Is Proteolytic Cleavage Essential for the Enhanced Activity of Hydra Pore-Forming Toxin, HALT-4?
Abstract
:1. Introduction
2. Results and Discussion
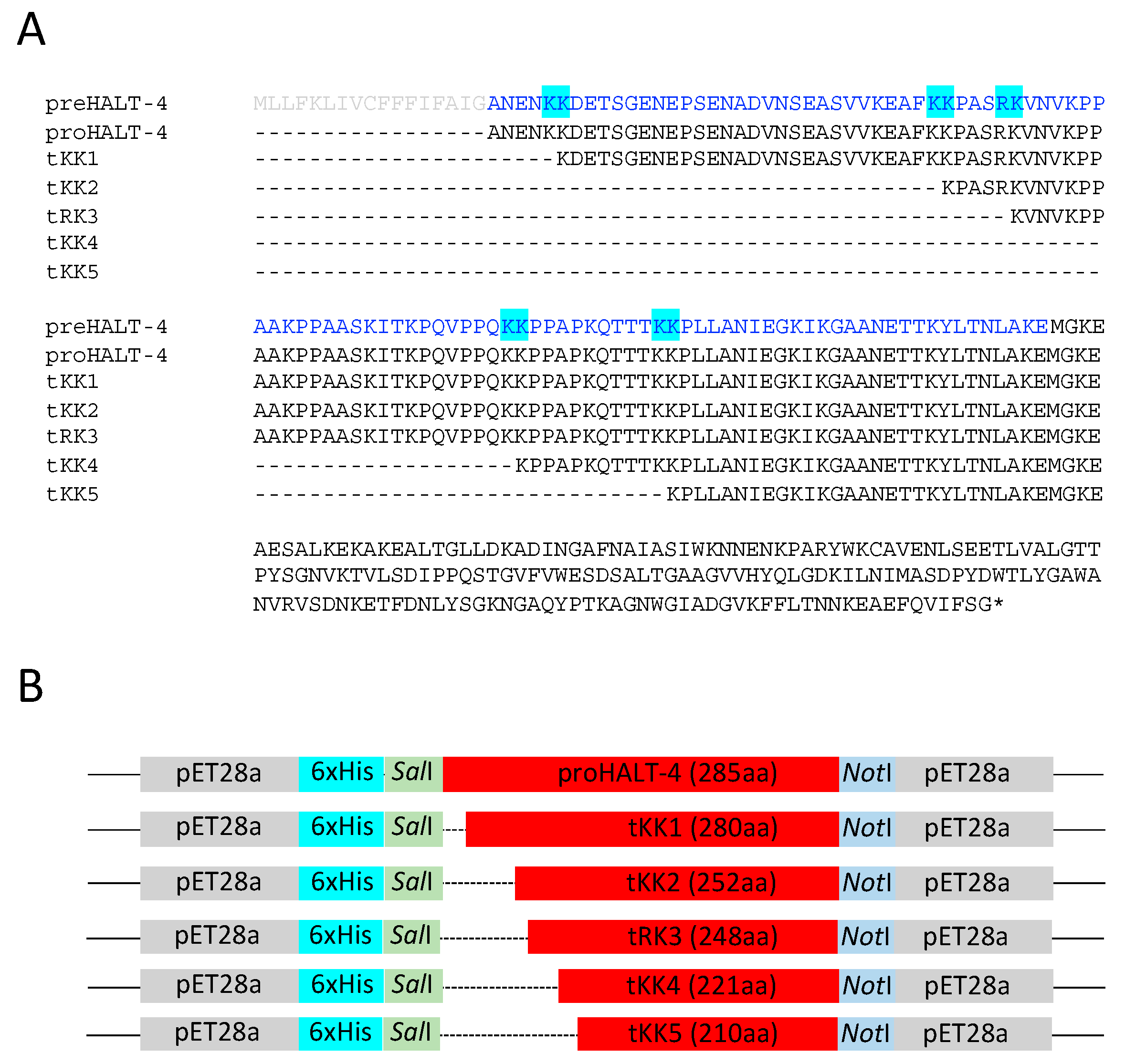
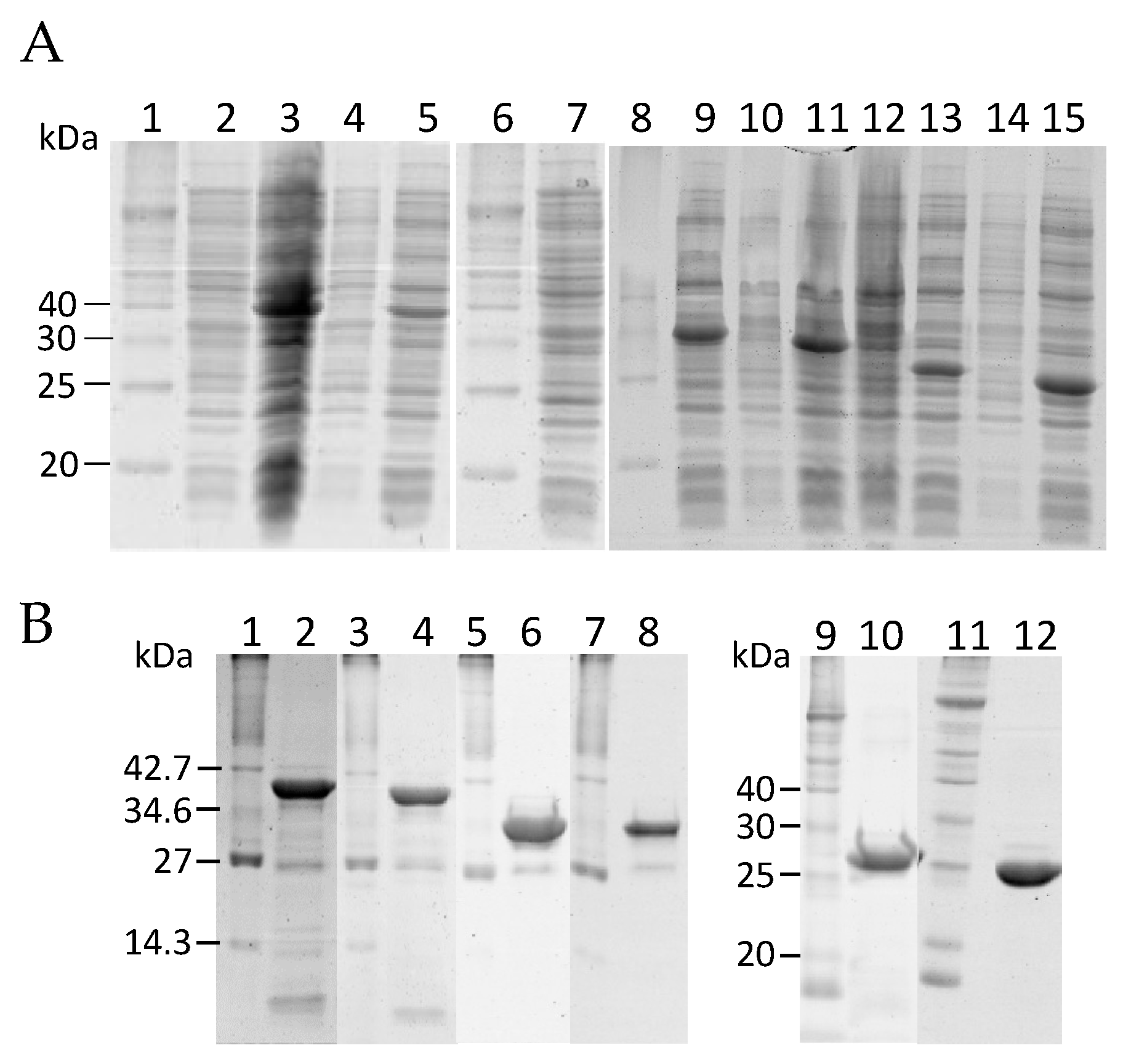
2.1. Construction of Truncated rHALT-4
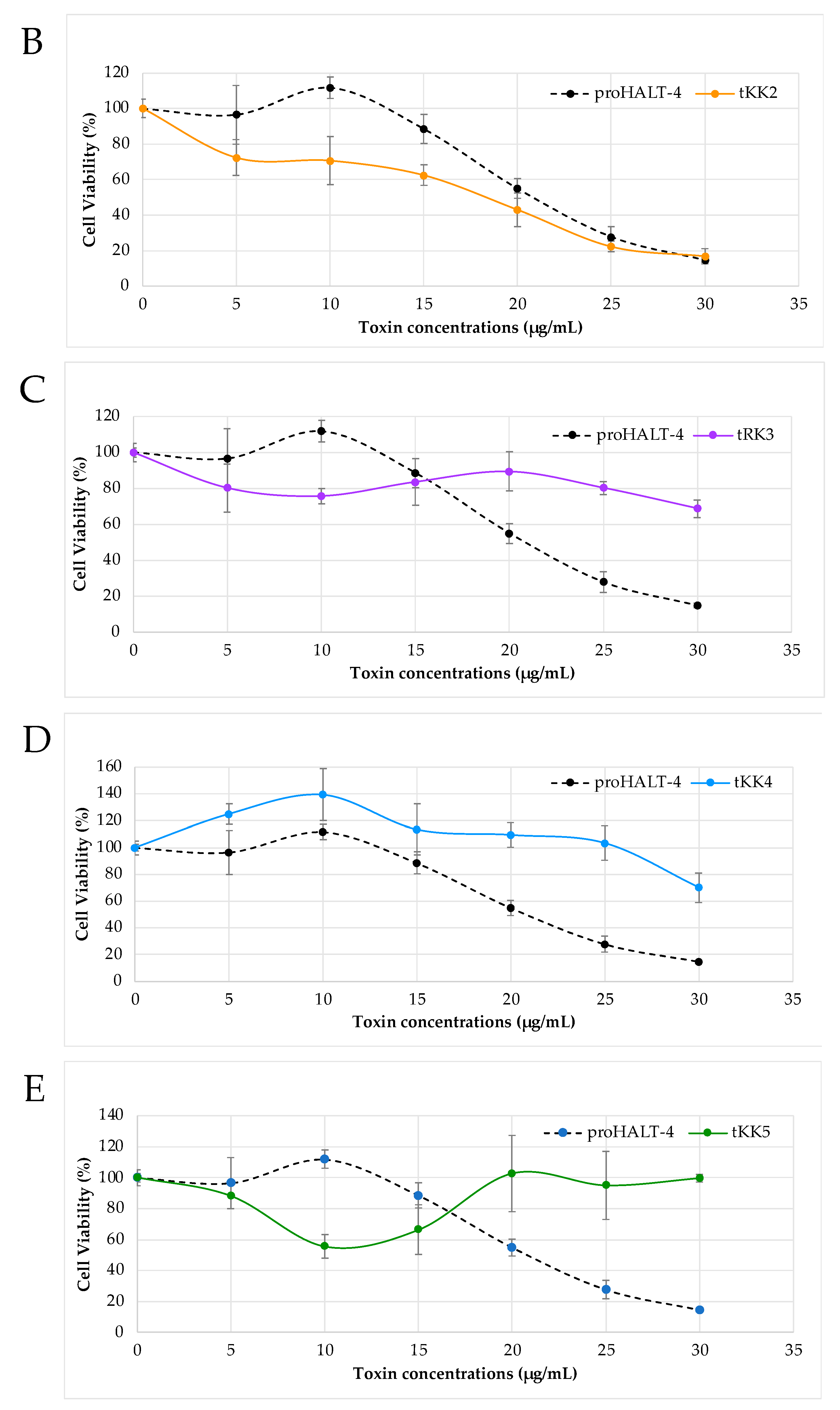
2.2. Cytotoxicity of Truncated rHALT-4
3. Conclusions
4. Materials and Methods
4.1. Construction of KK1, KK2, RK3, KK4 and KK5
4.2. Expression and Purification of KK1, KK2, RK3, KK4 and KK5
4.3. Cytotoxicity Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Glasser, E.; Rachamim, T.; Aharonovich, D.; Sher, D. Hydra actinoporin-like toxin-1, an unusual hemolysin from the nematocyst venom of Hydra magnipapillata which belongs to an extended gene family. Toxicon 2014, 91, 103–113. [Google Scholar] [CrossRef]
- Yap, W.Y.; Tan, K.J.S.X.; Hwang, J.S. Expansion of Hydra actinoporin-like toxin (HALT) gene family: Expression divergence and functional convergence evolved through gene duplication. Toxicon 2019, 170, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Rholam, M.; Nicolas, P.; Cohen, P. Precursors for peptide hormones share common secondary structures forming features at the proteolytic processing sites. FEBS Lett. 1986, 207, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rholam, M.; Brakch, N.; Germain, D.; Thomas, D.Y.; Fahy, C.; Bousetta, H.; Boileau, G.; Cohen, P. Role