One Size Fits All—Venomics of the Iberian Adder (Vipera seoanei, Lataste 1878) Reveals Low Levels of Venom Variation across Its Distributional Range
Abstract
:1. Introduction
2. Results
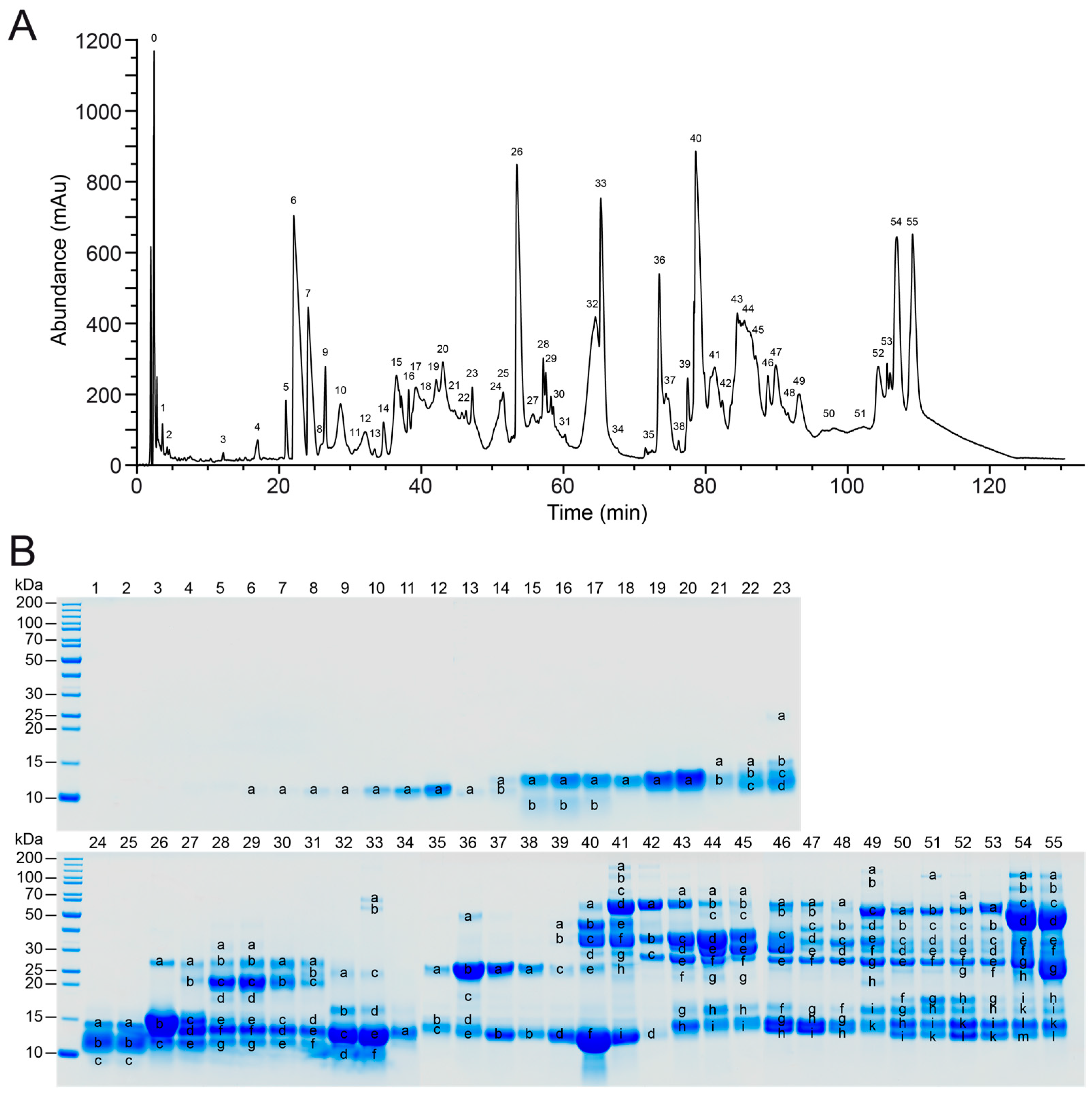
2.1. Protein Composition of V. seoanei Venom
2.2. Assessing Geographic Venom Variation
2.2.1. Analysis of the SDS-PAGE Profiles
2.2.2. Band Analysis and NMDS
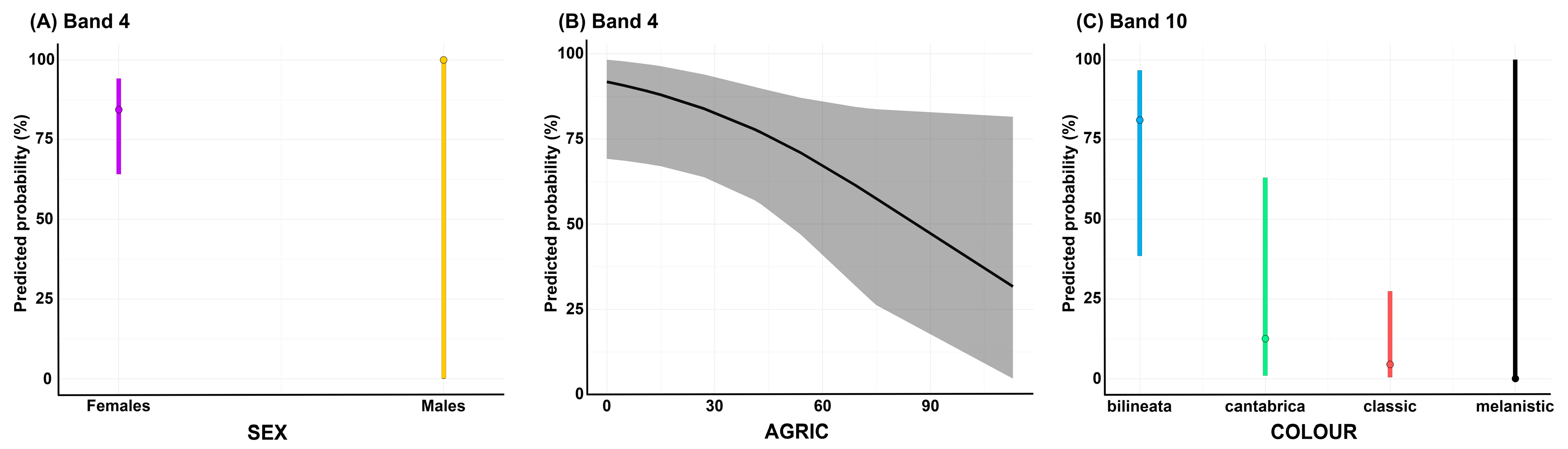
2.2.3. Regression Analysis
3. Discussion
3.1. Protein Composition of V. seoanei Venom
3.2. Low Levels of Geographic Variation
3.3. Correlates of Venom Variation
4. Conclusions
5. Materials and Methods
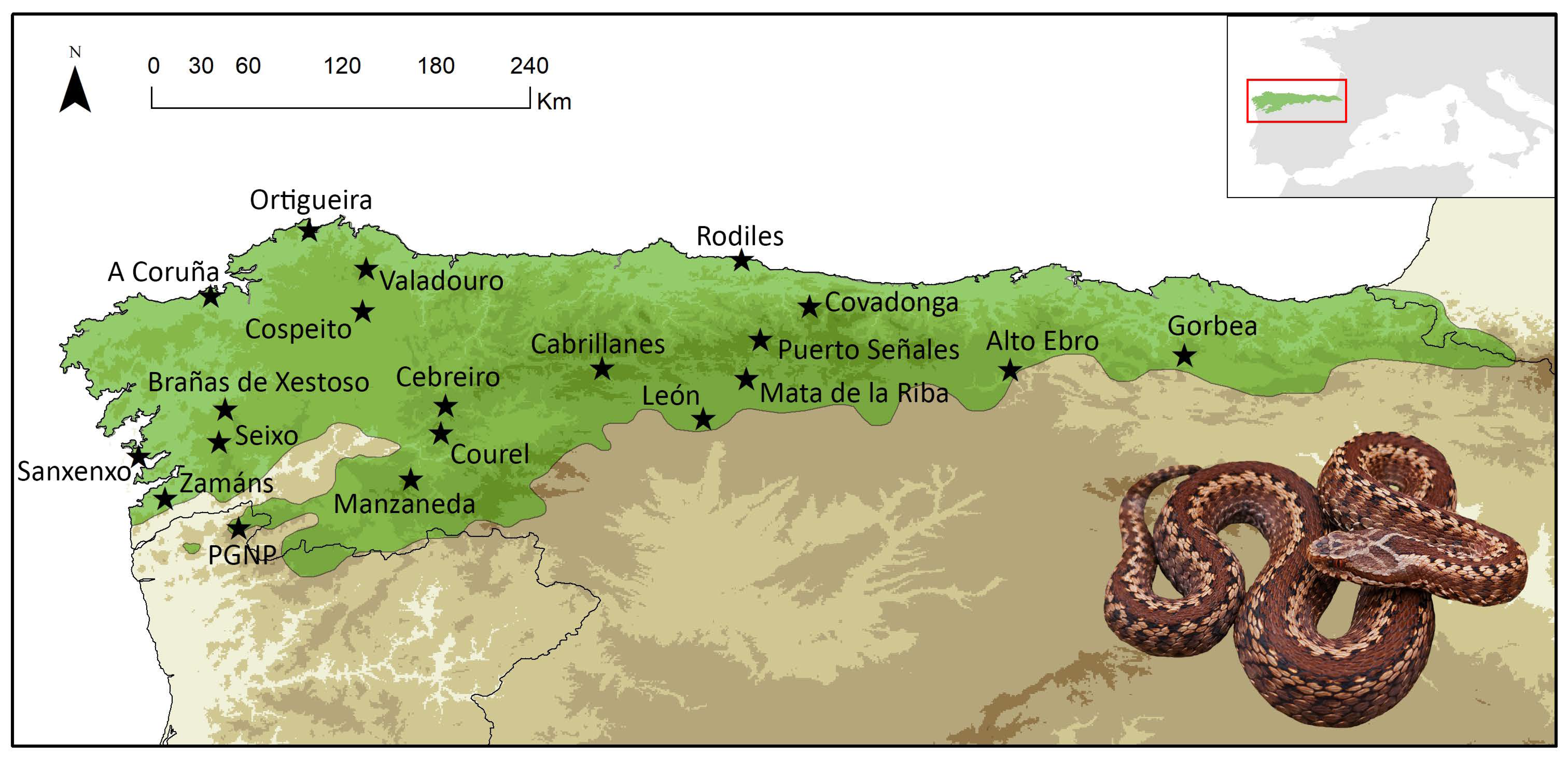
5.1. Sampling
5.2. Bottom-Up Venomics
5.2.1. Venom Fractionation through RP-HPLC
5.2.2. SDS-PAGE Profiling
5.2.3. Tryptic Digestion
5.2.4. Mass Spectrometry
5.2.5. Relative Quantification of the Venom Pool Proteome
5.3. Non-Metric Multidimensional Scaling
5.4. Predictors
5.5. Regression Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.S.; Cheung, R.C.F.; Xia, L.; Wong, J.H.; Ng, T.B.; Chan, W.Y. Snake venom toxins: Toxicity and medicinal applications. Appl. Microbiol. Biotechnol. 2016, 100, 6165–6181. [Google Scholar] [CrossRef] [PubMed]
- Arbuckle, K. Evolutionary context of venom in animals. In Evolution of Venomous Animals and Their Toxins; Gopalakrishnokone, P., Malhotra, A., Eds.; Springer: Dordrecht, Germany, 2017; pp. 3–31. [Google Scholar]
- Fry, B.G.; Vidal, N.; van der Weerd, L.; Kochva, E.; Renjifo, C. Evolution and diversification of the Toxicofera reptile venom system. J. Proteom. 2009, 72, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Kerkkamp, H.M.I.; Casewell, N.R.; Vonk, F.J. Evolution of the snake venom delivery system. In Evolution of Venomous Animals and Their Toxins; Gopalakrishnokone, P., Malhotra, A., Eds.; Springer: Dordrecht, Germany, 2017; pp. 303–316. [Google Scholar]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chippaux, J.P.; Williams, V.; White, J. Snake venom variability: Methods of study, results and interpretation. Toxicon 1991, 29, 1279–1303. [Google Scholar] [CrossRef]
- Casewell, N.R.; Jackson, T.N.; Laustsen, A.H.; Sunagar, K. Causes and consequences of snake venom variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef]
- von Reumont, B.M.; Anderluh, G.; Antunes, A.; Ayvazyan, N.; Beis, D.; Caliskan, F.; Crnković, A.; Damm, M.; Dutertre, S.; Ellgaard, L.; et al. Modern venomics—Current insights, novel methods, and future perspectives in biological and applied animal venom research. GigaScience 2022, 11, giac048. [Google Scholar] [CrossRef]
- Fry, B.G.; Casewell, N.R.; Wüster, W.; Vidal, N.; Young, B.; Jackson, T.N. The structural and functional diversification of the Toxicofera reptile venom system. Toxicon 2012, 60, 434–448. [Google Scholar] [CrossRef]
- Jackson, T.N.; Sunagar, K.; Undheim, E.A.; Koludarov, I.; Chan, A.H.; Sanders, K.; Ali, S.A.; Hendrikx, I.; Dunstan, N.; Fry, B.G. Venom down under: Dynamic evolution of Australian elapid snake toxins. Toxins 2013, 5, 2621–2655. [Google Scholar] [CrossRef] [Green Version]
