepA from the DON epimerization pathway. Microb. Biotechnol. 2018, 11, 1106–1111. [Google Scholar] [CrossRef] [PubMed]
- He, J.W.; Hassan, Y.I.; Perilla, N.; Li, X.Z.; Boland, G.J.; Zhou, T. Bacterial Epimerization as a Route for Deoxynivalenol Detoxification: The Influence of Growth and Environmental Conditions. Front. Microbiol. 2016, 7, 572. [Google Scholar] [CrossRef]
- Clark-Walker, G.D.; Weiller, G.F. The structure of the small mitochondrial DNA of Kluyveromyces thermotolerans is likely to reflect the ancestral gene order in fungi. J. Mol. Evol. 1994, 38, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Volkman, S.K.; Barry, A.E.; Lyons, E.J.; Nielsen, K.M.; Thomas, S.M.; Choi, M.; Thakore, S.S.; Day, K.P.; Wirth, D.F.; Hartl, D.L. Recent origin of Plasmodium falciparum from a single progenitor. Science 2001, 293, 482–484. [Google Scholar] [CrossRef]
- Jukes, T.H.; Osawa, S. Evolutionary changes in the genetic code. Comp. Biochem. Physiol. B Comp. Biochem. 1993, 106, 489–494. [Google Scholar] [CrossRef]
- Musa, M.M.; Phillips, R.S. Recent advances in alcohol dehydrogenase-catalyzed asymmetric production of hydrophobic alcohols. Catal. Sci. Technol. 2011, 1, 1311–1323. [Google Scholar] [CrossRef]
- Matsuda, T.; Yamanaka, R.; Nakamura, K. Recent progress in biocatalysis for asymmetric oxidation and reduction. Tetrahedron-Asymmetry 2009, 20, 513–557. [Google Scholar] [CrossRef]
- Duester, G. Involvement of alcohol dehydrogenase, short-chain dehydrogenase/reductase, aldehyde dehydrogenase, and cytochrome P450 in the control of retinoid signaling by activation of retinoic acid synthesis. Biochemistry 1996, 35, 12221–12227. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Xu, Y.; Nie, Y.; Mu, X.Q. Substrate-Specific Engineering of Amino Acid Dehydrogenase Superfamily for Synthesis of a Variety of Chiral Amines and Amino Acids. Catalysts 2022, 12, 380. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhang, R.Z.; Xu, Y.; Xiao, R.; Wang, L.; Zhou, X.T.; Liang, H.B.; Jiang, J.W. Efficient bioreduction of 2-hydroxyacetophenone to (S)-1-phenyl-1, 2-ethanediol through homologous expression of (S)-carbonyl reductase II in Candida parapsilosis CCTCC M203011. Process Biochem. 2016, 51, 1175–1182. [Google Scholar] [CrossRef]
- Zhang, R.Z.; Wang, L.; Xu, Y.; Liang, H.B.; Zhou, X.T.; Jiang, J.W.; Li, Y.H.; Xiao, R.; Ni, Y. In Situ expression of (R)-carbonyl reductase rebalancing an asymmetric pathway improves stereoconversion efficiency of racemic mixture to (S)-phenyl-1,2-ethanediol in Candida parapsilosis CCTCC M203011. Microb. Cell Fact. 2016, 15, 143. [Google Scholar] [CrossRef]
- Qin, H.M.; Yamamura, A.; Miyakawa, T.; Kataoka, M.; Nagai, T.; Kitamura, N.; Urano, N.; Maruoka, S.; Ohtsuka, J.; Nagata, K.; et al. Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding. Appl. Microbiol. Biot. 2014, 98, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Vinay, K.K.; Debayan, C.; Anju, C. Understanding (R) Specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 [CpCR]: Substrate Scope, Kinetic Studies and the Role of Zinc. Catalysts 2019, 9, 702. [Google Scholar] [CrossRef]
- Aggarwal, N.; Mandal, P.K.; Gautham, N.; Chadha, A. Expression, purification, crystallization and preliminary X-ray diffraction analysis of carbonyl reductase from Candida parapsilosis ATCC 7330. Acta Crystallogr. F 2013, 69, 313–315. [Google Scholar] [CrossRef]
