Toxic Peptides from the Mexican Scorpion Centruroides villegasi: Chemical Structure and Evaluation of Recognition by Human Single-Chain Antibodies
Abstract
:1. Introduction
2. Results
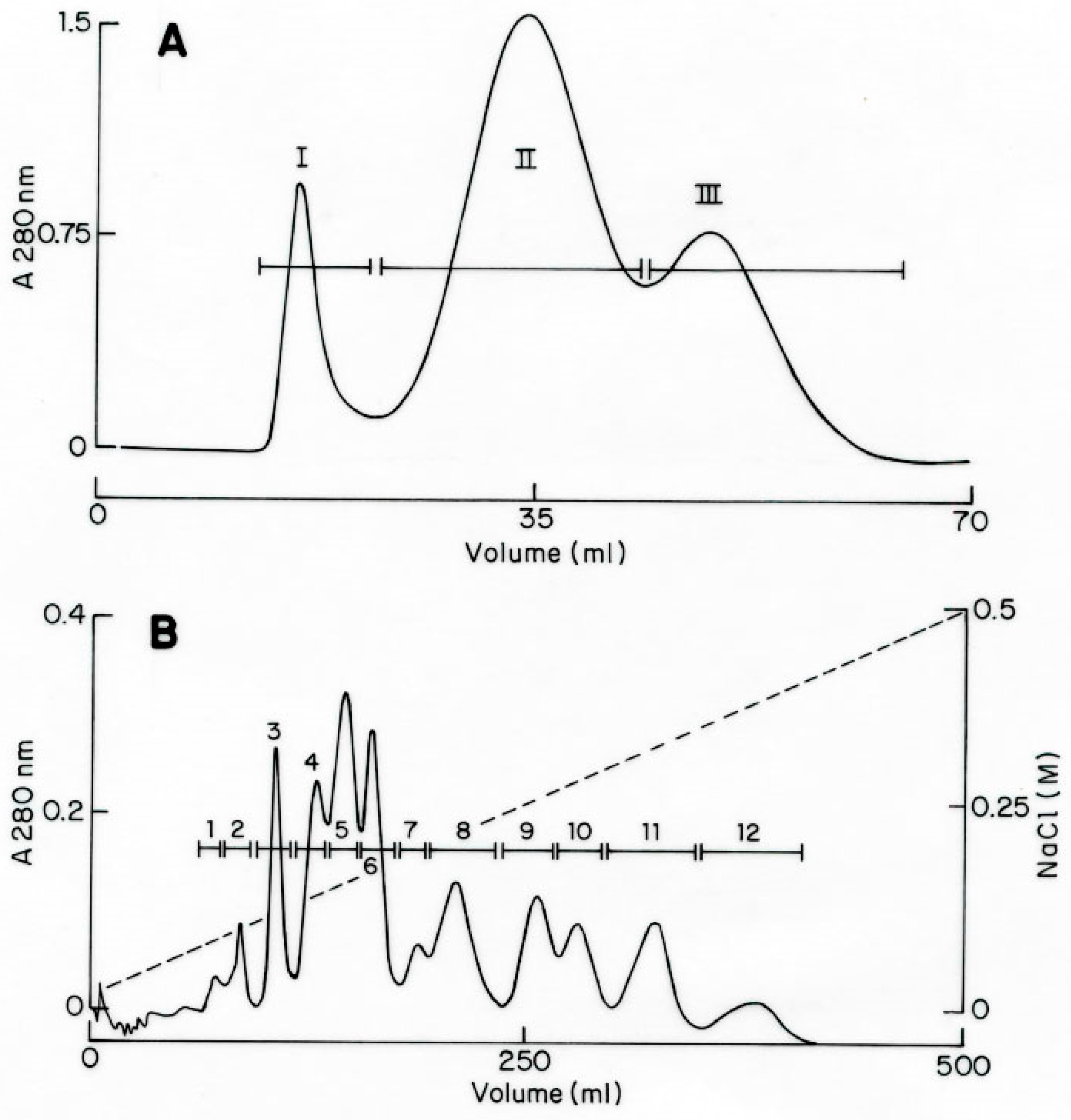
2.1. Purification and Sequencing of Toxic Peptides
2.2. Venom Toxicity and Amount of Pure Toxins
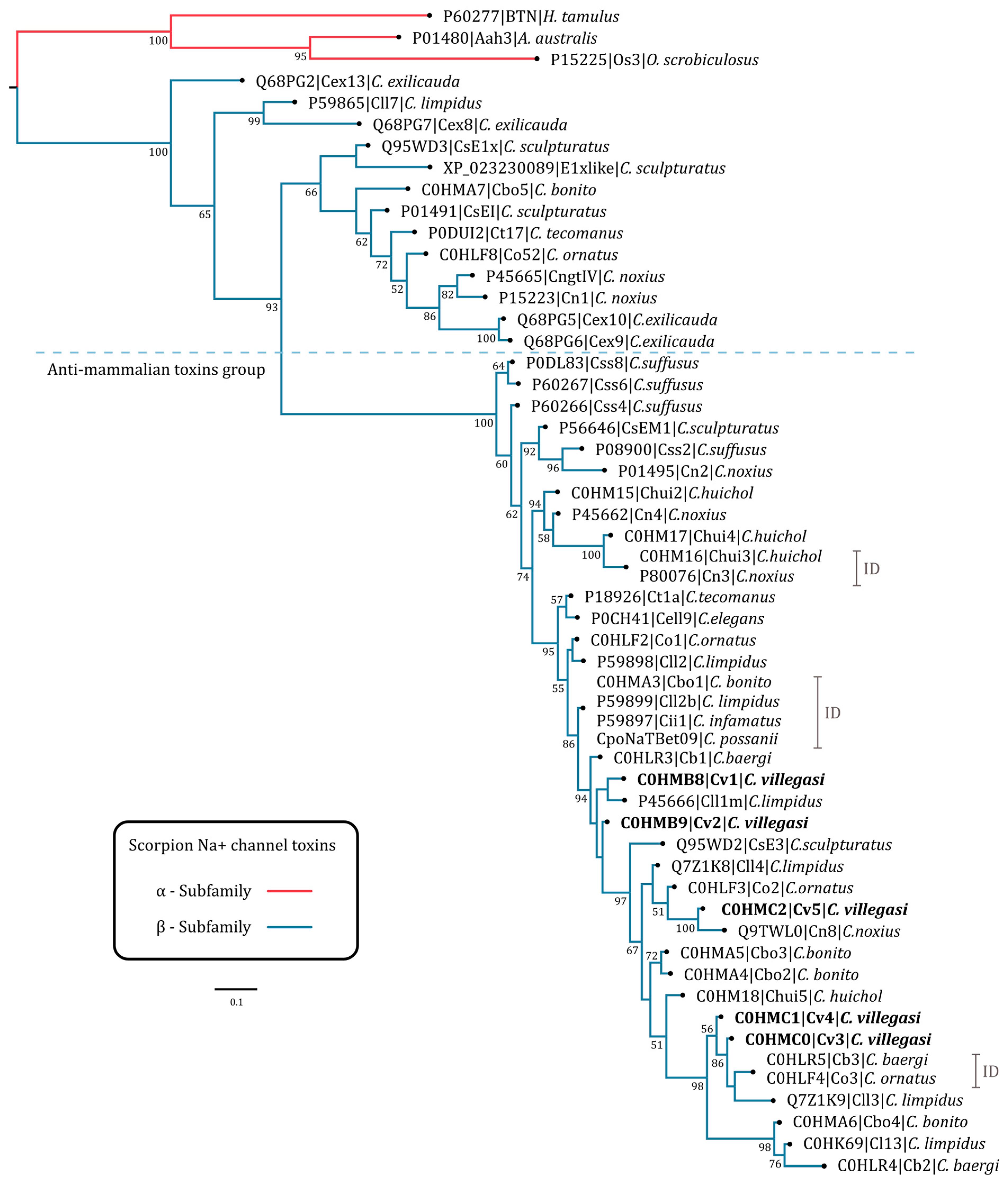
2.3. Phylogenetic Tree
2.4. Surface Plasmon Resonance Analysis
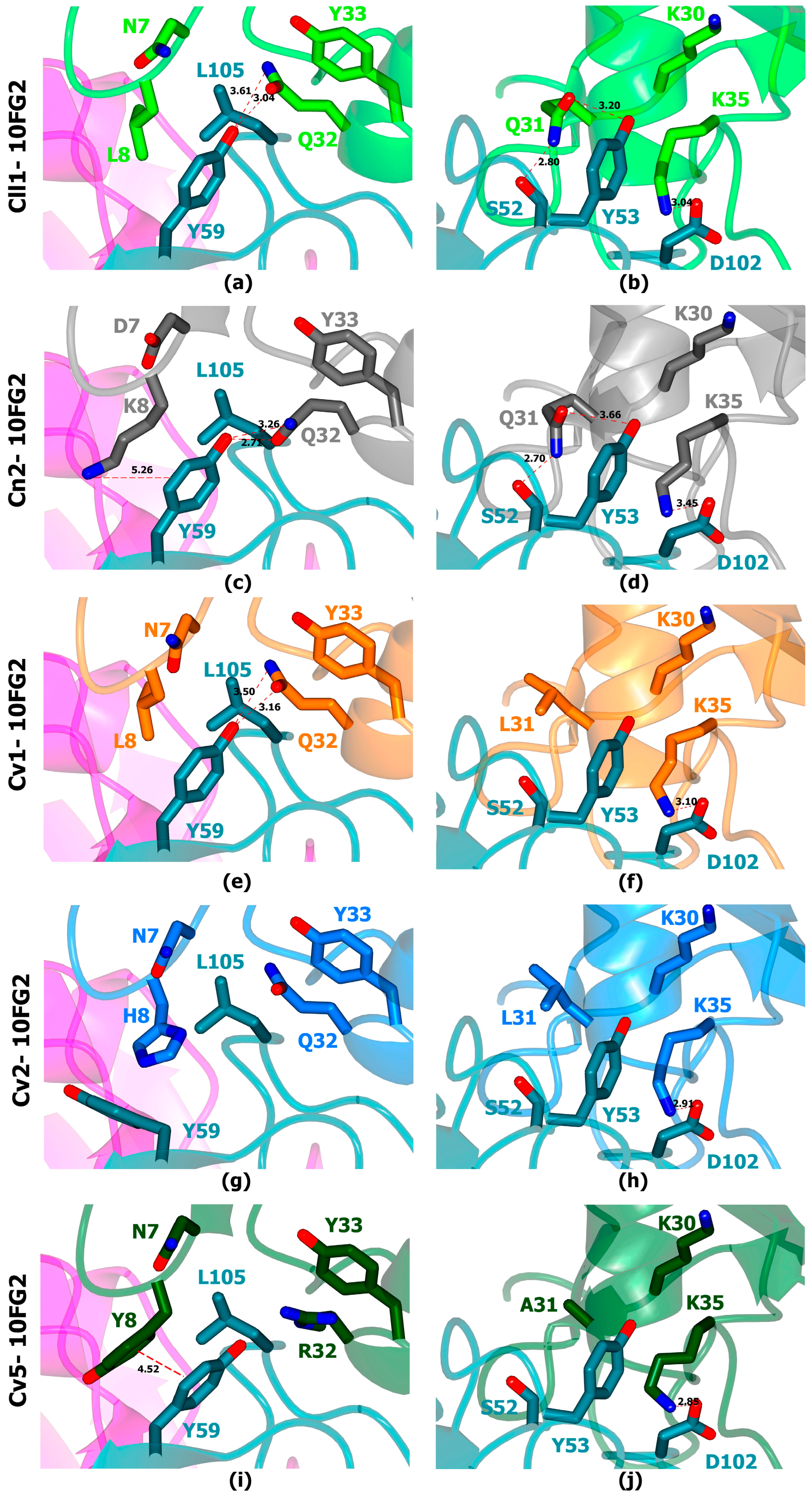
2.5. In Silico Analysis of the Toxin: scFv Structural Models Were Performed to Understand the Cross-Reactivity of scFv 10FG2 with the Three Toxins
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Venom Source, Lethality Tests, and Purification Procedure
5.2. Mass Spectrometry and Sequence Determination
5.3. Phylogenetic Analysis of the New Peptide-Toxins
5.4. Evaluation by Surface Plasmon Resonance (SPR)
5.5. In Silico Structural Analysis of Interactions of Cv1, Cv2, and Cv5 Toxins with scFv 10FG2
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chippaux, J.P. Emerging options for the management of scorpion stings. Drug Des. Dev. Ther. 2012, 6, 165–173. [Google Scholar] [CrossRef]
- Chippaux, J.P.; Celis, A.; Boyer, L.; Alagon, A. Factors involved in the resilience of incidence and decrease of mortality from scorpion stings in Mexico. Toxicon 2020, 188, 65–75. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Sola, M.; Jappe, E.C.; Oscoz, S.; Lauridsen, L.P.; Engmark, M. Biotechnological Trends in Spider and Scorpion Antivenom Development. Toxins 2016, 8, 226. [Google Scholar] [CrossRef]
- Boyer, L.; Degan, J.; Ruha, A.M.; Mallie, J.; Mangin, E.; Alagon, A. Safety of intravenous equine F(ab’)2: Insights following clinical trials involving 1534 recipients of scorpion antivenom. Toxicon 2013, 76, 386–393. [Google Scholar] [CrossRef]
- Bermudez-Mendez, E.; Fuglsang-Madsen, A.; Fons, S.; Lomonte, B.; Gutierrez, J.M.; Laustsen, A.H. Innovative Immunization Strategies for Antivenom Development. Toxins 2018, 10, 452. [Google Scholar] [CrossRef]