- Jackson, T.N.; Koludarov, I.; Ali, S.A.; Dobson, J.; Zdenek, C.N.; Dashevsky, D.; Op den Brouw, B.; Masci, P.P.; Nouwens, A.; Josh, P.; et al. Rapid radiations and the race to redundancy: An investigation of the evolution of Australian elapid snake venoms. Toxins 2016, 8, 309. [Google Scholar] [CrossRef] [Green Version]
- Damm, M.; Hempel, B.F.; Süssmuth, R.D. Old World Vipers—A review about snake venom proteomics of Viperinae and their variations. Toxins 2021, 13, 427. [Google Scholar] [CrossRef] [PubMed]
- Pla, D.; Sanz, L.; Sasa, M.; Acevedo, M.E.; Dwyer, Q.; Durban, J.; Pérez, A.; Rodríguez, Y.; Lomonte, B.; Calvete, J.J. Proteomic analysis of venom variability and ontogeny across the arboreal palm-pitvipers (genus Bothriechis). J. Proteom. 2017, 152, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, G.P.; Pessoa, L.A.; Portaro, F.C.; Maria de Fátima, D.F.; Tambourgi, D.V. Interspecific variation in venom composition and toxicity of Brazilian snakes from Bothrops genus. Toxicon 2008, 52, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Avella, I.; Calvete, J.J.; Sanz, L.; Wüster, W.; Licata, F.; Quesada-Bernat, S.; Rodríguez, Y.; Martínez-Freiría, F. Interpopulational variation and ontogenetic shift in the venom composition of Lataste’s viper (Vipera latastei, Boscá 1878) from northern Portugal. J. Proteom. 2022, 263, 104613. [Google Scholar] [CrossRef]
- Saldarriaga, M.M.; Otero, R.; Núñez, V.; Toro, M.F.; Dıaz, A.; Gutiérrez, J.M. Ontogenetic variability of Bothrops atrox and Bothrops asper snake venoms from Colombia. Toxicon 2003, 42, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Saviola, A.J.; Pla, D.; Sanz, L.; Castoe, T.A.; Calvete, J.J.; Mackessy, S.P. Comparative venomics of the Prairie Rattlesnake (Crotalus viridis viridis) from Colorado: Identification of a novel pattern of ontogenetic changes in venom composition and assessment of the immunoreactivity of the commercial antivenom CroFab®. J. Proteom. 2015, 121, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Malina, T.; Krecsák, L.; Westerström, A.; Szemán-Nagy, G.; Gyémánt, G.; M-Hamvas, M.; Rowan, E.G.; Harvey, A.L.; Warrell, D.A.; Pál, B.; et al. Individual variability of venom from the European adder (Vipera berus berus) from one locality in Eastern Hungary. Toxicon 2017, 135, 59–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menezes, M.C.; Furtado, M.F.; Travaglia-Cardoso, S.R.; Camargo, A.C.; Serrano, S.M. Sex-based individual variation of snake venom proteome among eighteen Bothrops jararaca siblings. Toxicon 2006, 47, 304–312. [Google Scholar] [CrossRef]
- Simizo, A.; Kitano, E.S.; Sant’Anna, S.S.; Grego, K.F.; Tanaka-Azevedo, A.M.; Tashima, A.K. Comparative gender peptidomics of Bothrops atrox venoms: Are there differences between them? J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20200055. [Google Scholar] [CrossRef]