- Qin, H.M.; Yamamura, A.; Miyakawa, T.; Kataoka, M.; Maruoka, S.; Ohtsuka, J.; Nagata, K.; Shimizu, S.; Tanokura, M. Crystal structure of conjugated polyketone reductase (CPR-C1) from Candida parapsilosis IFO 0708 complexed with NADPH. Proteins 2013, 81, 2059–2063. [Google Scholar] [CrossRef]
- Xu, Q.; Tao, W.Y.; Huang, H.; Li, S. Highly efficient synthesis of ethyl (S)-4-chloro-3-hydroxybutanoate by a novel carbonyl reductase from Yarrowia lipolytica and using mannitol or sorbitol as cosubstrate. Biochem. Eng. J. 2016, 106, 61–67. [Google Scholar] [CrossRef]
- Zhang, H.L.; Zhang, C.; Pei, C.H.; Han, M.N.; Li, W. Enantioselective synthesis of enantiopure chiral alcohols using carbonyl reductases screened from Yarrowia lipolytica. J. Appl. Microbiol. 2019, 126, 127–137. [Google Scholar] [CrossRef]
- Xu, Q.; Xu, X.; Huang, H.; Li, S. Efficient synthesis of (R)-2-chloro-1-phenylethol using a yeast carbonyl reductase with broad substrate spectrum and 2-propanol as cosubstrate. Biochem. Eng. J. 2015, 103, 277–285. [Google Scholar] [CrossRef]
- Cao, H.; Mi, L.; Ye, Q.; Zang, G.L.; Yan, M.; Wang, Y.; Zhang, Y.Y.; Li, X.M.; Xu, L.; Xiong, J.A.; et al. Purification and characterization of a novel NADH-dependent carbonyl reductase from Pichia stipitis involved in biosynthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate. Bioresour. Technol. 2011, 102, 1733–1739. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Yan, M.; Xu, L.; Cao, H.; Li, Z.; Chen, Y.; Li, S.; Ying, H. A novel carbonyl reductase from Pichia stipitis for the production of ethyl (S)-4-chloro-3-hydroxybutanoate. Biotechnol. Lett. 2009, 31, 537–542. [Google Scholar] [CrossRef] [PubMed]
- He, Y.C.; Tao, Z.C.; Zhang, X.; Yang, Z.X.; Xu, J.H. Highly efficient synthesis of ethyl (S)-4-chloro-3-hydroxybutanoate and its derivatives by a robust NADH-dependent reductase from E-coli CCZU-K14. Bioresour. Technol. 2014, 161, 461–464. [Google Scholar] [CrossRef]
- Suwa, Y.; Ohtsuka, J.; Miyakawa, T.; Imai, F.L.; Okai, M.; Sawano, Y.; Yasohara, Y.; Kataoka, M.; Shimizu, S.; Tanokura, M. Expression, purification, crystallization and preliminary X-ray analysis of carbonyl reductase S1 from Candida magnoliae. Acta Crystallogr. F 2012, 68, 540–542. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Fukae, T.; Yamamoto, T.; Kanamaru, S.; Matsuda, T. Purification and characterization of fluorinated ketone reductase from Geotrichum candidum NBRC 5767 (vol 76, pg 13, 2013). Biochem. Eng. J. 2015, 93, 309–310. [Google Scholar] [CrossRef]
- Han, M.N.; Wang, X.M.; Pei, C.H.; Zhang, C.; Xu, Z.D.; Zhang, H.L.; Li, W. Green and scalable synthesis of chiral aromatic alcohols through an efficient biocatalytic system. Microb. Biotechnol. 2021, 14, 444–452. [Google Scholar] [CrossRef]
- Zhang, R.; Ren, J.; Wang, Y.; Wu, Q.Q.; Wang, M.; Zhu, D.M. Isolation and characterization of a novel Rhodococcus strain with switchable carbonyl reductase and para-acetylphenol hydroxylase activities. J. Ind. Microbiol. Biot. 2013, 40, 11–20. [Google Scholar] [CrossRef]
- Xu, G.C.; Tang, M.H.; Ni, Y. Asymmetric synthesis of lipitor chiral intermediate using a robust carbonyl reductase at high substrate to catalyst ratio. J. Mol. Catal. B-Enzym. 2016, 123, 67–72. [Google Scholar] [CrossRef]