- Ponce-Saavedra, J.; Linares-Guillén, J.W.; Quijano-Ravell, A.F. Una nueva especie de alacrán del género Centruroides Marx (Scorpiones: Buthidae) de la costa Noroeste de México. Acta Zoológica Mex. 2022, 38, 1–24. [Google Scholar] [CrossRef]
- Gonzalez-Santillan, E.; Possani, L.D. North American scorpion species of public health importance with a reappraisal of historical epidemiology. Acta Trop. 2018, 187, 264–274. [Google Scholar] [CrossRef]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon Off. J. Int. Soc. Toxinol. 2015, 93, 125–135. [Google Scholar] [CrossRef]
- Garcia-Guerrero, I.A.; Carcamo-Noriega, E.; Gomez-Lagunas, F.; Gonzalez-Santillan, E.; Zamudio, F.Z.; Gurrola, G.B.; Possani, L.D. Biochemical characterization of the venom from the Mexican scorpion Centruroides ornatus, a dangerous species to humans. Toxicon 2020, 173, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Ramirez, I.V.; Riano-Umbarila, L.; Olamendi-Portugal, T.; Restano-Cassulini, R.; Possani, L.D.; Becerril, B. Biochemical, electrophysiological and immunological characterization of the venom from Centruroides baergi, a new scorpion species of medical importance in Mexico. Toxicon 2020, 184, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Martinez, H.; Olamendi-Portugal, T.; Restano-Cassulini, R.; Serrano-Posada, H.; Zamudio, F.; Possani, L.D.; Riano-Umbarila, L.; Becerril, B. Characterization of Four Medically Important Toxins from Centruroides huichol Scorpion Venom and Its Neutralization by a Single Recombinant Antibody Fragment. Toxins 2022, 14, 369. [Google Scholar] [CrossRef]
- Restano-Cassulini, R.; Olamendi-Portugal, T.; Riano-Umbarila, L.; Zamudio, F.Z.; Delgado-Prudencio, G.; Becerril, B.; Possani, L.D. Characterization of Sodium Channel Peptides Obtained from the Venom of the Scorpion Centruroides bonito. Toxins 2024, 16, 125. [Google Scholar] [CrossRef]