of amino acid sequences flanking dibasic cleavage sites in precursor proteolytic processing the importance of the first residue C-terminal of the cleavage site. Eur. J. Biochem. 1995, 227, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Podlesek, Z.; Macek, P. A common motif in proparts of Cnidarian toxins and nematocyst collagens and its putative role. Biochim. Biophys. Acta 2000, 1476, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Belmonte, G.; Menestrina, G.; Pederzolli, C.; Krizaj, I.; Gubensek, F.; Turk, T.; Macek, P. Primary and secondary structure of a pore-forming toxin from the sea anemone, Actinia equina L., and its association with lipid vesicles. Biochim. Biophys. Acta 1994, 1192, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Pungercar, J.; Strukelj, B.; Macek, P.; Gubensek, F. Cloning, sequencing, and expression of equinatoxin II. Biochem. Biophys. Res. Commun. 1996, 220, 437–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, S.J.; Oliva, A.A., Jr.; LaMendola, J.; Grens, A.; Bode, H.; Steiner, D.F. Conservation of the prohormone convertase gene family in metazoa: Analysis of cDNAs encoding a PC3-like protein from hydra. Proc. Natl. Acad. Sci. USA 1992, 89, 6678–6682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brakch, N.; Allemandou, F.; Cavadas, C.; Grouzmann, E.; Brunner, H.R. Dibasic cleavage site is required for sorting to the regulated secretory pathway for both pro- and neuropeptide Y. J. Neurochem. 