Two Novel Mosquitocidal Peptides Isolated from the Venom of the Bahia Scarlet Tarantula (Lasiodora klugi)
Abstract
1. Introduction
2. Results
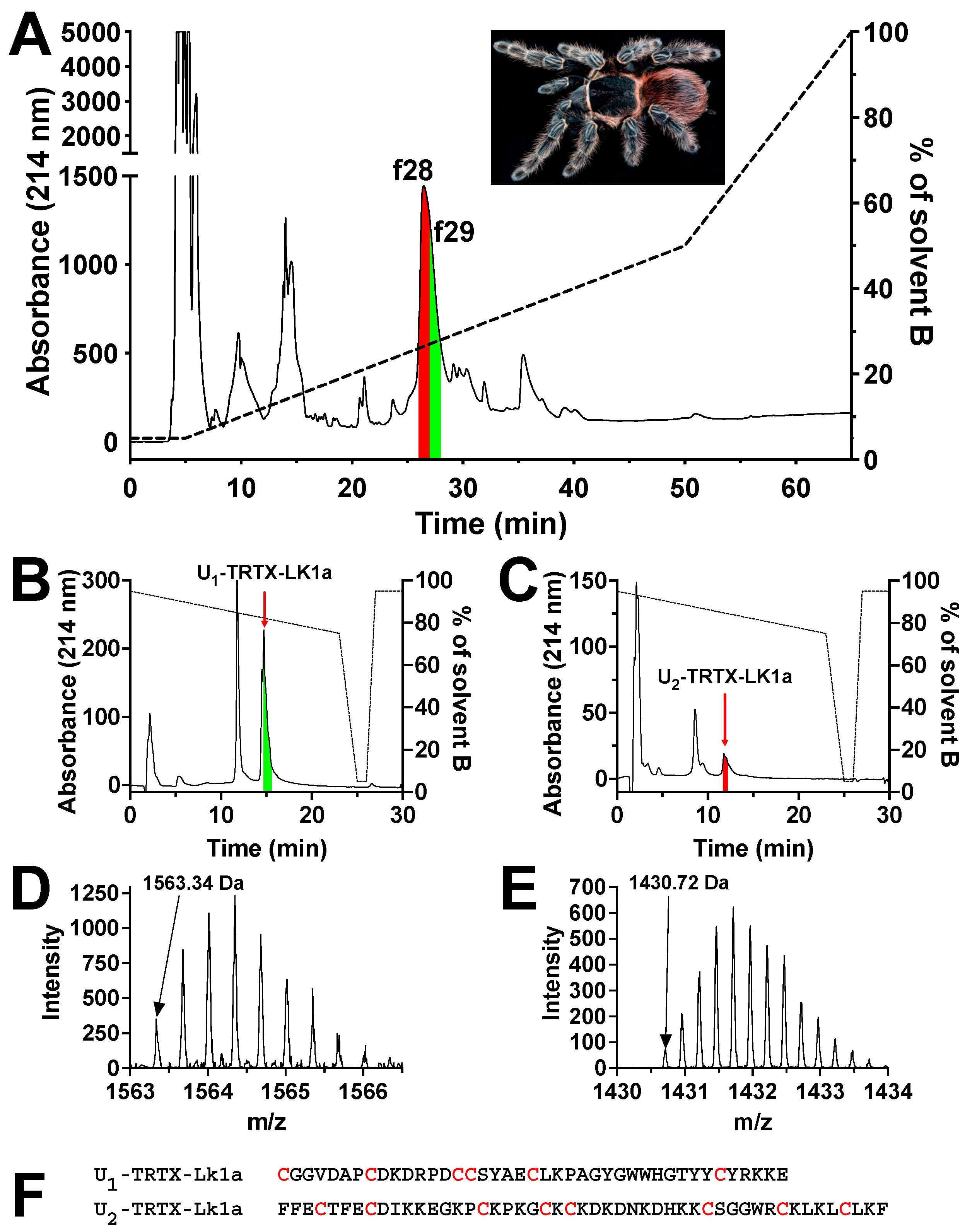
2.1. Isolation and Purification of Mosquitocidal Tarantula Venom Peptides
2.2. Mosquitocidal ACTIVITY
2.3. Primary Structure Determination for Mosquitocidal Venom Peptides
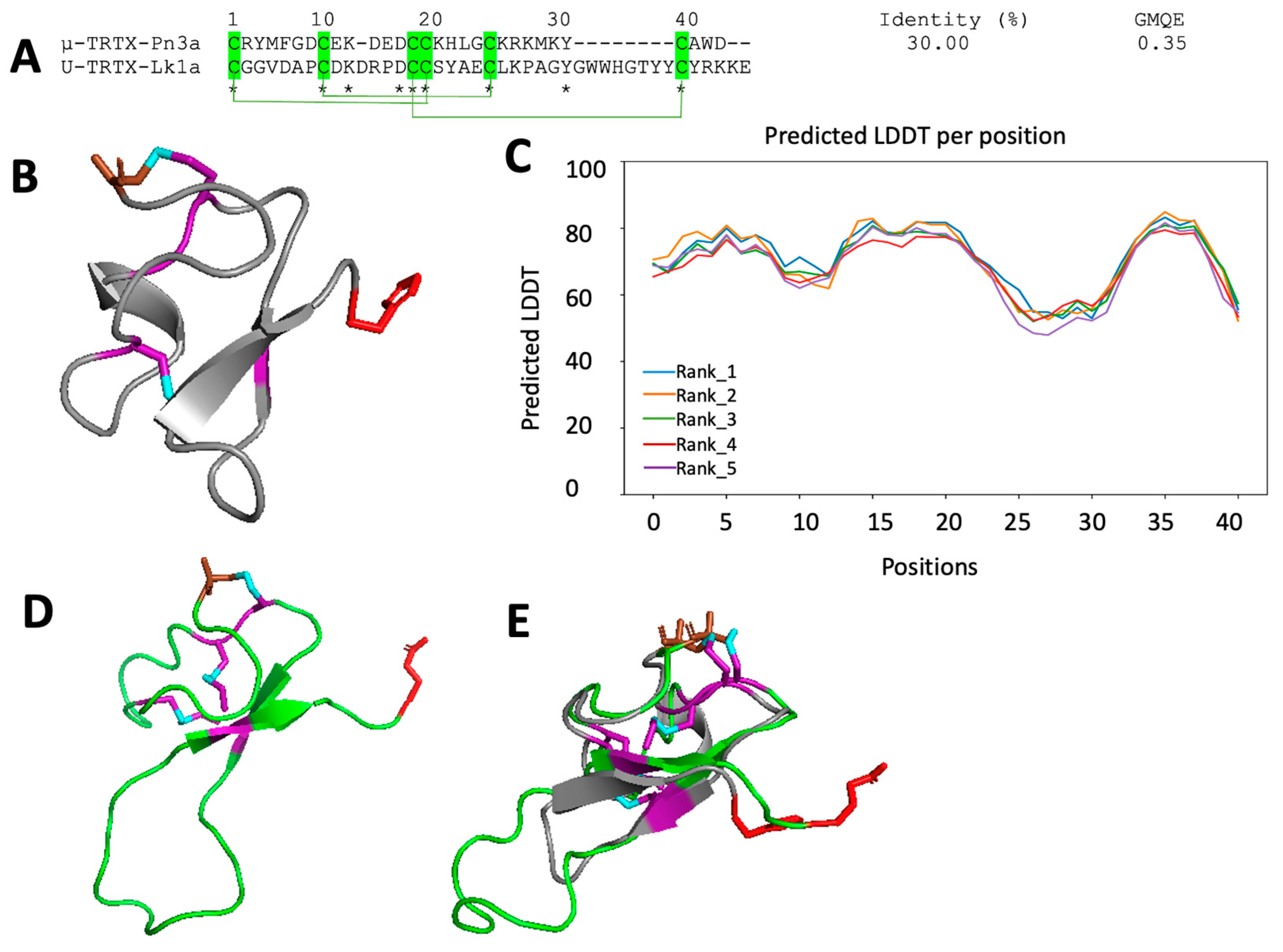
2.4. In Silico Structures of Isolated Peptides
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Rearing of Aedes aegypti
5.2. Venom Extraction
5.3. Mosquito Toxicity Bioassay
5.4. Peptide Isolation
5.5. Proteomics
5.6. Edman Sequencing
5.7. Structure Modeling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organisation (WHO) Fact Sheet: Yellow Fever. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/yellow-fever (accessed on 10 June 2022).
- World Health Organisation (WHO) Fact Sheet: Dengue and Severe Dengue. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 10 July 2022).