- Cestele, S.; Catterall, W.A. Molecular mechanisms of neurotoxin action on voltage-gated sodium channels. Biochimie 2000, 82, 883–892. [Google Scholar] [CrossRef]
- Catterall, W.A.; Goldin, A.L.; Waxman, S.G. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 2005, 57, 397–409. [Google Scholar] [CrossRef]
- Cestele, S.; Yarov-Yarovoy, V.; Qu, Y.; Sampieri, F.; Scheuer, T.; Catterall, W.A. Structure and function of the voltage sensor of sodium channels probed by a beta-scorpion toxin. J. Biol. Chem. 2006, 281, 21332–21344. [Google Scholar] [CrossRef]
- Gurevitz, M. Mapping of scorpion toxin receptor sites at voltage-gated sodium channels. Toxicon 2012, 60, 502–511. [Google Scholar] [CrossRef]
- Xia, Z.; He, D.; Wu, Y.; Kwok, H.F.; Cao, Z. Scorpion venom peptides: Molecular diversity, structural characteristics, and therapeutic use from channelopathies to viral infections and cancers. Pharmacol. Res. 2023, 197, 106978. [Google Scholar] [CrossRef]
- Bird, R.E.; Hardman, K.D.; Jacobson, J.W.; Johnson, S.; Kaufman, B.M.; Lee, S.M.; Lee, T.; Pope, S.H.; Riordan, G.S.; Whitlow, M. Single-chain antigen-binding proteins. Science 1988, 242, 423–426. [Google Scholar] [CrossRef]
- Baldazo-Monsivaiz, J.G.; Ponce-Saavedra, J.; Flores-Moreno, M. Una especie nueva de alacrán del género Centruroides de importancia médica (Scorpiones: Buthidae) del estado de Guerrero, México. Rev. Mex. Biodivers. 2013, 84, 100–116. [Google Scholar] [CrossRef]
- Riano-Umbarila, L.; Rodriguez-Rodriguez, E.R.; Santibanez-Lopez, C.E.; Guereca, L.; Uribe-Romero, S.J.; Gomez-Ramirez, I.V.; Carcamo-Noriega, E.N.; Possani, L.D.; Becerril, B. Updating knowledge on new medically important scorpion species in Mexico. Toxicon 2017, 138, 130–137. [Google Scholar] [CrossRef]
- Babin, D.R.; Watt, D.D.; Goos, S.M.; Mlejnek, R.V. Amino acid sequence of neurotoxin I from Centruroides sculpturatus Ewing. Arch. Biochem. Biophys. 1975, 166, 125–134. [Google Scholar] [CrossRef]
