Exploring the Biocontrol Capability of Non-Mycotoxigenic Strains of Penicillium expansum
Abstract
:1. Introduction
2. Results
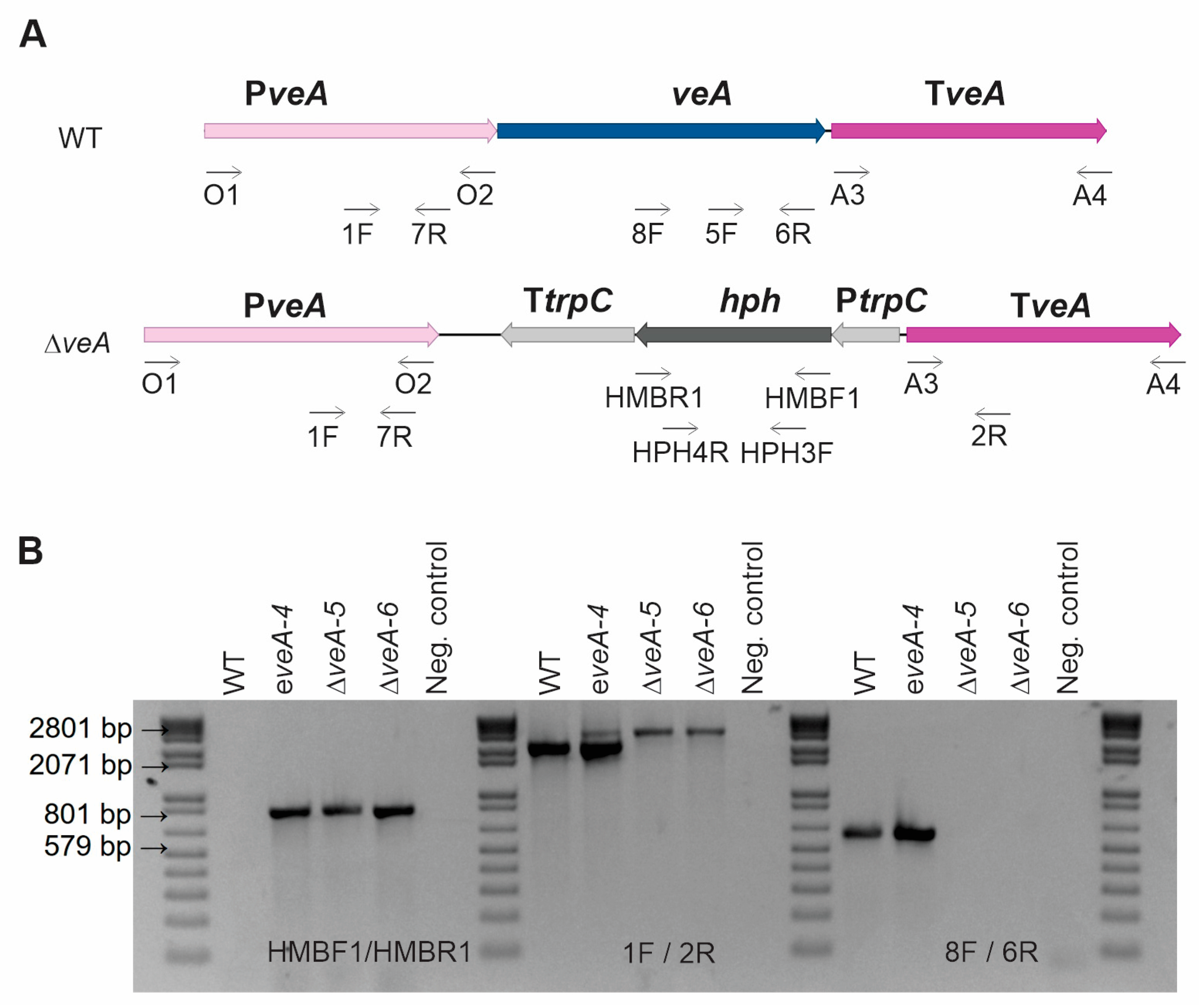
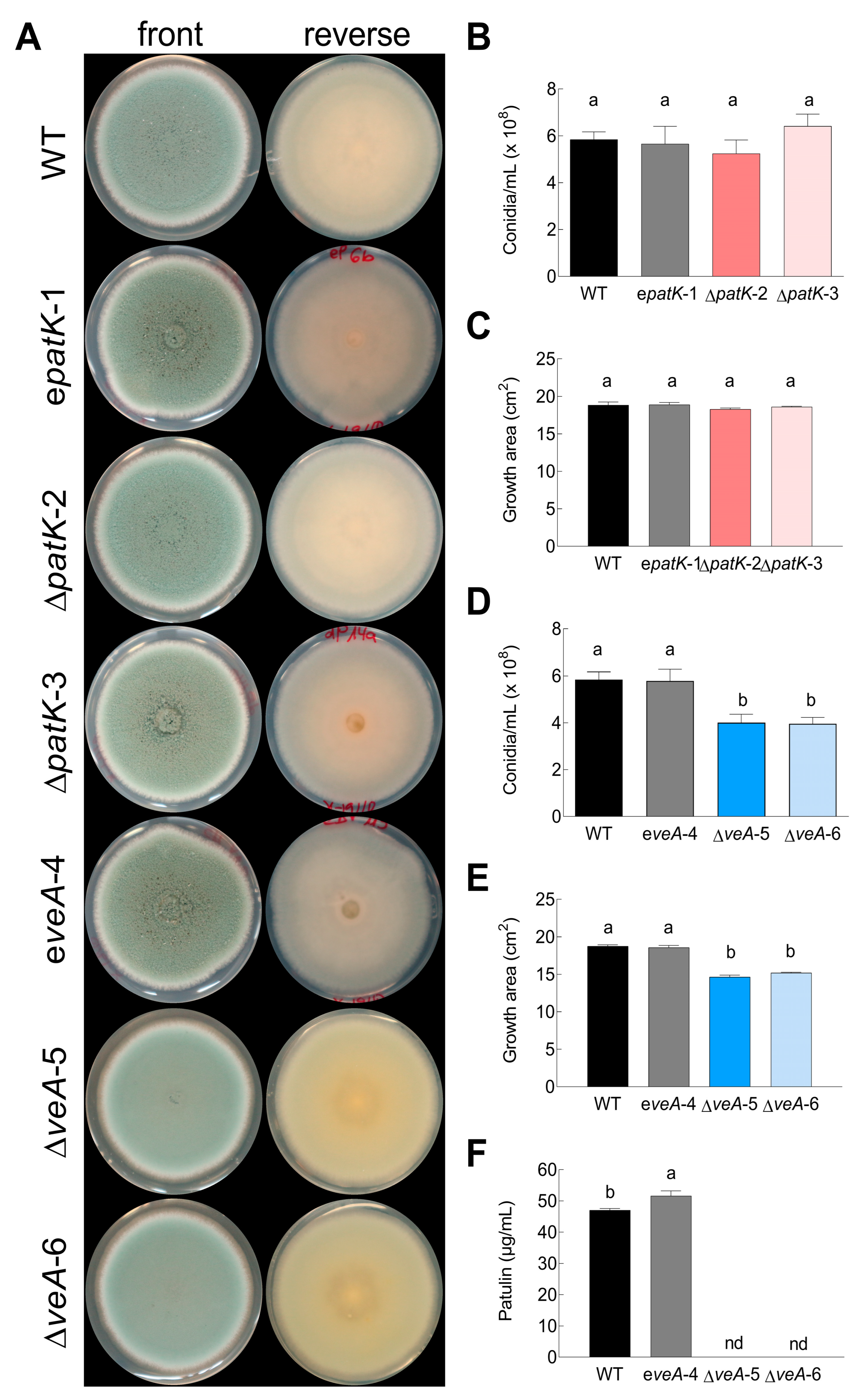
2.1. Generation and Characterization of ΔveA and ΔpatK Mutants
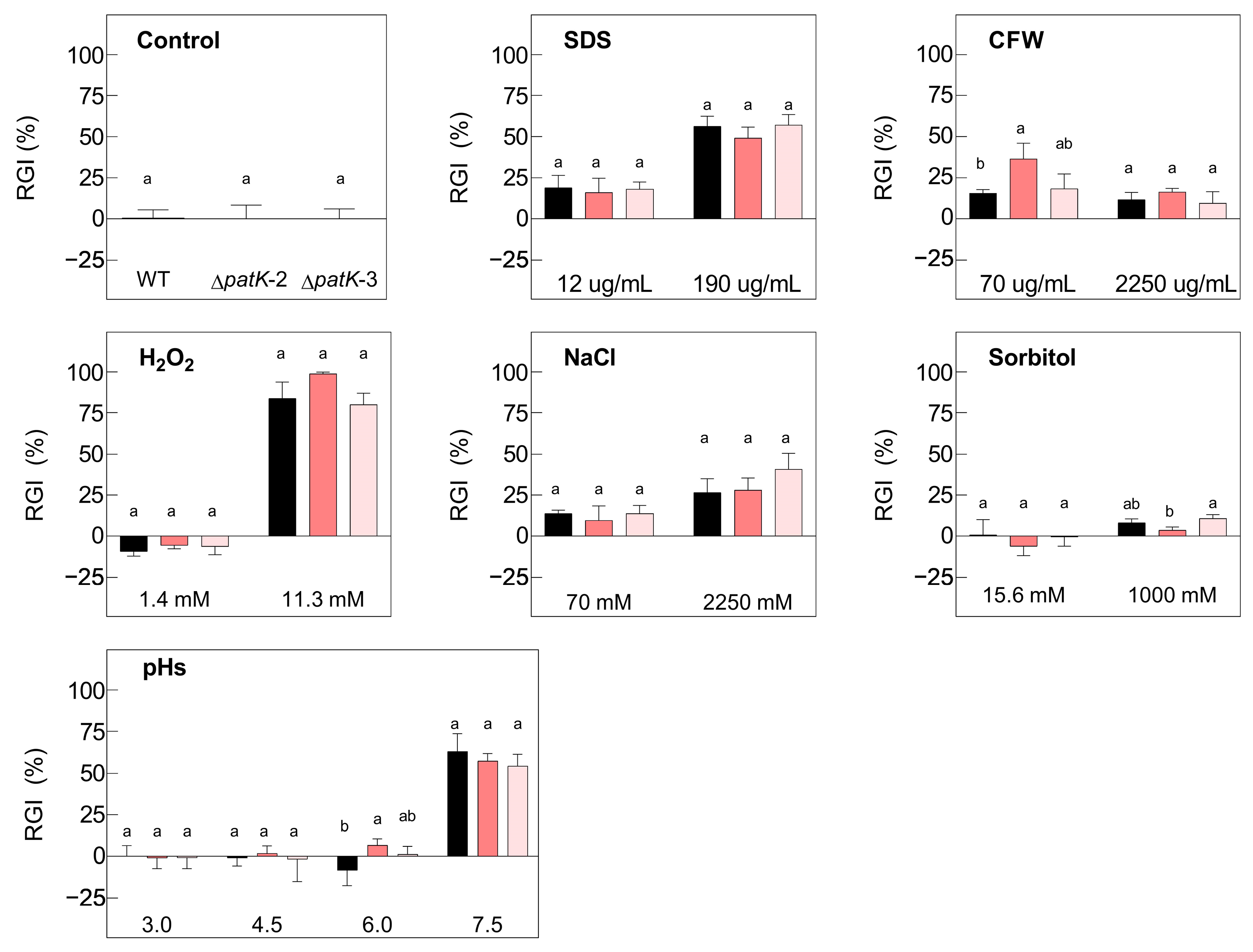
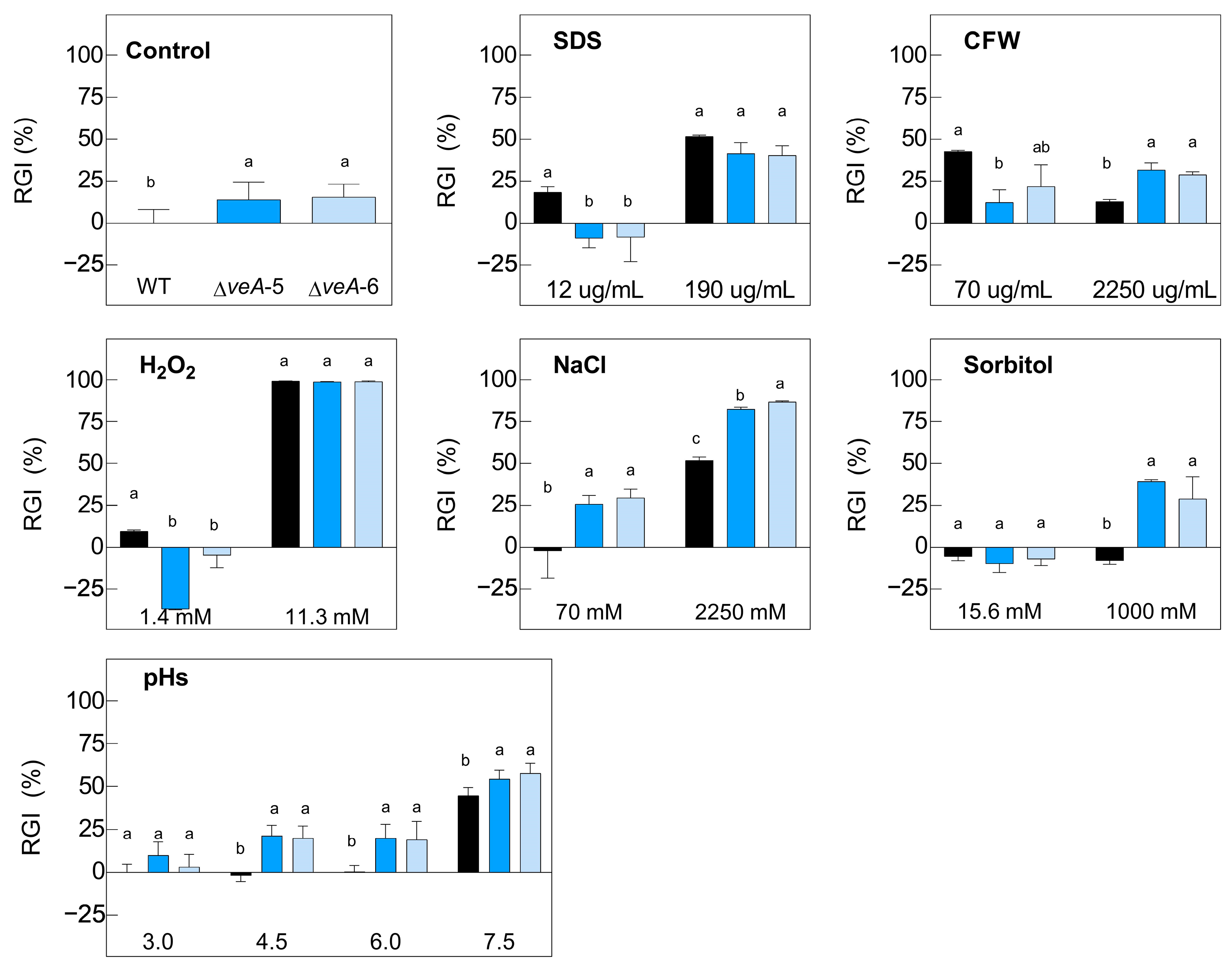
2.2. Effects of Different Stress Conditions on Growth of Knockout Mutants
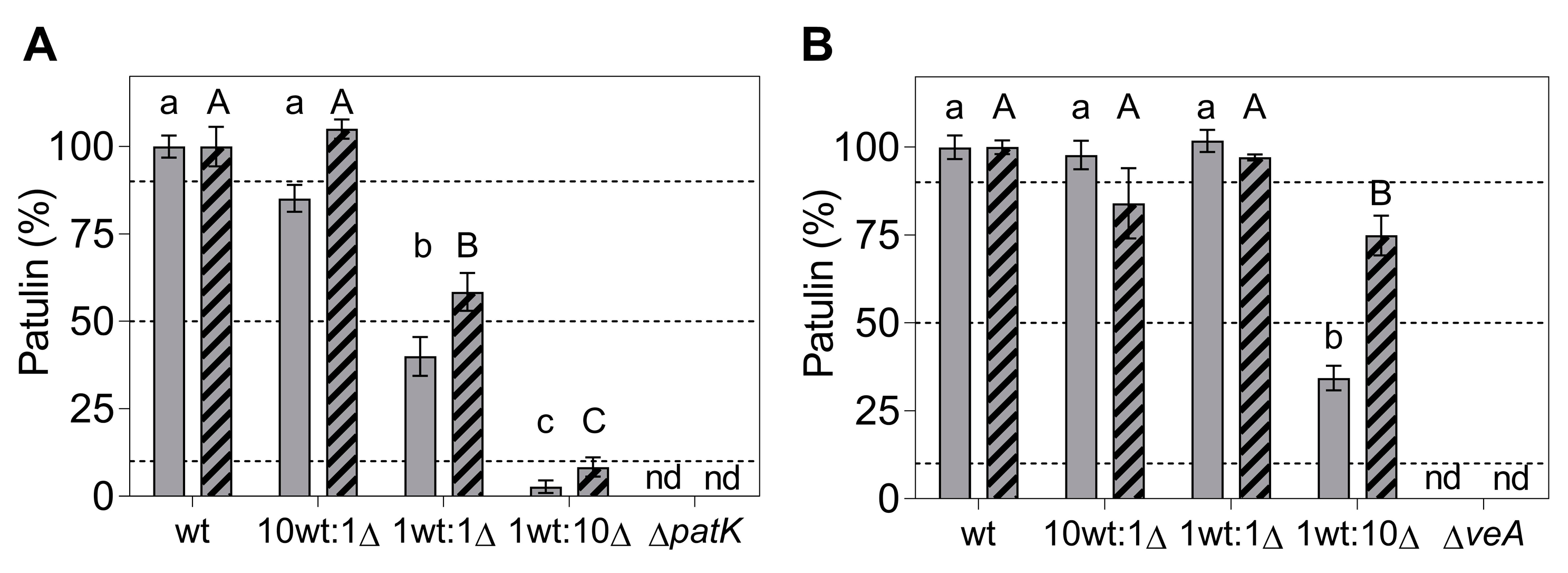
2.3. Effects of Gene Deletions on Competitiveness during In Vitro Growth
3. Discussion
4. Materials and Methods
4.1. Strains, Media, and Growth Conditions
4.2. Generation and Characterization of Mutant Strains
4.3. Macroscopic Morphology, Conidiation, and In Vitro Growth under Stress Conditions
4.4. In Vitro Competition Assays
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Samson, R.A.; Frisvad, J.C. Classification of Aspergillus, Penicillium, Talaromyces and Related Genera (Eurotiales): An Overview of Families, Genera, Subgenera, Sections, Series and Species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Luciano-Rosario, D.; Keller, N.P.; Jurick, W.M. Penicillium expansum: Biology, Omics, and Management Tools for a Global Postharvest Pathogen Causing Blue Mould of Pome Fruit. Mol. Plant Pathol. 2020, 21, 1391–1404. [Google Scholar] [CrossRef] [PubMed]