- Boldrini-França, J.; Corrêa-Netto, C.; Silva, M.M.; Rodrigues, R.S.; De La Torre, P.; Pérez, A.; Soares, A.M.; Zingali, R.B.; Nogueira, R.A.; Rodrigues, V.M.; et al. Snake venomics and antivenomics of Crotalus durissus subspecies from Brazil: Assessment of geographic variation and its implication on snakebite management. J. Proteom. 2010, 73, 1758–1776. [Google Scholar] [CrossRef] [PubMed]
- Neri-Castro, E.; Lomonte, B.; del Carmen Gutiérrez, M.; Alagón, A.; Gutiérrez, J.M. Intraspecies variation in the venom of the rattlesnake Crotalus simus from Mexico: Different expression of crotoxin results in highly variable toxicity in the venoms of three subspecies. J. Proteom. 2013, 87, 103–121. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Undheim, E.A.; Scheib, H.; Gren, E.C.; Cochran, C.; Person, C.E.; Koludarov, I.; Kelln, W.; Hayes, W.K.; King, G.F.; et al. Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake (Crotalus oreganus helleri): Biodiscovery, clinical and evolutionary implications. J. Proteom. 2014, 99, 68–83. [Google Scholar] [CrossRef]
- Zancolli, G.; Calvete, J.J.; Cardwell, M.D.; Greene, H.W.; Hayes, W.K.; Hegarty, M.J.; Herrmann, H.W.; Holycross, A.T.; Lannutti, D.I.; Mulley, J.F.; et al. When one phenotype is not enough: Divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc. R. Soc. B 2019, 286, 20182735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wüster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. R. Soc. B 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daltry, J.C.; Wüster, W.; Thorpe, R.S. Diet and snake venom evolution. Nature 1996, 379, 537–540. [Google Scholar] [CrossRef]
- Holding, M.L.; Strickland, J.L.; Rautsaw, R.M.; Hofmann, E.P.; Mason, A.J.; Hogan, M.P.; Nystrom, G.S.; Ellsworth, S.A.; Colston, T.J.; Borja, M.; et al. Phylogenetically diverse diets favor more complex venoms in North American pitvipers. Proc. Natl. Acad. Sci. USA 2021, 118, e2015579118. [Google Scholar] [CrossRef] [PubMed]
- Creer, S.; Malhotra, A.; Thorpe, R.S.; Stöcklin, R.S.; Favreau, P.S.; Hao Chou, W.S. Genetic and ecological correlates of intraspecific variation in pitviper venom composition detected using matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS) and isoelectric focusing. J. Mol. Evol. 2003, 56, 317–329. [Google Scholar] [CrossRef]
- Barua, A.; Mikheyev, A.S. Toxin expression in snake venom evolves rapidly with constant shifts in evolutionary rates. Proc. R. Soc. B 2020, 287, 20200613. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wagstaff, S.C.; Harrison, R.A.; Renjifo, C.; Wüster, W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 2011, 28, 2637–2649. [Google Scholar] [CrossRef] [Green Version]
- Holding, M.L.; Margres, M.J.; Rokyta, D.R.; Gibbs, H.L. Local prey community composition and genetic distance predict venom divergence among populations of the northern Pacific rattlesnake (Crotalus oreganus). J. Evol. Biol. 2018, 31, 1513–1528. [Google Scholar] [CrossRef]
