Edodin: A New Type of Toxin from Shiitake Mushroom (Lentinula edodes) That Inactivates Mammalian Ribosomes
Abstract
:1. Introduction
2. Results
2.1. Isolation of Edodin
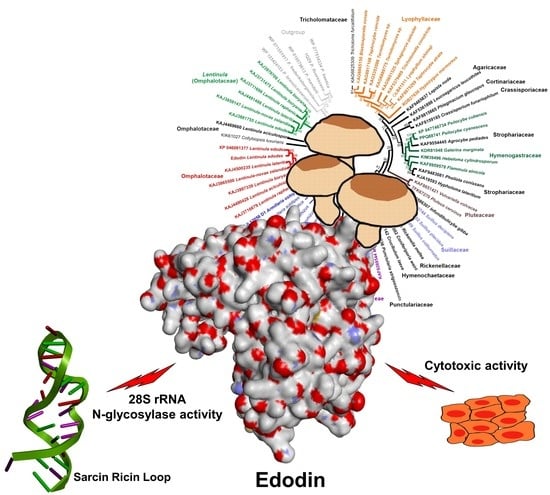
2.2. Enzymatic Activity of Edodin
2.3. Cytotoxicity of Edodin and Antifungal Activity Assay
2.4. Edodin Sequence
2.5. Prediction of Edodin Structure
2.6. Putative Active Site of Edodin
2.7. Homology of Edodin with Other Hypothetical Proteins
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Reagents, Cells, and Fungi
5.2. Methods
5.2.1. Purification of Edodin
5.2.2. Analytical Procedures
5.2.3. Cell-Free Protein Synthesis and Enzyme Activity Assays
5.2.4. Cell Viability and Antifungal Activity Assays
5.2.5. cDNA Synthesis, Cloning, and Sequencing
5.2.6. Edodin Cleavage and MALDI-ToF MS Analysis
5.2.7. Prediction of Edodin Structure
5.2.8. Sequence Alignment and Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Puri, M.; Kaur, I.; Perugini, M.; Gupta, R. Ribosome-inactivating proteins: Current status and biomedical applications. Drug Discov. Today 2012, 17, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon 2013, 67, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, A.; Bortolotti, M.; Maiello, S.; Battelli, M.G.; Polito, L. Ribosome-Inactivating Proteins from Plants: A Historical Overview. Molecules 2016, 21, 1627. [Google Scholar] [CrossRef] [PubMed]
- Robert, A.; Wiels, J. Shiga Toxins as Antitumor Tools. Toxins 2021, 13, 690. [Google Scholar] [CrossRef] [PubMed]
- Grela, P.; Szajwaj, M.; Horbowicz-Drozdzal, P.; Tchorzewski, M. How Ricin Damages the Ribosome. Toxins 2019, 11, 241. [Google Scholar] [CrossRef]
- Reyes, A.; Anne, J.; Mejia, A. Ribosome-inactivating proteins with an emphasis on bacterial RIPs and their potential medical applications. Future Microbiol. 2012, 7, 705–717. [Google Scholar] [CrossRef]
- Liu, R.; Yang, J.; Liu, W. Isolation and enzymatic characterization of lamjapin, the first ribosome-inactivating protein from cryptogamic algal plant (Laminaria japonica A). Eur. J. Biochem. 2002, 269, 4746–4752. [Google Scholar] [CrossRef] [PubMed]
- Landi, N.; Hussain, H.Z.F.; Pedone, P.V.; Ragucci, S.; Di Maro, A. Ribotoxic Proteins, Known as Inhibitors of Protein Synthesis, from Mushrooms and Other Fungi According to Endo’s Fragment Detection. Toxins 2022, 14, 403. [Google Scholar] [CrossRef] [PubMed]
- Citores, L.; Ragucci, S.; Russo, R.; Gay, C.; Chambery, A.; Di Maro, A.; Iglesias, R.; Ferreras, J.M. Structural and functional characterization of the cytotoxic protein ledodin, an atypical ribosome-inactivating protein from shiitake mushroom (Lentinula edodes). Protein Sci. 2023, 32, e4621. [Google Scholar] [CrossRef]
- Polito, L.; Bortolotti, M.; Battelli, M.G.; Calafato, G.; Bolognesi, A. Ricin: An Ancient Story for a Timeless Plant Toxin. Toxins 2019, 11, 324. [Google Scholar] [CrossRef] [PubMed]
- Olombrada, M.; Lazaro-Gorines, R.; Lopez-Rodriguez, J.; Martinez-del-Pozo, A.; Onaderra, M.; Maestro-Lopez, M.; Lacadena, J.; Gavilanes, J.; Garcia-Ortega, L. Fungal Ribotoxins: A Review of Potential Biotechnological Applications. Toxins 2017, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Olombrada, M.; Martínez-del-Pozo, A.; Medina, P.; Budia, F.; Gavilanes, J.; García-Ortega, L. Fungal ribotoxins: Natural protein-based weapons against insects. Toxicon 2014, 83, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Citores, L.; Ragucci, S.; Ferreras, J.M.; Di Maro, A.; Iglesias, R. Ageritin, a Ribotoxin from Poplar Mushroom (Agrocybe aegerita) with Defensive and Antiproliferative Activities. Acs Chem. Biol. 2019, 14, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Zhou, Y.K.; Ji, Z.L.; Chen, X.R. The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant Sci. 2018, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Citores, L.; Iglesias, R.; Ferreras, J.M. Antiviral Activity of Ribosome-Inactivating Proteins. Toxins 2021, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Polito, L.; Bortolotti, M.; Pedrazzi, M.; Bolognesi, A. Immunotoxins and Other Conjugates Containing Saporin-S6 for Cancer Therapy. Toxins 2011, 3, 697–720. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Stoppa, C.; Bolognesi, A. Large-scale chromatographic purification of ribosome-inactivating proteins. J. Chromatogr. 1987, 408, 235–243. [Google Scholar] [CrossRef]