- Centers for Disease Control and Prevention (CDC) Website: Areas with the Risk of Dengue. 2021. Available online: https://www.cdc.gov/dengue/areaswithrisk/index.html (accessed on 3 June 2022).
- Salam, N.; Mustafa, S.; Hafiz, A.; Chaudhary, A.A.; Deeba, F.; Parveen, S. Global prevalence and distribution of coinfection of malaria, dengue and chikungunya: A systematic review. BMC Public Health 2018, 18, 710. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation (WHO) Zika Virus. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/zika-virus (accessed on 23 January 2022).
- Ukpai, O.M.; Ekedo, C.M. Insecticide susceptibility status of Aedes aegypti in Umudike, Ikwuano LGA Abia state, Nigeria. Anim. Res. Int. 2018, 15, 3082–3089. [Google Scholar]
- Sene, N.M.; Mavridis, K.; Ndiaye, E.H.; Diagne, C.T.; Gaye, A.; Ngom, E.H.M.; Ba, Y.; Diallo, D.; Vontas, J.; Dia, I.; et al. Insecticide resistance status and mechanisms in Aedes aegypti populations from Senegal. PLoS Negl. Trop. Dis. 2021, 15, e0009393. [Google Scholar] [CrossRef] [PubMed]
- King, G.F. Tying pest insects in knots: The deployment of spider-venom-derived knottins as bioinsecticides. Pest Manag. Sci. 2019, 75, 2437–2445. [Google Scholar] [CrossRef] [PubMed]
- Agnarsson, I.; Coddington, J.A.; Kuntner, M. Systematics—Progress in the study of spider diversity and evolution. In Spider Research in the 21st Century; Penny, D., Ed.; Siri Scientific Press: Rochdale, UK, 2013; pp. 11–58. [Google Scholar]
- World Spider Catalog. Version 19.5. Natural History Museum Bern. 2023. Available online: http://wsc.nmbe.ch (accessed on 3 April 2023).
- Windley, M.J.; Herzig, V.; Dziemborowicz, S.A.; Hardy, M.C.; King, G.F.; Nicholson, G.M. Spider-venom peptides as bioinsecticides. Toxins 2012, 4, 191–227. [Google Scholar] [CrossRef]
- Langenegger, N.; Nentwig, W.; Kuhn-Nentwig, L. Spider venom: Components, modes of action, and novel strategies in transcriptomic and proteomic analyses. Toxins 2019, 11, 611. [Google Scholar] [CrossRef]
- Guo, S.; Herzig, V.; King, G.F. Dipteran toxicity assays for determining the oral insecticidal activity of venoms and toxins. Toxicon 2018, 150, 297–303. [Google Scholar] [CrossRef]
- Nyffeler, M.; Sunderland, K.D. Composition, abundance and pest control potential of spider communities in agroecosystems: A comparison of European and US studies. Agric. Ecosyst. Environ. 2003, 95, 579–612. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L.; Largiader, C.R.; Streitberger, K.; Chandru, S.; Baumann, T.; Kampfer, U.; Schaller, J.; Schurch, S.; Nentwig, W. Purification, cDNA structure and biological significance of a single insulin-like growth factor-binding domain protein (SIBD-1) identified in the hemocytes of the spider Cupiennius salei. Insect Biochem. Mol. Biol. 2011, 41, 891–901. [Google Scholar] [CrossRef]
- Forster, Y.M.; Reusser, S.; Forster, F.; Bienz, S.; Bigler, L. VenoMS—A website for the low molecular mass compounds in spider venoms. Metabolites 2020, 10, 327. [Google Scholar] [CrossRef]
- Lüddecke, T.; Herzig, V.; Von Reumont, B.M.; Andreas, V. The biology and evolution of spider venoms. Biol. Rev. 2022, 97, 163–178. [Google Scholar] [CrossRef]
- Pineda, S.S.; Chaumeil, P.-A.; Kunert, A.; Kaas, Q.; Thang, M.W.C.; Le, L.; Nuhn, M.; Herzig, V.; Saez, N.J.; Cristofori-Armstrong, B.; et al. ArachnoServer 3.0: An online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics 2018, 34, 1074–1076. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Zhang, Y.; Zeng, J.; Liang, S.; Tang, C.; Liu, Z. Purification and characterization of a novel insecticidal toxin, μ-sparatoxin-Hv2, from the venom of the spider Heteropoda venatoria. Toxins 2018, 10, 233. [Google Scholar] [CrossRef]
- Alvarado, D.; Cardoso-Arenas, S.; Corrales-Garcia, L.-L.; Clement, H.; Arenas, I.; Montero-Dominguez, P.A.; Olamendi-Portugal, T.; Zamudio, F.; Csoti, A.; Borrego, J.; et al. A novel insecticidal spider peptide that affects the mammalian voltage-gated ion channel hKv1.5. Front. Pharmacol. 2021, 11, 563858. [Google Scholar] [CrossRef] [PubMed]
- Windley, M.J.; Vetter, I.; Lewis, R.J.; Nicholson, G.M. Lethal effects of an insecticidal spider venom peptide involve positive allosteric modulation of insect nicotinic acetylcholine receptors. Neuropharmacology 2017, 127, 224–242. [Google Scholar] [CrossRef]
- Dunlop, J.A. Geological history and phylogeny of Chelicerata. Arthropod Struct. Dev. 2010, 39, 124–142. [Google Scholar] [CrossRef] [PubMed]
- Waddington, J.; Rudkin, D.M.; Dunlop, J.A. A new mid-silurian aquatic scorpion-one step closer to land? Biol. Lett. 2015, 11, 20140815. [Google Scholar] [CrossRef]
- The Scorpion Files. Available online: https://www.ntnu.no/ub/scorpion-files/ (accessed on 30 November 2022).