- Saviola, A.J.; Gandara, A.J.; Bryson, R.W., Jr.; Mackessy, S.P. Venom phenotypes of the Rock Rattlesnake (Crotalus lepidus) and the Ridge-nosed Rattlesnake (Crotalus willardi) from México and the United States. Toxicon 2017, 138, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Sousa, L.F.; Portes-Junior, J.A.; Nicolau, C.A.; Bernardoni, J.L.; Nishiyama, M.Y., Jr.; Amazonas, D.R.; Freitas-de-Sousa, L.A.; Mourão, R.H.V.; Chalkidis, H.M.; Valente, R.H.; et al. Functional proteomic analyses of Bothrops atrox venom reveals phenotypes associated with habitat variation in the Amazon. J. Proteom. 2017, 159, 32–46. [Google Scholar] [CrossRef]
- Massey, D.J.; Calvete, J.J.; Sánchez, E.E.; Sanz, L.; Richards, K.; Curtis, R.; Boesen, K. Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from Southern Arizona. J. Proteom. 2012, 75, 2576–2587. [Google Scholar] [CrossRef] [PubMed]
- Schield, D.R.; Adams, R.H.; Card, D.C.; Corbin, A.B.; Jezkova, T.; Hales, N.R.; Meik, J.M.; Perry, B.W.; Spencer, C.L.; Smith, L.L.; et al. Cryptic genetic diversity, population structure, and gene flow in the Mojave rattlesnake (Crotalus scutulatus). Mol. Phylogenet. Evol. 2018, 127, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Strickland, J.L.; Smith, C.F.; Mason, A.J.; Schield, D.R.; Borja, M.; Castañeda-Gaytán, G.; Spencer, C.L.; Smith, L.L.; Trápaga, A.; Bouzid, N.M.; et al. Evidence for divergent patterns of local selection driving venom variation in Mojave Rattlesnakes (Crotalus scutulatus). Sci. Rep. 2018, 8, 17622. [Google Scholar] [CrossRef] [Green Version]
- Ferquel, E.; De Haro, L.; Jan, V.; Guillemin, I.; Jourdain, S.; Teynié, A.; d’Alayer, J.; Choumet, V. Reappraisal of Vipera aspis venom neurotoxicity. PLoS ONE 2007, 2, e1194. [Google Scholar] [CrossRef] [Green Version]
- Malina, T.; Babocsay, G.; Krecsák, L.; Erdész, C. Further clinical evidence for the existence of neurotoxicity in a population of the European adder (Vipera berus berus) in eastern Hungary: Second authenticated case. Wilderness Environ. Med. 2013, 24, 378–383. [Google Scholar] [CrossRef] [Green Version]
- Di Nicola, M.R.; Pontara, A.; Kass, G.E.N.; Kramer, N.I.; Avella, I.; Pampena, R.; Mercuri, S.R.; Dorne, J.L.M.; Paolino, G. Vipers of major clinical relevance in Europe: Taxonomy, venom composition, toxicology and clinical management of human bites. Toxicology 2021, 453, 152724. [Google Scholar] [CrossRef]
- Paolino, G.; Di Nicola, M.R.; Pontara, A.; Didona, D.; Moliterni, E.; Mercuri, S.R.; Grano, M.; Borgianni, N.; Kumar, R.; Pampena, R. Vipera snakebite in Europe: A systematic review of a neglected disease. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 2247–2260. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Snakebite Information and Data Platform. 2020. Available online: https://www.who.int/teams/control-of-neglected-tropical-diseases/snakebite-envenoming/snakebite-information-and-data-platform/overview#tab=tab_1,2020 (accessed on 19 January 2022).