- Ragucci, S.; Bulgari, D.; Landi, N.; Russo, R.; Clemente, A.; Valletta, M.; Chambery, A.; Gobbi, E.; Faoro, F.; Di Maro, A. The Structural Characterization and Antipathogenic Activities of Quinoin, a Type 1 Ribosome-Inactivating Protein from Quinoa Seeds. Int. J. Mol. Sci. 2021, 22, 8964. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.K.; Rybak, S.M.; Winkler, G.; Meade, H.M.; McGray, P.; Youle, R.J.; Ackerman, E.J. Comparison of RNases and toxins upon injection into Xenopus oocytes. J. Biol. Chem. 1991, 266, 21208–21214. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Wool, I.G. The site of action of alpha-sarcin on eukaryotic ribosomes—The sequence at the alpha-sarcin cleavage site in 28 S-ribosomal ribonucleic-acid. J. Biol. Chem. 1982, 257, 9054–9060. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Mitsui, K.; Motizuki, M.; Tsurugi, K. The Mechanism of Action of Ricin and Related Toxic Lectins on Eukaryotic Ribosomes—The Site and the Characteristics of the Modification in 28-S Ribosomal-RNA Caused by the Toxins. J. Biol. Chem. 1987, 262, 5908–5912. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, R.; Citores, L.; Ragucci, S.; Russo, R.; Di Maro, A.; Ferreras, J.M. Biological and antipathogenic activities of ribosome-inactivating proteins from Phytolacca dioica L. Biochim. Biophys. Acta-Gen. Subj. 2016, 1860, 1256–1264. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Valbonesi, P.; Bonora, E.; Gorini, P.; Bolognesi, A.; Stirpe, F. Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: Effect on DNA, RNA and poly(A). Nucleic Acids Res. 1997, 25, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Battelli, M.G.; Stirpe, F. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta 1993, 1154, 237–282. [Google Scholar] [CrossRef] [PubMed]
- Monteuuis, G.; Wong, J.; Bailey, C.; Schmitz, U.; Rasko, J. The changing paradigm of intron retention: Regulation, ramifications and recipes. Nucleic Acids Res. 2019, 47, 11497–11513. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.; Wang, J.; Cong, Q.; Kinch, L.; Schaeffer, R.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Choy, W.; Karttunen, M. AlphaFold2: A Role for Disordered Protein/Region Prediction? Int. J. Mol. Sci. 2022, 23, 4591. [Google Scholar] [CrossRef]
- Liang, J.; Han, Q.; Tan, Y.; Ding, H.; Li, J. Current Advances on Structure-Function Relationships o Pyridoxal 5′-Phosphate-Dependent Enzymes. Front. Mol. Biosci. 2019, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Momany, C.; Levdikov, V.; Blagova, L.; Lima, S.; Phillips, R. Three-dimensional structure of kynureninase from Pseudomonas fluorescens. Biochemistry 2004, 43, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Percudani, R.; Peracchi, A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003, 4, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Fesko, K.; Suplatov, D.; Svedas, V. Bioinformatic analysis of the fold type I PLP-dependent enzymes reveals determinants of reaction specificity in L-threonine aldolase from Aeromonas jandaei. FEBS Open Bio 2018, 8, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.; Ng, T. Ribonucleases of different origins with a wide spectrum of medicinal applications. Biochim. Biophys. Acta-Rev. Cancer 2011, 1815, 65–74. [Google Scholar] [CrossRef]