- Meves, H.; Rubly, N.; Watt, D.D. Effect of toxins isolated from the venom of the scorpion Centruroides sculpturatus on the Na currents of the node of Ranvier. Pflugers Arch. 1982, 393, 56–62. [Google Scholar] [CrossRef]
- Valdez-Velazquez, L.L.; Romero-Gutierrez, M.T.; Delgado-Enciso, I.; Dobrovinskaya, O.; Melnikov, V.; Quintero-Hernandez, V.; Ceballos-Magana, S.G.; Gaitan-Hinojosa, M.A.; Coronas, F.I.; Puebla-Perez, A.M.; et al. Comprehensive analysis of venom from the scorpion Centruroides tecomanus reveals compounds with antimicrobial, cytotoxic, and insecticidal activities. Toxicon Off. J. Int. Soc. Toxinol. 2016, 118, 95–103. [Google Scholar] [CrossRef]
- Schiavon, E.; Pedraza-Escalona, M.; Gurrola, G.B.; Olamendi-Portugal, T.; Corzo, G.; Wanke, E.; Possani, L.D. Negative-shift activation, current reduction and resurgent currents induced by beta-toxins from Centruroides scorpions in sodium channels. Toxicon 2012, 59, 283–293. [Google Scholar] [CrossRef]
- Riano-Umbarila, L.; Contreras-Ferrat, G.; Olamendi-Portugal, T.; Morelos-Juarez, C.; Corzo, G.; Possani, L.D.; Becerril, B. Exploiting cross-reactivity to neutralize two different scorpion venoms with one single chain antibody fragment. J. Biol. Chem. 2011, 286, 6143–6151. [Google Scholar] [CrossRef]
- Riano-Umbarila, L.; Gomez-Ramirez, I.V.; Ledezma-Candanoza, L.M.; Olamendi-Portugal, T.; Rodriguez-Rodriguez, E.R.; Fernandez-Taboada, G.; Possani, L.D.; Becerril, B. Generation of a Broadly Cross-Neutralizing Antibody Fragment against Several Mexican Scorpion Venoms. Toxins 2019, 11, 32. [Google Scholar] [CrossRef]
- Riano-Umbarila, L.; Ledezma-Candanoza, L.M.; Serrano-Posada, H.; Fernandez-Taboada, G.; Olamendi-Portugal, T.; Rojas-Trejo, S.; Gomez-Ramirez, I.V.; Rudino-Pinera, E.; Possani, L.D.; Becerril, B. Optimal Neutralization of Centruroides noxius Venom Is Understood through a Structural Complex between Two Antibody Fragments and the Cn2 Toxin. J. Biol. Chem. 2016, 291, 1619–1630. [Google Scholar] [CrossRef]