- Freitas, I.; Ursenbacher, S.; Mebert, K.; Zinenko, O.; Schweiger, S.; Wüster, W.; Brito, J.C.; Crnobrnja-Isailović, J.; Halpern, B.; Fahd, S.; et al. Evaluating taxonomic inflation: Towards evidence-based species delimitation in Eurasian vipers (Serpentes: Viperinae). Amphib-Reptil. 2020, 41, 285–311. [Google Scholar] [CrossRef]
- Martínez-Freiría, F.; Brito, J.C. Vipera seoanei (Lataste, 1879). In Fauna Ibérica, Vol. 10, Reptiles, 2nd ed.; Ramos, M.A., Alba, J., Bellés, X., Gosálbez, J., Guerra, A., Macpherson, E., Serrano, J., Templado, J., Eds.; Museo Nacional de Ciencias Naturales, CSIC: Madrid, Spain, 2014; pp. 942–957. [Google Scholar]
- Martínez-Freiría, F.; Velo-Antón, G.; Brito, J.C. Trapped by climate: Interglacial refuge and recent population expansion in the endemic Iberian adder Vipera seoanei. Divers. Distrib. 2015, 21, 331–344. [Google Scholar] [CrossRef]
- Lucchini, N.; Kaliontzopoulou, A.; Aguado-Val, G.; Martínez-Freiría, F. Sources of intraspecific morphological variation in Vipera seoanei: Allometry, sex, and colour phenotype. Amphib.-Reptil. 2020, 42, 1–16. [Google Scholar] [CrossRef]
- Martínez-Freiría, F.; Brito, J.C. Integrating classical and spatial multivariate analyses for assessing morphological variability in the endemic Iberian viper Vipera seoanei. J. Zoolog. Syst. Evol. Res. 2013, 51, 122–131. [Google Scholar] [CrossRef]
- Martínez-Freiría, F.; i de Lanuza, G.P.; Pimenta, A.A.; Pinto, T.; Santos, X. Aposematism and crypsis are not enough to explain dorsal polymorphism in the Iberian adder. Acta Oecol. 2017, 85, 165–173. [Google Scholar] [CrossRef]
- Braña, F.; Bea, A.; Saint Girons, H. Composición de la dieta y ciclos de alimentación en Vipera seoanei Lataste, 1879. Variaciones en relación con la edad y el ciclo reproductor. Munibe Cienc. Nat. 1988, 40, 19–27. [Google Scholar]
- Espasandín, I.; Galán, P.; Martínez-Freiría, F. Sex, size and eco-geographic factors affect the feeding ecology of the Iberian adder, Vipera seoanei. Amphib.-Reptil. 2022, 1, 1–16. [Google Scholar] [CrossRef]
- Galán, P. Segregación ecológica en una comunidad de ofidios. Doñana. Acta Vertebr. 1988, 15, 59–78. [Google Scholar]
- Luiselli, L.M.; Agrimi, U. Composition and variation of the diet of Vipera aspis francisciredi in relation to age and reproductive stage. Amphib.-Reptil. 1991, 12, 137–144. [Google Scholar] [CrossRef]
- Brito, J.C. Feeding ecology of Vipera latastei in northern Portugal ontogenetic shifts, prey size and seasonal variations. Herpetol. J. 2004, 14, 13–19. [Google Scholar]
- Tomović, L.; Anđelković, M.; Golubović, A.; Arsovski, D.; Ajtić, R.; Sterijovski, B.; Nikolić, S.; Crnobrnja-Isailović, J.; Lakušić, M.; Bonnet, X. Dwarf vipers on a small island: Body size, diet and fecundity correlates. Biol. J. Linn. 2022, 137, 267–279. [Google Scholar] [CrossRef]
- Santos, X.; Pleguezuelos, J.M.; Brito, J.C.; Llorente, G.A.; Parellada, X.; Fahd, S. Prey availability drives geographic dietary differences of a Mediterranean predator, the Lataste’s viper (Vipera latastei). Herpetol. J. 2008, 18, 16–22. [Google Scholar]
- Detrait, J.; Bea, A.; Saint Girons, H.; Choumet, V. Les variations geographiques du venin de Vipera seoanei Lataste (1879). Bull. Soc. Zool. Fr. 1990, 115, 277–285. [Google Scholar]
- Archundia, I.G.; de Roodt, A.R.; Ramos-Cerrillo, B.; Chippaux, J.P.; Olguín-Pérez, L.; Alagón, A.; Stock, R.P. Neutralization of Vipera and Macrovipera venoms by two experimental polyvalent antisera: A study of paraspecificity. Toxicon 2011, 57, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Olaoba, O.T.; Dos Santos, P.K.; Selistre-de-Araujo, H.S.; de Souza, D.H.F. Snake venom metalloproteinases (SVMPs): A structure-function update. Toxicon X 2020, 7, 100052. [Google Scholar] [CrossRef]
- Požek, K.; Leonardi, A.; Pungerčar, J.; Rao, W.; Gao, Z.; Liu, S.; Laustsen, A.H.; Bakija, A.T.; Reberšek, K.; Podgornik, H.; et al. Genomic Confirmation of the P-IIIe Subclass of Snake Venom Metalloproteinases and Characterisation of Its First Member, a Disintegrin-Like/Cysteine-Rich Protein. Toxins 2022, 14, 232. [Google Scholar] [CrossRef] [PubMed]
- Eble, J.A. Structurally robust and functionally highly versatile—C-type lectin (-related) proteins in snake venoms. Toxins 2019, 11, 136. [Google Scholar] [CrossRef] [Green Version]
- Latinović, Z.; Leonardi, A.; Šribar, J.; Sajevic, T.; Žužek, M.C.; Frangež, R.; Halassy, B.; Trampuš-Bakija, A.; Pungerčar, J.; Križaj, I. Venomics of Vipera berus berus to explain differences in pathology elicited by Vipera ammodytes ammodytes envenomation: Therapeutic implications. J. Proteom. 2016, 146, 34–47. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Primers 2017, 3, 17063. [Google Scholar] [CrossRef]