- Santibáñez-López, C.E.; Francke, O.F.; Ureta, C.; Possani, L.D. Scorpions from Mexico: From species diversity to venom complexity. Toxins 2016, 8, 2. [Google Scholar] [CrossRef]
- Zlotkin, E. Scorpion venoms. In Comprehensive Molecular Insect Science; Gilbert, L.I., Iatrou, K., Gill, S.S., Eds.; Elsevier B.V.: Oxford, UK, 2005; pp. 173–220. [Google Scholar]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef]
- Fitches, E.C.; Bell, H.A.; Powell, M.E.; Back, E.; Sargiotti, C.; Weaver, R.J.; Gatehouse, J.A. Insecticidal activity of scorpion toxin (ButaIT) and snowdrop lectin (GNA) containing fusion proteins towards pest species of different orders. Pest Manag. Sci. 2010, 66, 74–83. [Google Scholar] [CrossRef]
- Juichi, H.; Miyashita, M.; Nakagawa, Y.; Miyagawa, H. Isolation and characterization of the insecticidal, two-domain toxin LaIT3 from the Liocheles australasiae scorpion venom. Biosci. Biotech. Biochem. 2017, 83, 2183–2189. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, Y.; Miyashita, M.; Abdel-Wahab, M.; Sarhan, M.; Nakagawa, Y.; Miyagawa, H. Isolation and characterization of insecticidal toxins from the venom of the North African scorpion, Buthacus leptochelys. Toxins 2019, 11, 236. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez-Guzmán, M.J.; Jiménez-Vargas, J.M.; Possani, L.D.; Zamudio, F.; Orozco-Gutiérrez, G.; Oceguera-Contreras, E.; Enríquez-Vara, J.N.; Vazquez-Vuelvas, O.F.; García-Villalvazo, P.E.; Valdez-Velázquez, L.L. Biochemical characterization and insecticidal activity of isolated peptides from the venom of the scorpion Centruroides tecomanus. Toxicon 2022, 206, 90–102. [Google Scholar] [CrossRef]
- Saez, N.J.; Herzig, V. Versatile spider venom peptides and their medical and agricultural applications. Toxicon 2019, 158, 109–126. [Google Scholar] [CrossRef]
- Wang, C.; St. Leger, R.J. A scorpion neurotoxin increases the potency of a fungal insecticide. Nat. Biotechnol. 2007, 25, 1455–1456. [Google Scholar] [CrossRef] [PubMed]
- Lovett, B.; Bilgo, E.; Millogo, S.A.; Ouattarra, A.; Sare, I.; Gnambani, E.J.; Dabire, R.K. Transgenic Metarhizium rapidly kills mosquitoes in a malaria-endemic region in Burkina Faso. Science 2019, 364, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Bomgardner, M.M. Spider venom: An insecticide whose time has come. Chem. Eng. News 2017, 95, 30–31. Available online: https://cen.acs.org/articles/95/i11/Spider-venom-insecticide-whose-time.html (accessed on 15 June 2022).