- Palou, L. Chapter 2—Penicillium digitatum, Penicillium italicum (Green Mold, Blue Mold). In Postharvest Decay; Bautista-Baños, S., Ed.; Academic Press: San Diego, CA, USA, 2014; pp. 45–102. ISBN 978-0-12-411552-1. [Google Scholar]
- Chain, E.; Florey, H.W.; Gardner, A.D.; Heatley, N.G.; Jennings, M.A.; Orr-Ewing, J.; Sanders, A.G. Penicillin as a Chemotherapeutic Agent. Lancet 1940, 236, 226–228. [Google Scholar] [CrossRef]
- Frisvad, J. A Critical Review of Producers of Small Lactone Mycotoxins: Patulin, Penicillic Acid and Moniliformin. World Mycotoxin J. 2018, 11, 73–100. [Google Scholar] [CrossRef]
- Capcarova, M.; Zbynovska, K.; Kalafova, A.; Bulla, J.; Bielik, P. Environment Contamination by Mycotoxins and Their Occurrence in Food and Feed: Physiological Aspects and Economical Approach. J. Environ. Sci. Health Part B 2016, 51, 236–244. [Google Scholar] [CrossRef]
- Tannous, J.; Keller, N.P.; Atoui, A.; El Khoury, A.; Lteif, R.; Oswald, I.P.; Puel, O. Secondary Metabolism in Penicillium expansum: Emphasis on Recent Advances in Patulin Research. Crit. Rev. Food Sci. Nutr. 2018, 58, 2082–2098. [Google Scholar] [CrossRef]