- Trinidad-Porfirio, B.E.; Morales-Perez, A.; Nava-Aguilera, E.; Flores-Moreno, M.; Morales-Nava, L.; Garcia-Leyva, J.; Silva-Dominguez, R.; Cortes-Guzman, A.J.; Fernandez-Salas, I.; Andersson, N. Occurrence of scorpion sting and associated factors in a highly marginalized municipality in Guerrero, Mexico: A cross-sectional study. PLoS Negl. Trop. Dis. 2023, 17, e0011271. [Google Scholar] [CrossRef]
- Ledsgaard, L.; Jenkins, T.P.; Davidsen, K.; Krause, K.E.; Martos-Esteban, A.; Engmark, M.; Rordam Andersen, M.; Lund, O.; Laustsen, A.H. Antibody Cross-Reactivity in Antivenom Research. Toxins 2018, 10, 393. [Google Scholar] [CrossRef]
- Sorensen, C.V.; Ledsgaard, L.; Wildenauer, H.H.K.; Dahl, C.H.; Ebersole, T.W.; Bohn, M.F.; Ljungars, A.; Jenkins, T.P.; Laustsen, A.H. Cross-reactivity trends when selecting scFv antibodies against snake toxins using a phage display-based cross-panning strategy. Sci. Rep. 2023, 13, 10181. [Google Scholar] [CrossRef]
- Rivera-de-Torre, E.; Lampadariou, S.; Moiniche, M.; Bohn, M.F.; Kazemi, S.M.; Laustsen, A.H. Discovery of broadly-neutralizing antibodies against brown recluse spider and Gadim scorpion sphingomyelinases using consensus toxins as antigens. Protein Sci. 2024, 33, e4901. [Google Scholar] [CrossRef] [PubMed]
- Azzazy, H.M.; Highsmith, W.E., Jr. Phage display technology: Clinical applications and recent innovations. Clin. Biochem. 2002, 35, 425–445. [Google Scholar] [CrossRef] [PubMed]
- Boder, E.T.; Midelfort, K.S.; Wittrup, K.D. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl. Acad. Sci. USA 2000, 97, 10701–10705. [Google Scholar] [CrossRef] [PubMed]