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 2008, 52, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Deuis, J.; Dekan, Z.; Wingerd, J.; Smith, J.J.; Munasinghe, N.R.; Bhola, R.F.; Imlach, W.L.; Herzig, V.; Armstrong, D.A.; Rosengren, K.J.; et al. Pharmacological characterisation of the highly NaV1.7 selective spider venom peptide Pn3a. Sci. Rep. 2017, 7, 40883. [Google Scholar] [CrossRef]
- Pallaghy, P.K.; Nielsen, K.J.; Craik, D.J.; Norton, R.S. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994, 3, 1833–1839. [Google Scholar] [CrossRef]
- Corzo, G.; Bernard, C.; Clement, H.; Peigneur, S.; Odell, G.; Tytgat, J.; Possani, L.D.; Alago´n, A. Insecticidal peptides from the theraphosid spider Brachypelma albiceps: An NMR-based model of Ba2. Biochim. Biophys. Acta 2009, 1794, 1190–1196. [Google Scholar] [CrossRef]
- Wang, X.H.; Connor, M.; Smith, R.; Maciejewski, M.W.; Howden, M.E.H.; Nicholson, G.M.; Christie, M.J.; King, G.F. Discovery and characterization of a family of insecticidal neurotoxins with a rare vicinal disulfide bond. Nat. Struct. Biol. 2000, 7, 505–513. [Google Scholar] [CrossRef]
- Herzig, V.; Ikonomopoulou, M.; Smith, J.J.; Dziemborowicz, S.; Gilchrist, J.; Kuhn-Nentwig, L.; Oliveira Rezende, F.; Andrade Moreira, L.; Nicholson, G.M.; Bosmans, F.; et al. Molecular basis of the remarkable species selectivity of an insecticidal sodium channel toxin from African spider Augacephalus ezendami. Sci. Rep. 2016, 6, 29538–29549. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Herzig, V.; Ikonomopoulou, M.P.; Dziemborowicz, S.; Bosmans, F.; Nicholson, G.M.; King, G.F. Insect-active toxins with prosmiscuous pharmacology from the African theraphosid spider Monocentropus balfouri. Toxins 2017, 9, 155. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Lu, H.L.; King, G.F.; St. Leger, R.J. Construction of a hypervirulent and specific mycoinsecticide for locust control. Sci. Rep. 2014, 4, 7345. [Google Scholar] [CrossRef] [PubMed]
- Bilgo, E.; Lovett, B.; Fang, W.; Bende, N.; King, G.F.; Dabate, A.; Leger, R.J. Improved efficacy of an arthropod toxin expressing fungus against insecticide-resistant malaria-vector mosquitoes. Sci. Rep. 2017, 7, 3433. [Google Scholar] [CrossRef] [PubMed]
- Accoti, A.; Engdahl, C.S.; Dimopoulos, G. Discovery of novel entomopathogenic fungi for mosquito-borne disease control. Front. Fungal Biol. 2021, 2, 637234. [Google Scholar] [CrossRef]
- Herzig, V.; Bende, N.S.; Alam, M.S.; Tedford, W.H.; Kennedy, R.M.; King, G.F. Methods for deployment of spider venom peptides as bioinsecticides. In Advances in Insect Physiology: Insect Midgut and Insecticidal Proteins for Insect Control; Dhadialla, T.S., Gills, S.S., Eds.; Academic Press: London, UK, 2014; pp. 389–411. [Google Scholar]
- Pineda, S.S.; Chin, Y.K.; Undheim, E.A.B.; Senff, S.; Mobli, M.; Dauly, C.; Escoubas, P.; Nicholson, G.M.; Kaas, Q.; Guo, S.; et al. Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene. Proc. Natl. Acad. Sci. USA 2020, 117, 11399–11408. [Google Scholar] [CrossRef]
- Kolmar, H. Natural and engineered cystine knot miniproteins for diagnostic and therapeutic applications. Curr. Pharm. Des. 2011, 17, 4329–4336. [Google Scholar] [CrossRef]
- Herzig, V.; King, G.F. The cystine knot is responsible for the exceptional stability of the insecticidal spider toxin ω-hexatoxin-Hv1a. Toxins 2015, 7, 4366–4380. [Google Scholar] [CrossRef]
- Shu, Q.; Lu, S.-Y.; Gu, X.-C.; Liang, S.-P. The structure of spider toxin huwentoxin-II with unique disulfide linkage: Evidence for structural evolution. Protein Sci. 2002, 11, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Hill, J.M.; Little, M.J.; Nicholson, G.M.; King, G.F.; Alewood, P.F. Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif. Proc. Natl. Acad. Sci. USA 2011, 108, 10478–10483. [Google Scholar] [CrossRef]