- Filtenborg, O.; Frisvad, J.C.; Thrane, U. Moulds in Food Spoilage. Int. J. Food Microbiol. 1996, 33, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Baert, K.; Devlieghere, F.; Flyps, H.; Oosterlinck, M.; Ahmed, M.M.; Rajković, A.; Verlinden, B.; Nicolaï, B.; Debevere, J.; De Meulenaer, B. Influence of Storage Conditions of Apples on Growth and Patulin Production by Penicillium expansum. Int. J. Food Microbiol. 2007, 119, 170–181. [Google Scholar] [CrossRef]
- Tannous, J.; El Khoury, R.; Snini, S.P.; Lippi, Y.; El Khoury, A.; Atoui, A.; Lteif, R.; Oswald, I.P.; Puel, O. Sequencing, Physical Organization and Kinetic Expression of the Patulin Biosynthetic Gene Cluster from Penicillium expansum. Int. J. Food Microbiol. 2014, 189, 51–60. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Zhang, Z.; Qin, G.; Chen, T.; Tian, S. Molecular Basis and Regulation of Pathogenicity and Patulin Biosynthesis in Penicillium Expansum. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3416–3438. [Google Scholar] [CrossRef]
- Ballester, A.R.; Marcet-Houben, M.; Levin, E.; Sela, N.; Selma, C.; Carmona, L.; Wisniewski, M.; Droby, S.; González-Candelas, L.; Gabaldón, T. Genome, Transcriptome, and Functional Analyses of Penicillium expansum Provide New Insights into Secondary Metabolism and Pathogenicity. Mol. Plant Microbe Interact. 2015, 28, 232–248. [Google Scholar] [CrossRef]