- Olamendi-Portugal, T.; Bartok, A.; Zamudio-Zuniga, F.; Balajthy, A.; Becerril, B.; Panyi, G.; Possani, L.D. Isolation, chemical and functional characterization of several new K(+)-channel blocking peptides from the venom of the scorpion Centruroides tecomanus. Toxicon 2016, 115, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Noble, J.E.; Bailey, M.J.A. Chapter 8 Quantitation of Protein. In Methods in Enzymology; Burgess, R.R., Deutscher, M.P., Eds.; Elsevier: New York, NY, USA, 2009; Volume 463, pp. 73–95. [Google Scholar]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Torreri, P.; Ceccarini, M.; Macioce, P.; Petrucci, T.C. Biomolecular interactions by Surface Plasmon Resonance technology. Ann. Dell’istituto Super. Sanita 2005, 41, 437–441. [Google Scholar]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallographica. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Tina, K.G.; Bhadra, R.; Srinivasan, N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007, 35, W473–W476. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riaño-Umbarila, L.; Olamendi-Portugal, T.; Romero-Moreno, J.A.; Delgado-Prudencio, G.; Zamudio, F.Z.; Becerril, B.; Possani, L.D. Toxic Peptides from the Mexican Scorpion Centruroides villegasi: Chemical Structure and Evaluation of Recognition by Human Single-Chain Antibodies. Toxins 2024, 16, 301. https://doi.org/10.3390/toxins16070301
Riaño-Umbarila L, Olamendi-Portugal T, Romero-Moreno JA, Delgado-Prudencio G, Zamudio FZ, Becerril B, Possani LD. Toxic Peptides from the Mexican Scorpion Centruroides villegasi: Chemical Structure and Evaluation of Recognition by Human Single-Chain Antibodies. Toxins. 2024; 16(7):301. https://doi.org/10.3390/toxins16070301
Chicago/Turabian StyleRiaño-Umbarila, Lidia, Timoteo Olamendi-Portugal, José Alberto Romero-Moreno, Gustavo Delgado-Prudencio, Fernando Z. Zamudio, Baltazar Becerril, and Lourival D. Possani. 2024. "Toxic Peptides from the Mexican Scorpion Centruroides villegasi: Chemical Structure and Evaluation of Recognition by Human Single-Chain Antibodies" Toxins 16, no. 7: 301. https://doi.org/10.3390/toxins16070301