- Warrell, D.A. Epidemiology, clinical features and management of snake bites in Central and South America. In Venomous Reptiles of the Western Hemisphere, 2nd ed.; Campbell, J.R., Lamar, W.W., Eds.; Cornell University Press: Ithaca, NY, USA, 2004; pp. 709–761. [Google Scholar]
- Kini, R.M. Serine proteases affecting blood coagulation and fibrinolysis from snake venoms. Pathophysiol. Haemos. Thromb. 2005, 34, 200–204. [Google Scholar] [CrossRef]
- Lazarovici, P.; Marcinkiewicz, C.; Lelkes, P.I. From snake venom’s disintegrins and C-type lectins to anti-platelet drugs. Toxins 2019, 11, 303. [Google Scholar] [CrossRef] [Green Version]
- Marcinkiewicz, C. Functional characteristic of snake venom disintegrins: Potential therapeutic implication. Curr. Pharm. Des. 2005, 11, 815–827. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Rucavado, A.; Escalante, T. Snake venom metalloproteinases: Biological roles and participation in the pathophysiology of envenomation. In Evolution of Venomous Animals and their Toxins, 1st ed.; Mackessy, S.P., Ed.; CRC Press: Boca Ratón, FL, USA, 2017; pp. 115–130. [Google Scholar]
- Ramos, O.H.P.; Selistre-de-Araujo, H.S. Snake venom metalloproteases—Structure and function of catalytic and disintegrin domains. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006, 142, 328–346. [Google Scholar] [CrossRef] [PubMed]
- Jan, V.M.; Guillemin, I.; Robbe-Vincent, A.; Choumet, V. Phospholipase A2 diversity and polymorphism in European viper venoms: Paradoxical molecular evolution in Viperinae. Toxicon 2007, 50, 1140–1161. [Google Scholar] [CrossRef]
- Rouault, M.; Rash, L.D.; Escoubas, P.; Boilard, E.; Bollinger, J.; Lomonte, B.; Maurin, T.; Guillaume, C.; Canaan, S.; Deregnaucourt, C.; et al. Neurotoxicity and other pharmacological activities of the snake venom phospholipase A2 OS2: The N-terminal region is more important than enzymatic activity. Biochemistry 2006, 45, 5800–5816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.Y.; Clemetson, K.J. Snake venom L-amino acid oxidases. Toxicon 2002, 40, 659–665. [Google Scholar] [CrossRef]
- Morjen, M.; Honoré, S.; Bazaa, A.; Abdelkafi-Koubaa, Z.; Ellafi, A.; Mabrouk, K.; Kovacic, H.; El Ayeb, M.; Marrakchi, N.; Luis, J. PIVL, a snake venom Kunitz-type serine protease inhibitor, inhibits in vitro and in vivo angiogenesis. Microvasc. Res. 2014, 95, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K.; Mackessy, S.P.; Dutta, S. Characterization of a Kunitz-type protease inhibitor peptide (Rusvikunin) purified from Daboia russelii russelii venom. Int. J. Biol. Macromol. 2014, 67, 154–162. [Google Scholar] [CrossRef]
- Ferreira, I.G.; Pucca, M.B.; de Oliveira, I.S.; Cerni, F.A.; da Silva Jacob, B.D.C.; Arantes, E.C. Snake venom vascular endothelial growth factors (svVEGFs): Unravelling their molecular structure, functions, and research potential. Cytokine Growth Factor Rev. 2021, 60, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Lecht, S.; Chiaverelli, R.A.; Gerstenhaber, J.; Calvete, J.J.; Lazarovici, P.; Casewell, N.R.; Harrison, R.A.; Lelkes, P.I.; Marcinkiewicz, C. Anti-angiogenic activities of snake venom CRISP isolated from Echis carinatus sochureki. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Fry, B.G.; Jackson, T.N.; Casewell, N.R.; Undheim, E.A.; Vidal, N.; Ali, S.A.; King, G.F.; Vasudevan, K.; Vasconcelos, V.; et al. Molecular evolution of vertebrate neurotrophins: Co-option of the highly conserved nerve growth factor gene into the advanced snake venom arsenal. PLoS ONE 2013, 8, e81827. [Google Scholar] [CrossRef]
- Oliveira, A.L.; Viegas, M.F.; da Silva, S.L.; Soares, A.M.; Ramos, M.J.; Fernandes, P.A. The chemistry of snake venom and its medicinal potential. Nat. Rev. Chem. 2022, 6, 451–469. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, E.P.; Rautsaw, R.M.; Strickland, J.L.; Holding, M.L.; Hogan, M.P.; Mason, A.J.; Rokyta, D.R.; Parkinson, C.L. Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Sci. Rep. 2018, 8, 15534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rautsaw, R.M.; Hofmann, E.P.; Margres, M.J.; Holding, M.L.; Strickland, J.L.; Mason, A.J.; Rokyta, D.R.; Parkinson, C.L. Intraspecific sequence and gene expression variation contribute little to venom diversity in sidewinder rattlesnakes (Crotalus cerastes). Proc. R. Soc. B 2019, 286, 20190810. [Google Scholar] [CrossRef] [PubMed]
- Margres, M.J.; Patton, A.; Wray, K.P.; Hassinger, A.T.; Ward, M.J.; Lemmon, E.M.; Lemmon, A.R.; Rokyta, D.R. Tipping the scales: The migration–selection balance leans toward selection in snake venoms. Mol. Biol. Evol. 2019, 36, 271–282. [Google Scholar] [CrossRef]