- Amaike, S.; Keller, N.P. Distinct Roles for VeA and LaeA in Development and Pathogenesis of Aspergillus flavus. Eukaryot. Cell 2009, 8, 1051–1060. [Google Scholar] [CrossRef]
- Calvo, A.M. The VeA Regulatory System and Its Role in Morphological and Chemical Development in Fungi. Fungal Genet. Biol. 2008, 45, 1053–1061. [Google Scholar] [CrossRef]
- Estiarte, N.; Lawrence, C.B.; Sanchis, V.; Ramos, A.J.; Crespo-Sempere, A. LaeA and VeA Are Involved in Growth Morphology, Asexual Development, and Mycotoxin Production in Alternaria alternata. Int. J. Food Microbiol. 2016, 238, 153–164. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, H.; Sumarah, M.W.; Gao, Q.; Wang, D.; Zhang, Y. veA Gene Acts as a Positive Regulator of Conidia Production, Ochratoxin a Biosynthesis, and Oxidative Stress Tolerance in Aspergillus niger. J. Agric. Food Chem. 2018, 66, 13199–13208. [Google Scholar] [CrossRef]
- Puel, O.; Galtier, P.; Oswald, I.P. Biosynthesis and Toxicological Effects of Patulin. Toxins 2010, 2, 613–631. [Google Scholar] [CrossRef]
- Drusch, S.; Aumann, J. Mycotoxins in Fruits: Microbiology, Occurrence, and Changes during Fruit Processing. Adv. Food Nutr. Res. 2005, 50, 33–78. [Google Scholar] [CrossRef]