2002, 81, 1166–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feliciangeli, S.; Kitabgi, P.; Bidard, J.N. The role of dibasic residues in prohormone sorting to the regulated secretory pathway. A study with proneurotensin. J. Biol. Chem. 2001, 276, 6140–6150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brakch, N.; Cohen, P.; Boileau, G. Processing of human prosomatostatin in AtT-20 cells: S-28 and S-14 are generated in different secretory pathways. Biochem. Biophys. Res. Commun. 1994, 205, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Valle, A.; Hervis, Y.P.; Socas, L.B.; Canet, L.; Faheem, M.; Barbosa, J.A.; Lanio, M.E.; Pazos, I.F. The multigene families of actinoporins (part II): Strategies for heterologous production in Escherichia coli. Toxicon 2016, 118, 64–81. [Google Scholar] [CrossRef] [PubMed]
- Uechi, G.; Toma, H.; Arakawa, T.; Sato, Y. Molecular cloning and functional expression of hemolysin from the sea anemone Actineria villosa. Protein Expr. Purif. 2005, 40, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Kristan, K.C.; Viero, G.; Serra, M.D.; Maček, P.; Anderluh, G. Molecular mechanism of pore formation by actinoporins. Toxicon 2009, 54, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Ortega, J.; García-Linares, S.; Rivera-de-Torre, E.; Gavilanes, J.G.; Martínez-del-Pozo, Á.; Slotte, J.P. Differential effect of bilayer thickness on sticholysin activity. Langmuir 2017, 33, 11018–11027. [Google Scholar] [CrossRef] [PubMed]

| Primer | Nucleotide Sequence a |
|---|---|
| KK1HALT4_FW | 5′-CAGGTCGACAAAAAGACGAAACCTCAGGT-3′ |
| KK2HALT4_FW | 5′-CGCGTCGACAAAAACCTGCATCACGAAAA-3′ |
| RK3HALT4_FW | 5′-CGCGTCGACGAAAAGTCAATGTAAAACCC-3′ |
| KK4HALT4_FW | 5′-CGCGTCGACAAAAACCACCCGCACCAAAA-3′ |
| KK5HALT4_FW | 5′-GGCGTCGACAAAAACCTTTGTTAGCTAAT-3′ |
| HALT4_RV | 5′-GTTGCGGCCGCTTACCCAGAAAAAATTAC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yap, W.Y.; Hwang, J.S. Is Proteolytic Cleavage Essential for the Enhanced Activity of Hydra Pore-Forming Toxin, HALT-4? Toxins 2023, 15, 396. https://doi.org/10.3390/toxins15060396
Yap WY, Hwang JS. Is Proteolytic Cleavage Essential for the Enhanced Activity of Hydra Pore-Forming Toxin, HALT-4? Toxins. 2023; 15(6):396. https://doi.org/10.3390/toxins15060396
Chicago/Turabian StyleYap, Wei Yuen, and Jung Shan Hwang. 2023. "Is Proteolytic Cleavage Essential for the Enhanced Activity of Hydra Pore-Forming Toxin, HALT-4?" Toxins 15, no. 6: 396. https://doi.org/10.3390/toxins15060396