- Margres, M.J.; McGivern, J.J.; Seavy, M.; Wray, K.P.; Facente, J.; Rokyta, D.R. Contrasting modes and tempos of venom expression evolution in two snake species. Genetics 2015, 199, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, H.L.; Sanz, L.; Pérez, A.; Ochoa, A.; Hassinger, A.T.; Holding, M.L.; Calvete, J.J. The molecular basis of venom resistance in a rattlesnake-squirrel predator-prey system. Mol. Ecol. 2020, 29, 2871–2888. [Google Scholar] [CrossRef]
- Poran, N.S.; Coss, R.G.; Benjamini, E.L.I. Resistance of California ground squirrels (Spermophilus beecheyi) to the venom of the northern Pacific rattlesnake (Crotalus viridis oreganus): A study of adaptive variation. Toxicon 1987, 25, 767–777. [Google Scholar] [CrossRef]
- van Thiel, J.; Khan, M.A.; Wouters, R.M.; Harris, R.J.; Casewell, N.R.; Fry, B.G.; Kini, R.M.; Mackessy, S.P.; Vonk, F.J.; Wüster, W.; et al. Convergent evolution of toxin resistance in animals. Biol. Rev. 2022, 97, 1823–1843. [Google Scholar] [CrossRef]
- Forsman, A. Variation in sexual size dimorphism and maximum body size among adder populations: Effects of prey size. J. Anim. Ecol. 1991, 60, 253–267. [Google Scholar] [CrossRef]
- Shine, R. Intersexual dietary divergence and the evolution of sexual dimorphism in snakes. Am. Nat. 1991, 138, 103–122. [Google Scholar] [CrossRef]
- McCue, M.D. Prey envenomation does not improve digestive performance in western diamondback rattlesnakes (Crotalus atrox). J. Exp. Zool. A Ecol. Gen. Physiol. 2007, 307, 568–577. [Google Scholar] [CrossRef]
- Calvete, J.J.; Bonilla, F.; Granados-Martínez, S.; Sanz, L.; Lomonte, B.; Sasa, M. Venomics of the Duvernoy’s gland secretion of the false coral snake Rhinobothryum bovallii (Andersson, 1916) and assessment of venom lethality towards synapsid and diapsid animal models. J. Proteom. 2020, 225, 103882. [Google Scholar] [CrossRef]
- Modahl, C.M.; Mrinalini; Frietze, S.; Mackessy, S.P. Adaptive evolution of distinct prey-specific toxin genes in rear-fanged snake venom. Proc. R. Soc. B 2018, 285, 20181003. [Google Scholar] [CrossRef] [PubMed]
- Braña, F. Vipera seoanei Lataste, 1879. In Fauna Ibérica Vol. 10, Reptiles; Salvador, A., Ed.; Museo Nacional de Ciencias Naturales, CSIC: Madrid, Spain, 1998; pp. 489–497. [Google Scholar]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Guangcan, S.; Yong, C.; Zhenlin, C.; Chao, L.; Shangtong, L.; Hao, C.; Meng-Qiu, D. How to use open-pFind in deep proteomics data analysis?—A protocol for rigorous identification and quantitation of peptides and proteins from mass spectrometry data. Biophys. Rep. 2021, 7, 207–226. [Google Scholar] [CrossRef]
- Altschul, S.F.; Wootton, J.C.; Gertz, E.M.; Agarwala, R.; Morgulis, A.; Schäffer, A.A.; Yu, Y.K. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005, 272, 5101–5109. [Google Scholar] [CrossRef]
- Calderón-Celis, F.; Cid-Barrio, L.; Encinar, J.R.; Sanz-Medel, A.; Calvete, J.J. Absolute venomics: Absolute quantification of intact venom proteins through elemental mass spectrometry. J. Proteom. 2017, 164, 33–42. [Google Scholar] [CrossRef]
- Calvete, J.J. Next-generation snake venomics: Protein-locus resolution through venom proteome decomplexation. Expert Rev. Proteom. 2014, 11, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Eichberg, S.; Sanz, L.; Calvete, J.J.; Pla, D. Constructing comprehensive venom proteome reference maps for integrative venomics. Expert Rev. Proteom. 2015, 12, 557–573. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Zancolli, G.; Sanz, L.; Calvete, J.J.; Wüster, W. Venom on-a-chip: A fast and efficient method for comparative venomics. Toxins 2017, 9, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minchin, P.R. An evaluation of relative robustness of techniques for ecological orderings. Vegetatio 1987, 71, 145–156. [Google Scholar] [CrossRef]
- Kruskal, J.B. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 1964, 29, 1–27. [Google Scholar] [CrossRef]
- Martínez-Freiría, F.; Sillero, N.; Lizana, M.; Brito, J.C. GIS-based niche models identify environmental correlates sustaining a contact zone between three species of European vipers. Divers. Distrib. 2008, 14, 452–461. [Google Scholar] [CrossRef]
- Chamorro, D.; Martínez-Freiría, F.; Real, R.; Muñoz, A.R. Understanding parapatry: How do environment and competitive interactions shape Iberian vipers’ distributions? J. Biogeogr. 2021, 48, 1322–1335. [Google Scholar] [CrossRef]
- Mira, A.; Marques, C.C.; Santos, S.M.; Rosário, I.T.; Mathias, M.L. Environmental determinants of the distribution of the Cabrera vole (Microtus cabrerae) in Portugal: Implications for conservation. Mamm. Biol. 2008, 73, 102–110. [Google Scholar] [CrossRef]