- Notardonato, I.; Gianfagna, S.; Castoria, R.; Ianiri, G.; Curtis, F.D.; Russo, M.V.; Avino, P. Critical Review of the Analytical Methods for Determining the Mycotoxin Patulin in Food Matrices. Rev. Anal. Chem. 2021, 40, 144–160. [Google Scholar] [CrossRef]
- Pinedo, C.; Wright, S.A.I.; Collado, I.G.; Goss, R.J.M.; Castoria, R.; Hrelia, P.; Maffei, F.; Durán-Patrón, R. Isotopic Labeling Studies Reveal the Patulin Detoxification Pathway by the Biocontrol Yeast Rhodotorula kratochvilovae LS11. J. Nat. Prod. 2018, 81, 2692–2699. [Google Scholar] [CrossRef]
- Berni, E.; Montagna, I.; Restivo, F.M.; Degola, F. Ochratoxin A Control in Meat Derivatives: Intraspecific Biocompetition between Penicillium nordicum Strains. J. Food Qual. 2017, 2017, 8370106. [Google Scholar] [CrossRef]
- Álvarez, M.; Núñez, F.; Delgado, J.; Andrade, M.J.; Rodríguez, M.; Rodríguez, A. Competitiveness of Three Biocontrol Candidates against Ochratoxigenic Penicillium nordicum under Dry-Cured Meat Environmental and Nutritional Conditions. Fungal Biol. 2021, 125, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Bartholomew, H.P.; Luciano-Rosario, D.; Bradshaw, M.J.; Gaskins, V.L.; Peng, H.; Fonseca, J.M.; Jurick, W.M. Avirulent Isolates of Penicillium chrysogenum to Control the Blue Mold of Apple Caused by P. expansum. Microorganisms 2023, 11, 2792. [Google Scholar] [CrossRef] [PubMed]
