Combinatory Effects of Acrylamide and Deoxynivalenol on In Vitro Cell Viability and Cytochrome P450 Enzymes of Human HepaRG Cells
Abstract
1. Introduction
2. Results
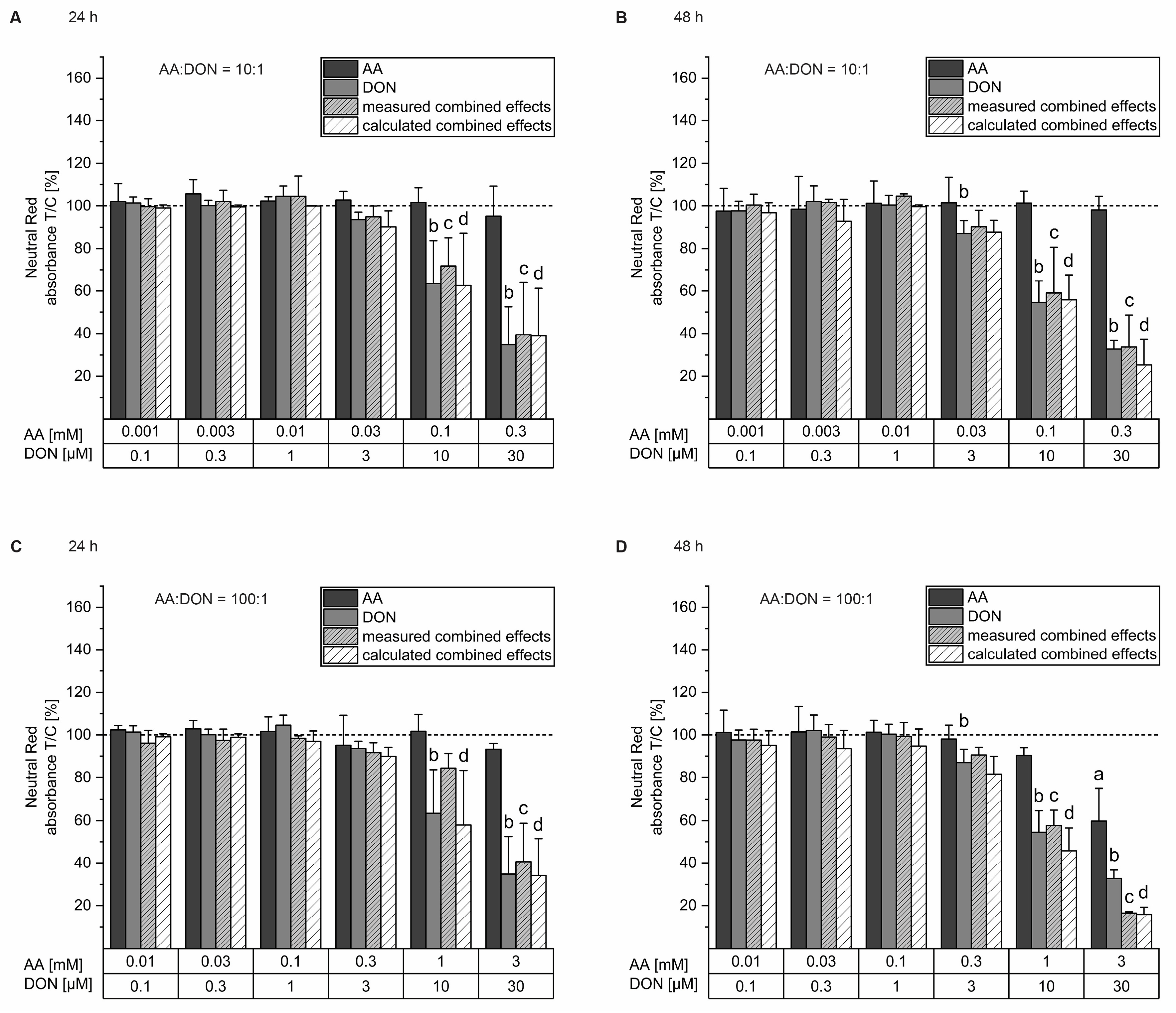