- Chen, J.; Xu, Y.; San, M.; Cao, Z.; Li, W.; Wu, Y.; Chen, Z. Cloning and genomic characterization of a natural insecticidal peptide LaIT1 with unique DDH structural fold. J. Biochem. Mol. Toxicol. 2015, 29, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, H.D.; Hussain, M.; Parry, R.; Etebari, K.; Hedges, L.M.; Zang, G.; Schulz, B.M.; Asgari, S. Human blood microRNA hsa-miR-21-5p induces vitellogenin in the mosquito Aedes aegypti. Commun. Biol. 2021, 4, 856. [Google Scholar] [CrossRef]
- Herzig, V.; Hodgson, W.C. Intersexual variations in the pharmacological properties of Coremiocnemis tropix (Araneae, Theraphosidae) spider venom. Toxicon 2009, 53, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.; Pimenta, A.M.C.; Bemquerer, M.P.; Santoro, M.M.; Beirao, P.S.L.; Lima, M.E.; Figueiredo, S.G.; Bloch, C., Jr.; Vasconcelos, E.A.R.; Campos, F.A.P.; et al. Comparison of the partial proteomes of the venoms of Brazilian spiders of the genus Phoneutria. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006, 142, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Badgett, M.J.; Boyes, B.; Orlando, R. The Separation and quantitation of peptides with and without oxidation of methionine and deamidation of asparagine using hydrophilic interaction liquid chromatography with mass spectrometry (HILIC-MS). J. Am. Soc. Mass Spectrom. 2017, 28, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.A.; Mayhew, M.L.; Jin, J.; Herzig, V.; Undheim, E.A.B.; Sombke, A.; Fry, G.B.; Meritt, D.J.; King, G.F. The assassin bug Pristhesancus plagipennis produces two distinct venoms in separate gland lumens. Nat. Comm. 2018, 9, 755–765. [Google Scholar] [CrossRef]
- Wang, X.-H.; Smith, R.; Fletcher, J.I.; Wilson, H.; Wood, C.J.; Howden, M.E.; King, G.F. Structure function studies of v-atracotoxin, a potent antagonist of insect voltage-gated calcium channels. Eur. J. Biochem. 2001, 264, 488–494. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Meier, M.; Mirdita, M.; Vöhringer, H.; Haunsberger, S.J.; Söding, J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinform. 2019, 20, 473–488. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, J.; Walker, A.A.; Perdomo, H.D.; Guo, S.; Nixon, S.A.; Vetter, I.; Okoh, H.I.; Shehu, D.M.; Shuaibu, M.N.; Ndams, I.S.; et al. Two Novel Mosquitocidal Peptides Isolated from the Venom of the Bahia Scarlet Tarantula (Lasiodora klugi). Toxins 2023, 15, 418. https://doi.org/10.3390/toxins15070418
Ahmed J, Walker AA, Perdomo HD, Guo S, Nixon SA, Vetter I, Okoh HI, Shehu DM, Shuaibu MN, Ndams IS, et al. Two Novel Mosquitocidal Peptides Isolated from the Venom of the Bahia Scarlet Tarantula (Lasiodora klugi). Toxins. 2023; 15(7):418. https://doi.org/10.3390/toxins15070418
Chicago/Turabian StyleAhmed, Jamila, Andrew A. Walker, Hugo D. Perdomo, Shaodong Guo, Samantha A. Nixon, Irina Vetter, Hilary I. Okoh, Dalhatu M. Shehu, Mohammed N. Shuaibu, Iliya S. Ndams, and et al. 2023. "Two Novel Mosquitocidal Peptides Isolated from the Venom of the Bahia Scarlet Tarantula (Lasiodora klugi)" Toxins 15, no. 7: 418. https://doi.org/10.3390/toxins15070418
APA StyleAhmed, J., Walker, A. A., Perdomo, H. D., Guo, S., Nixon, S. A., Vetter, I., Okoh, H. I., Shehu, D. M., Shuaibu, M. N., Ndams, I. S., King, G. F., & Herzig, V. (2023). Two Novel Mosquitocidal Peptides Isolated from the Venom of the Bahia Scarlet Tarantula (Lasiodora klugi). Toxins, 15(7), 418. https://doi.org/10.3390/toxins15070418