- Requena, E.; Carreras, M.; Espeso, E.A.; Larena, I. A Role for Penicillium rubens Strain 212 Xylanolytic System in Biocontrol of Fusarium Wilt Disease in Tomato Plants. Eur. J. Plant Pathol. 2023, 167, 621–635. [Google Scholar] [CrossRef]
- De Cal, A.; Redondo, C.; Sztejnberg, A.; Melgarejo, P. Biocontrol of Powdery Mildew by Penicillium oxalicum in Open-Field Nurseries of Strawberries. Biol. Control 2008, 47, 103–107. [Google Scholar] [CrossRef]
- Cebrián, E.; Núñez, F.; Álvarez, M.; Roncero, E.; Rodríguez, M. Biocontrol of Ochratoxigenic Penicillium nordicum in Dry-Cured Fermented Sausages by Debaryomyces hansenii and Staphylococcus xylosus. Int. J. Food Microbiol. 2022, 375, 109744. [Google Scholar] [CrossRef]
- Agirman, B.; Erten, H. Biocontrol Ability and Action Mechanisms of Aureobasidium pullulans GE17 and Meyerozyma guilliermondii KL3 against Penicillium digitatum DSM2750 and Penicillium expansum DSM62841 Causing Postharvest Diseases. Yeast 2020, 37, 437–448. [Google Scholar] [CrossRef] [PubMed]
- EPA EPA. US Environmental Protection Agency Office of Pesticide Programs. Biopesticide Registration Action Document: Aspergillus Flavus AF36. PC Code 006456. 2003. Available online: https://www3.epa.gov/pesticides/chem_search/reg_actions/registration/decision_PC-006456_3-Jul-03.pdf (accessed on 15 March 2022).