- Sillero, N.; Brito, J.C.; Skidmore, A.K.; Toxopeus, A.G. Biogeographical patterns derived from remote sensing variables: The amphibians and reptiles of the Iberian Peninsula. Amphib.-Reptil. 2009, 30, 185–206. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Palumbi, S.R. The polymerase chain reaction. In Molecular Systematics; Hillis, D., Moritz, C., Mable, B.K., Eds.; Sinauer Associates: Sunderland, MA, USA, 1996; pp. 205–247. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- ESRI. ArcGIS Desktop: Release 10.5; Environmental Systems Research Institute: Redlands, CA, USA, 2016. [Google Scholar]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2022; Available online: https://www.R-project.org (accessed on 9 November 2022).
- Moran, P.A. Notes on continuous stochastic phenomena. Biometrika 1950, 37, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Cramér, H. Mathematical Methods of Statistics; Princeton University Press: Princeton, NJ, USA, 1999. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2009. [Google Scholar]
- Oksanen, J. Design Decisions and Implementation Details in Vegan. Vignette of the Package Vegan. Available online: https://cran.r-project.org/web/packages/vegan/vignettes/decision-vegan.pdf (accessed on 9 November 2022).
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed; Sage Publications: Washington, DC, USA, 2018. [Google Scholar]
- Lüdecke, D. ggeffects: Tidy data frames of marginal effects from regression models. J. Open Source Softw. 2018, 3, 772. [Google Scholar] [CrossRef] [Green Version]

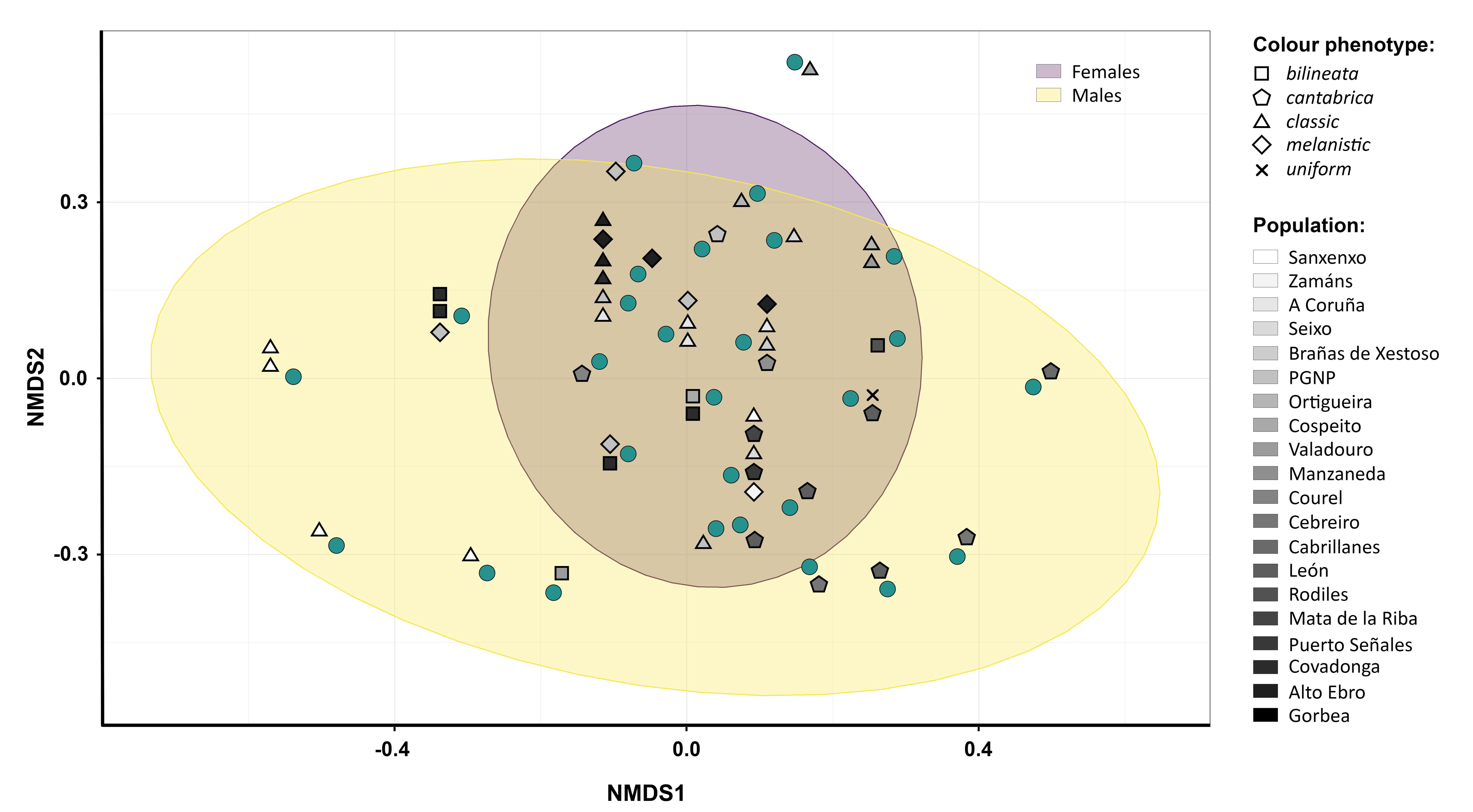
| NMDS1 | NMDS2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Predictor | Sum Sq | Df | F | p | Sum Sq | Df | F | p |
| SVL | 0.263 | 1 | 6.351 | 0.016 | <0.001 | 1 | 0.013 | 0.911 |
| SEX | 0.081 | 1 | 1.794 | 0.187 | 0.205 | 1 | 5.626 | 0.022 |
| COLOUR | 0.397 | 3 | 3.312 | 0.028 | 0.324 | 3 | 2.983 | 0.041 |
| POPULATION | 1.538 | 19 | 3.508 | 0.001 | 1.032 | 19 | 1.779 | 0.079 |
| GEN1 | 0.029 | 1 | 0.635 | 0.429 | <0.001 | 1 | <0.001 | 0.997 |
| GEN2 | 0.015 | 1 | 0.322 | 0.573 | 0.016 | 1 | 0.403 | 0.529 |
| BIO1 | 0.061 | 1 | 1.334 | 0.254 | 0.001 | 1 | 0.031 | 0.861 |
| BIO5 | 0.145 | 1 | 3.314 | 0.075 | 0.123 | 1 | 3.379 | 0.072 |
| BIO12 | 0.133 | 1 | 3.019 | 0.089 | <0.001 | 1 | 0.009 | 0.925 |
| BIO14 | 0.002 | 1 | 0.048 | 0.823 | 0.015 | 1 | 0.363 | 0.549 |
| AGRIC | 0.016 | 1 | 0.345 | 0.559 | 0.102 | 1 | 2.646 | 0.111 |
| FOREST | 0.201 | 1 | 4.702 | 0.035 | 0.178 | 1 | 4.793 | 0.033 |
| MOOR | 0.002 | 1 | 0.051 | 0.823 | 0.004 | 1 | 0.107 | 0.746 |
| PASTURE | <0.001 | 1 | <0.001 | 0.981 | <0.001 | 1 | 0.011 | 0.912 |
| Response | Predictor | Sum Sq | Df | F | p | Res. Sum Sq | Res. Df |
|---|---|---|---|---|---|---|---|
| NMDS1 | SVL | 0.084 | 1 | 2.142 | 0.151 | 1.641 | 42 |
| COLOUR | 0.208 | 3 | 1.773 | 0.167 | |||
| FOREST | 0.021 | 1 | 0.536 | 0.468 | |||
| NMDS2 | SEX | 0.136 | 1 | 4.255 | 0.045 | 1.347 | 42 |
| COLOUR | 0.223 | 3 | 2.317 | 0.089 | |||
| FOREST | 0.084 | 1 | 2.611 | 0.114 |
| Response | Predictors | LR χ2 | Df | p |
|---|---|---|---|---|
| Band 4 | SEX | 5.544 | 1 | 0.018 |
| AGRIC | 4.068 | 1 | 0.044 | |
| Band 8 | SVL | 0.529 | 1 | 0.467 |
| COLOUR | 3.638 | 3 | 0.303 | |
| BIO5 | 0.059 | 1 | 0.807 | |
| BIO12 | 0.000 | 1 | 0.996 | |
| Band 9 | SEX * | 20.679 | 1 | <0.001 |
| GEN2 * | 16.784 | 1 | <0.001 | |
| Band 10 | COLOUR | 16.612 | 3 | <0.001 |
| BIO5 | 2.387 | 1 | 0.122 | |
| FOREST | 3.765 | 1 | 0.052 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avella, I.; Damm, M.; Freitas, I.; Wüster, W.; Lucchini, N.; Zuazo, Ó.; Süssmuth, R.D.; Martínez-Freiría, F. One Size Fits All—Venomics of the Iberian Adder (Vipera seoanei, Lataste 1878) Reveals Low Levels of Venom Variation across Its Distributional Range. Toxins 2023, 15, 371. https://doi.org/10.3390/toxins15060371
Avella I, Damm M, Freitas I, Wüster W, Lucchini N, Zuazo Ó, Süssmuth RD, Martínez-Freiría F. One Size Fits All—Venomics of the Iberian Adder (Vipera seoanei, Lataste 1878) Reveals Low Levels of Venom Variation across Its Distributional Range. Toxins. 2023; 15(6):371. https://doi.org/10.3390/toxins15060371
Chicago/Turabian StyleAvella, Ignazio, Maik Damm, Inês Freitas, Wolfgang Wüster, Nahla Lucchini, Óscar Zuazo, Roderich D. Süssmuth, and Fernando Martínez-Freiría. 2023. "One Size Fits All—Venomics of the Iberian Adder (Vipera seoanei, Lataste 1878) Reveals Low Levels of Venom Variation across Its Distributional Range" Toxins 15, no. 6: 371. https://doi.org/10.3390/toxins15060371
APA StyleAvella, I., Damm, M., Freitas, I., Wüster, W., Lucchini, N., Zuazo, Ó., Süssmuth, R. D., & Martínez-Freiría, F. (2023). One Size Fits All—Venomics of the Iberian Adder (Vipera seoanei, Lataste 1878) Reveals Low Levels of Venom Variation across Its Distributional Range. Toxins, 15(6), 371. https://doi.org/10.3390/toxins15060371