- Chen, R.J.; Zheng, G.W.; Ni, Y.; Zeng, B.B.; Xu, J.H. Efficient synthesis of an epsilon-hydroxy ester in a space-time yield of 1580 gL−1d−1 by a newly identified reductase RhCR. Tetrahedron-Asymmetry 2014, 25, 1501–1504. [Google Scholar] [CrossRef]
- Chen, X.H.; Wei, P.; Wang, X.T.; Zong, M.H.; Lou, W.Y. A Novel Carbonyl Reductase with Anti-Prelog Stereospecificity from Acetobacter sp CCTCC M209061: Purification and Characterization. PLoS ONE 2014, 9, e94543. [Google Scholar] [CrossRef] [PubMed]
- Weckbecker, A.; Hummel, W. Cloning, expression, and characterization of an (R)-specific alcohol dehydrogenase from Lactobacillus kefir. Biocatal. Biotransfor. 2006, 24, 380–389. [Google Scholar] [CrossRef]
- Li, X.Z.; Hassan, Y.I.; Lepp, D.; Zhu, Y.; Zhou, T. 3-keto-DON, but Not 3-epi-DON, Retains the in Planta Toxicological Potential after the Enzymatic Biotransformation of Deoxynivalenol. Int. J. Mol. Sci. 2022, 23, 7230. [Google Scholar] [CrossRef]
- Qu, R.; Jiang, C.M.; Wu, W.Q.; Pang, B.; Lei, S.Z.; Lian, Z.Y.; Shao, D.Y.; Jin, M.L.; Shi, J.L. Conversion of DON to 3-epi-DON in vitro and toxicity reduction of DON in vivo by Lactobacillus rhamnosus. Food Funct. 2019, 10, 2785–2796. [Google Scholar] [CrossRef]
- Koesoema, A.A.; Standley, D.M.; Senda, T.; Matsuda, T. Impact and relevance of alcohol dehydrogenase enantioselectivities on biotechnological applications. Appl. Microbiol. Biot. 2020, 104, 2897–2909. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhang, J.; Liu, Y.; Guo, Y.; Tang, Y.; Zhang, Q.; Ma, Q.; Ji, C.; Zhao, L. A quinoprotein dehydrogenase from Pelagibacterium halotolerans ANSP101 oxidizes deoxynivalenol to 3-keto-deoxynivalenol. Food Control. 2022, 136, 108834. [Google Scholar] [CrossRef]


| Strains | Conversion Rate/% | Products | Carbonyl Reductase Enzyme Activity in the Literature | |
|---|---|---|---|---|
| 3-epi-DON | DON | |||
| Yarrowia lipolytica GDMCC 2.187 | 99.63 ± 0.05 | 42.98 ± 0.30 | 56.65 ± 0.23 | 96 U/mg, 85 U/mg [53] |
| Candida parapsilosis ACCC 20221 | 86.59 ± 0.10 | 51.25 ± 0.76 | 35.34 ± 0.79 | 1.27 U/mg [46,47] |
| Pichia pastoris GS115 | 58.60 ± 3.88 | 34.94 ± 2.20 | 23.66 ± 1.69 | 224.4 U/mg [55]; 27 U/mg [56] |
| Saccharomycopsis lipolytica GDMCC 2.197 | 37.13 ± 2.67 | / | 37.13 ± 2.67 | 96 U/mg, 85 U/mg [53] |
| Candida parapsilosis GDMCC 2.190 | 30.21 ± 2.63 | / | 30.21 ± 2.63 | 1.27 U/mg [46,47]; |
| Rhodococcus opacus PD 630 | / | / | / | 110 U/mg [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Y.; Xiao, D.; Liu, C. Two-Step Epimerization of Deoxynivalenol by Quinone-Dependent Dehydrogenase and Candida parapsilosis ACCC 20221. Toxins 2023, 15, 286. https://doi.org/10.3390/toxins15040286
Tang Y, Xiao D, Liu C. Two-Step Epimerization of Deoxynivalenol by Quinone-Dependent Dehydrogenase and Candida parapsilosis ACCC 20221. Toxins. 2023; 15(4):286. https://doi.org/10.3390/toxins15040286
Chicago/Turabian StyleTang, Yuqian, Dingna Xiao, and Chendi Liu. 2023. "Two-Step Epimerization of Deoxynivalenol by Quinone-Dependent Dehydrogenase and Candida parapsilosis ACCC 20221" Toxins 15, no. 4: 286. https://doi.org/10.3390/toxins15040286
APA StyleTang, Y., Xiao, D., & Liu, C. (2023). Two-Step Epimerization of Deoxynivalenol by Quinone-Dependent Dehydrogenase and Candida parapsilosis ACCC 20221. Toxins, 15(4), 286. https://doi.org/10.3390/toxins15040286