- EPA EPA. US Environmental Protection Agency Office of Pesticide Programs. BIOPESTICIDE REGISTRATION ACTION DOCUMENT Aspergillus Flavus (NRRL 21882) (PC Code 006500). 2004. Available online: https://www3.epa.gov/pesticides/chem_search/reg_actions/registration/decision_PC-006500_24-Mar-04.pdf (accessed on 15 March 2022).
- Senghor, L.A.; Ortega-Beltran, A.; Atehnkeng, J.; Callicott, K.A.; Cotty, P.J.; Bandyopadhyay, R. The Atoxigenic Biocontrol Product Aflasafe SN01 Is a Valuable Tool to Mitigate Aflatoxin Contamination of Both Maize and Groundnut Cultivated in Senegal. Plant Dis. 2019, 104, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Mauro, A.; Garcia-Cela, E.; Pietri, A.; Cotty, P.J.; Battilani, P. Biological Control Products for Aflatoxin Prevention in Italy: Commercial Field Evaluation of Atoxigenic Aspergillus flavus Active Ingredients. Toxins 2018, 10, 30. [Google Scholar] [CrossRef]
- Spadola, G.; Giannelli, G.; Magagnoli, S.; Lanzoni, A.; Albertini, M.; Nicoli, R.; Ferrari, R.; Burgio, G.; Restivo, F.M.; Degola, F. Validation and Ecological Niche Investigation of a New Fungal Intraspecific Competitor as a Biocontrol Agent for the Sustainable Containment of Aflatoxins on Maize Fields. JoF 2022, 8, 425. [Google Scholar] [CrossRef]
- Castellá, G.; Bragulat, M.R.; Cigliano, R.A.; Cabañes, F.J. Transcriptome Analysis of Non-Ochratoxigenic Aspergillus carbonarius Strains and Interactions between Some Black Aspergilli Species. Int. J. Food Microbiol. 2020, 317, 108498. [Google Scholar] [CrossRef]
- Llobregat, B.; González-Candelas, L.; Ballester, A.-R. Ochratoxin a Defective Aspergillus carbonarius Mutants as Potential Biocontrol Agents. Toxins 2022, 14, 745. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, R.J.; Andersson, J.A.; Kristensen, M.B.; Giese, H. Efficient Four Fragment Cloning for the Construction of Vectors for Targeted Gene Replacement in Filamentous Fungi. BMC Mol. Biol. 2008, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- El Hajj Assaf, C.; Snini, S.; Tadrist, S.; Bailly, S.; Naylies, C.; Oswald, I.P.; Lorber, S.; Puel, O. Impact of veA on the Development, Aggressiveness, Dissemination and Secondary Metabolism of Penicillium expansum. Mol. Plant Pathol. 2018, 19, 1971–1983. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Sempere, A.; Marín, S.; Sanchis, V.; Ramos, A.J. VeA and LaeA Transcriptional Factors Regulate Ochratoxin A Biosynthesis in Aspergillus carbonarius. Int. J. Food Microbiol. 2013, 166, 479–486. [Google Scholar] [CrossRef]
- Duran, R.M.; Cary, J.W.; Calvo, A.M. Production of Cyclopiazonic Acid, Aflatrem, and Aflatoxin by Aspergillus flavus Is Regulated by veA, a Gene Necessary for Sclerotial Formation. Appl. Microbiol. Biotechnol. 2007, 73, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Brown, D.W.; Kleigrewe, K.; Bok, J.W.; Keller, N.P.; Humpf, H.-U.; Tudzynski, B. FfVel1 and FfLae1, Components of a Velvet-like Complex in Fusarium fujikuroi, Affect Differentiation, Secondary Metabolism and Virulence: Identification of a Velvet-like Complex in F. fujikuroi. Mol. Microbiol. 2010, 77, 972–994. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, S.; Pacher, S.; Hedtke, M.; Kniemeyer, O.; Fischer, R. A Phosphorylation Code of the Aspergillus nidulans Global Regulator VelvetA (VeA) Determines Specific Functions: Light Regulation in A. nidulans. Mol. Microbiol. 2016, 99, 909–924. [Google Scholar] [CrossRef]