- Niego, A.; Rapior, S.; Thongklang, N.; Raspe, O.; Jaidee, W.; Lumyong, S.; Hyde, K. Macrofungi as a Nutraceutical Source: Promising Bioactive Compounds and Market Value. J. Fungi 2021, 7, 397. [Google Scholar] [CrossRef]
- Kobayashi, H.; Kumagai, F.; Itagaki, T.; Inokuchi, N.; Koyama, T.; Iwama, M.; Ohgi, K.; Irie, M. Amino acid sequence of a nuclease (nuclease Le1) from Lentinus edodes. Biosci. Biotechnol. Biochem. 2000, 64, 948–957. [Google Scholar] [CrossRef]
- Kobayashi, H.; Hara, J.; Itagaki, T.; Inokuchi, N.; Koyama, T.; Sanda, A.; Iwama, M.; Ohgi, K.; Irie, M. Relationship of two ribonucleases with molecular masses of 45 kDa and 37 kDa from the culture medium Lentinus edodes. Biol. Pharm. Bull. 2000, 23, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Qin, Y.; Achenbach, J.; Li, C.; Kijek, J.; Spahn, C.; Nierhaus, K. EF-G and EF4: Translocation and back-translocation on the bacterial ribosome. Nat. Rev. Microbiol. 2014, 12, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, A.; Polito, L.; Scicchitano, V.; Orrico, C.; Pasquinelli, G.; Musiani, S.; Santi, S.; Riccio, M.; Bortolotti, M.; Battelli, M. Endocytosis and intracellular localisation of type 1 ribosome-inactivating protein saporin-S6. J. Biol. Regul. Homeost. Agents 2012, 26, 97–109. [Google Scholar] [PubMed]
- Polito, L.; Bortolotti, M.; Pedrazzi, M.; Mercatelli, D.; Battelli, M.G.; Bolognesi, A. Apoptosis and necroptosis induced by stenodactylin in neuroblastoma cells can be completely prevented through caspase inhibition plus catalase or necrostatin-1. Phytomedicine 2016, 23, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, K.; Hudak, K. Phylogeny and domain architecture of plant ribosome inactivating proteins. Phytochemistry 2022, 202, 113337. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wen, D.; Song, X.; Su, P.; Lou, J.; Yao, D.; Zhang, C. Evolution and natural selection of ribosome-inactivating proteins in bacteria, fungi, and plants. Int. J. Biol. Macromol. 2023, 248, 125929. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, H.; Kono, H.; Sakurai, H.; Tokimoto, K. Comparison of C-S lyase in Lentinus edodes and Allium sativum. Biosci. Biotechnol. Biochem. 2002, 66, 2560–2566. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lei, X.; Chen, L.; Bian, Y.; Yang, H.; Ibrahim, S.; Huang, W. A novel cysteine desulfurase influencing organosulfur compounds in Lentinula edodes. Sci. Rep. 2015, 5, 10047. [Google Scholar] [CrossRef] [PubMed]
- Kalb, V.F.J.; Bernlohr, R.W. A new spectrophotometric assay for protein in cell extracts. Anal. Biochem. 1977, 82, 10. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.; Wong, L.; Jennings, P. The solvent in CNBr cleavage reactions determines the fragmentation efficiency of ketosteroid isomerase fusion proteins used in the production of recombinant peptides. Protein Expr. Purif. 2003, 28, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.; Hon, G.; Chandonia, J.; Brenner, S. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Citores, L.; Ragucci, S.; Gay, C.C.; Russo, R.; Chambery, A.; Di Maro, A.; Iglesias, R.; Ferreras, J.M. Edodin: A New Type of Toxin from Shiitake Mushroom (Lentinula edodes) That Inactivates Mammalian Ribosomes. Toxins 2024, 16, 185. https://doi.org/10.3390/toxins16040185
Citores L, Ragucci S, Gay CC, Russo R, Chambery A, Di Maro A, Iglesias R, Ferreras JM. Edodin: A New Type of Toxin from Shiitake Mushroom (Lentinula edodes) That Inactivates Mammalian Ribosomes. Toxins. 2024; 16(4):185. https://doi.org/10.3390/toxins16040185
Chicago/Turabian StyleCitores, Lucía, Sara Ragucci, Claudia C. Gay, Rosita Russo, Angela Chambery, Antimo Di Maro, Rosario Iglesias, and José M. Ferreras. 2024. "Edodin: A New Type of Toxin from Shiitake Mushroom (Lentinula edodes) That Inactivates Mammalian Ribosomes" Toxins 16, no. 4: 185. https://doi.org/10.3390/toxins16040185