- Kato, N.; Brooks, W.; Calvo, A.M. The Expression of Sterigmatocystin and Penicillin Genes in Aspergillus nidulans Is Controlled by veA, a Gene Required for Sexual Development. Eukaryot. Cell 2003, 2, 1178–1186. [Google Scholar] [CrossRef]
- Lewis, M.H.; Carbone, I.; Luis, J.M.; Payne, G.A.; Bowen, K.L.; Hagan, A.K.; Kemerait, R.; Heiniger, R.; Ojiambo, P.S. Biocontrol Strains Differentially Shift the Genetic Structure of Indigenous Soil Populations of Aspergillus flavus. Front. Microbiol. 2019, 10, 1738. [Google Scholar] [CrossRef]
- Bhatnagar-Mathur, P.; Sunkara, S.; Bhatnagar-Panwar, M.; Waliyar, F.; Sharma, K.K. Biotechnological Advances for Combating Aspergillus flavus and Aflatoxin Contamination in Crops. Plant Sci. 2015, 234, 119–132. [Google Scholar] [CrossRef]
- Dorner, J.; Lamb, M. Development and Commercial Use of Afla-Guard®, an Aflatoxin Biocontrol Agent. Mycotoxin Res. 2006, 22, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Sempere, A.; López-Pérez, M.; Martínez-Culebras, P.V.; González-Candelas, L. Development of a Green Fluorescent Tagged Strain of Aspergillus carbonarius to Monitor Fungal Colonization in Grapes. Int. J. Food Microbiol. 2011, 148, 135–140. [Google Scholar] [CrossRef] [PubMed]
- López-Pérez, M.; Ballester, A.-R.; González-Candelas, L. Identification and Functional Analysis of Penicillium digitatum Genes Putatively Involved in Virulence towards Citrus Fruit. Mol. Plant Pathol. 2015, 16, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Gerin, D.; González-Candelas, L.; Ballester, A.-R.; Pollastro, S.; De Miccolis Angelini, R.; Faretra, F. Functional Characterization of the Alb1 Orthologue Gene in the Ochratoxigenic Fungus Aspergillus carbonarius (AC49 Strain). Toxins 2018, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Kahm, M.; Hasenbrink, G.; Lichtenberg-Fraté, H.; Ludwig, J.; Kschischo, M. grofit: Fitting Biological Growth Curves with R. J. Stat. Softw. 2010, 33, 1–21. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Zong, Y.; Shang, Y.; Zhang, Z.; Xu, X.; Wang, X.; Long, M.; Tian, S. Dissection of patulin biosynthesis, spatial control and regulation mechanism in Penicillium expansum. Environ. Microbiol. 2019, 21, 1124–1139. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llobregat, B.; González-Candelas, L.; Ballester, A.-R. Exploring the Biocontrol Capability of Non-Mycotoxigenic Strains of Penicillium expansum. Toxins 2024, 16, 52. https://doi.org/10.3390/toxins16010052
Llobregat B, González-Candelas L, Ballester A-R. Exploring the Biocontrol Capability of Non-Mycotoxigenic Strains of Penicillium expansum. Toxins. 2024; 16(1):52. https://doi.org/10.3390/toxins16010052
Chicago/Turabian StyleLlobregat, Belén, Luis González-Candelas, and Ana-Rosa Ballester. 2024. "Exploring the Biocontrol Capability of Non-Mycotoxigenic Strains of Penicillium expansum" Toxins 16, no. 1: 52. https://doi.org/10.3390/toxins16010052
APA StyleLlobregat, B., González-Candelas, L., & Ballester, A. -R. (2024). Exploring the Biocontrol Capability of Non-Mycotoxigenic Strains of Penicillium expansum. Toxins, 16(1), 52. https://doi.org/10.3390/toxins16010052