2.1. Cell Viability Testing
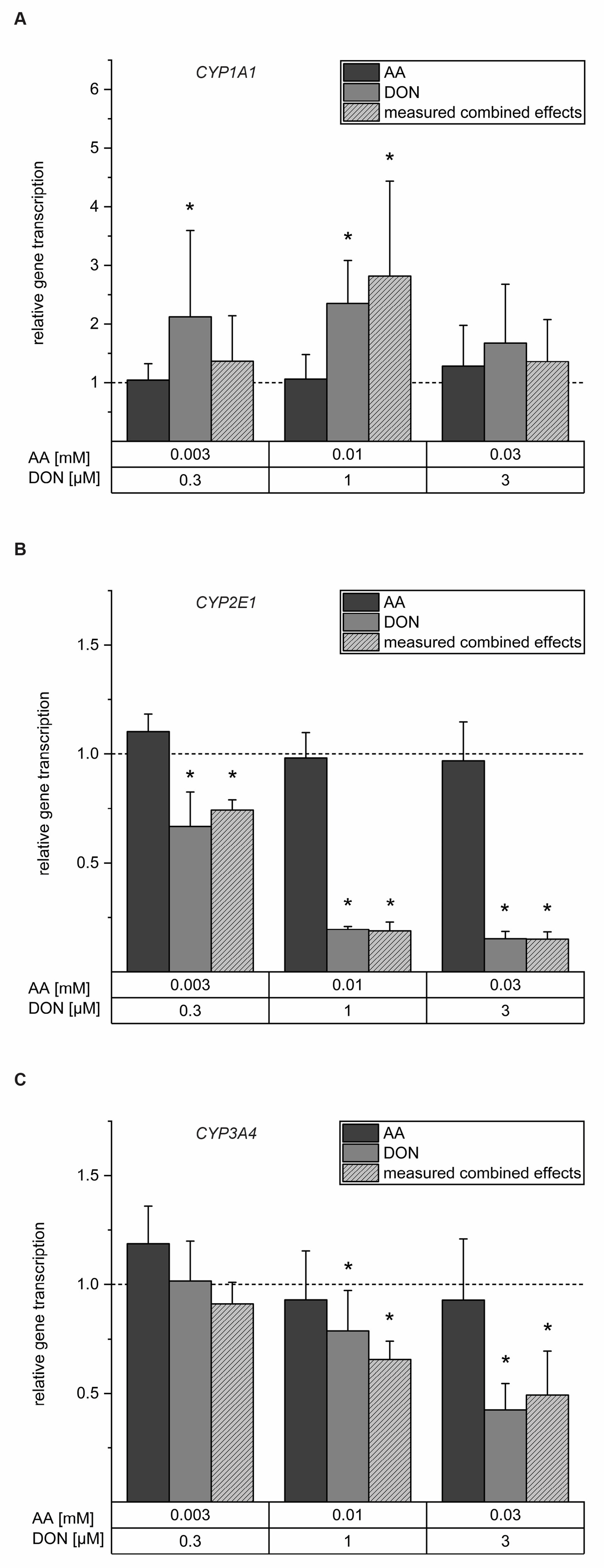
2.2. Interferences with Gene Transcription of Selected CYP Enzymes
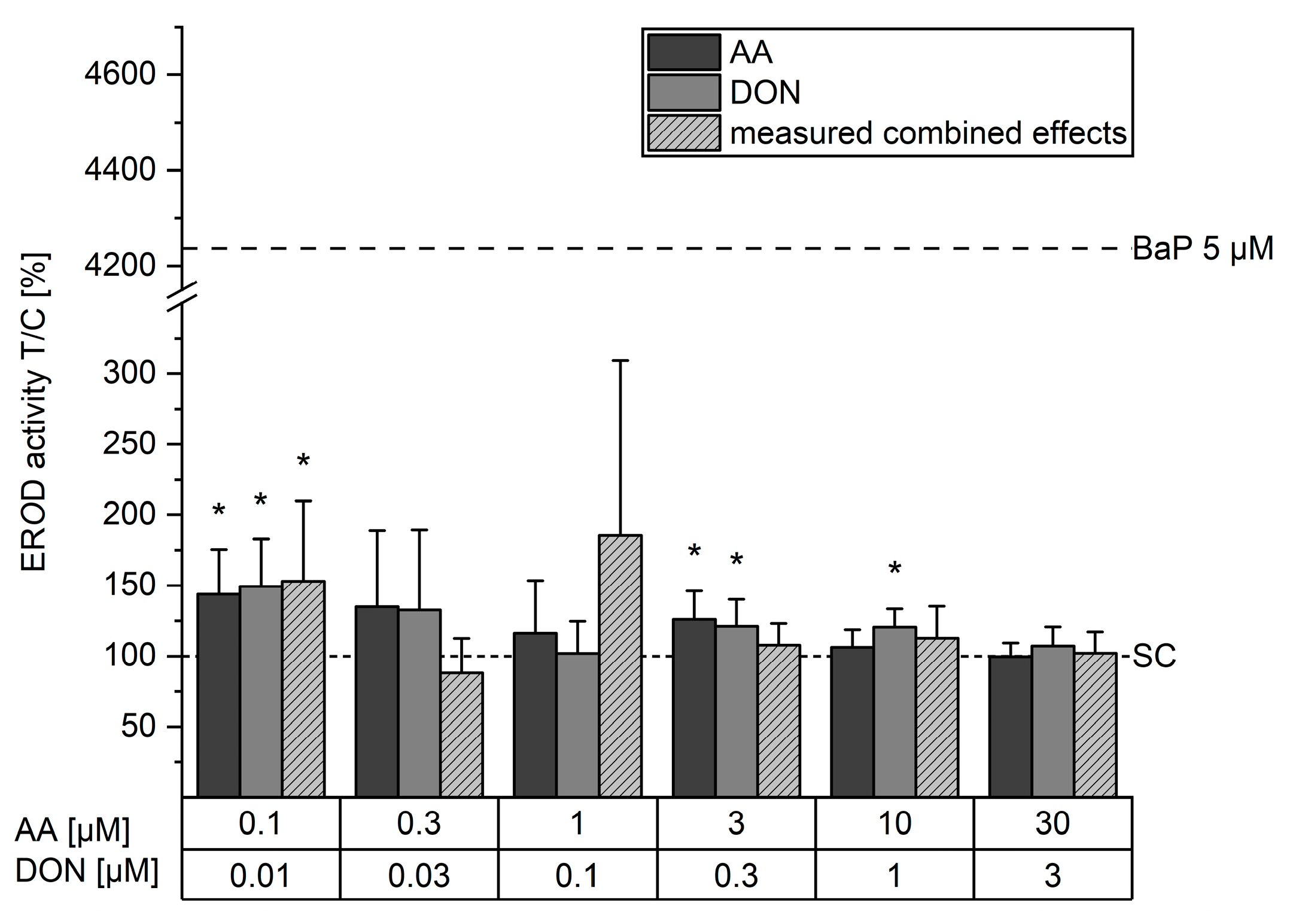
2.3. Effects on EROD Activity
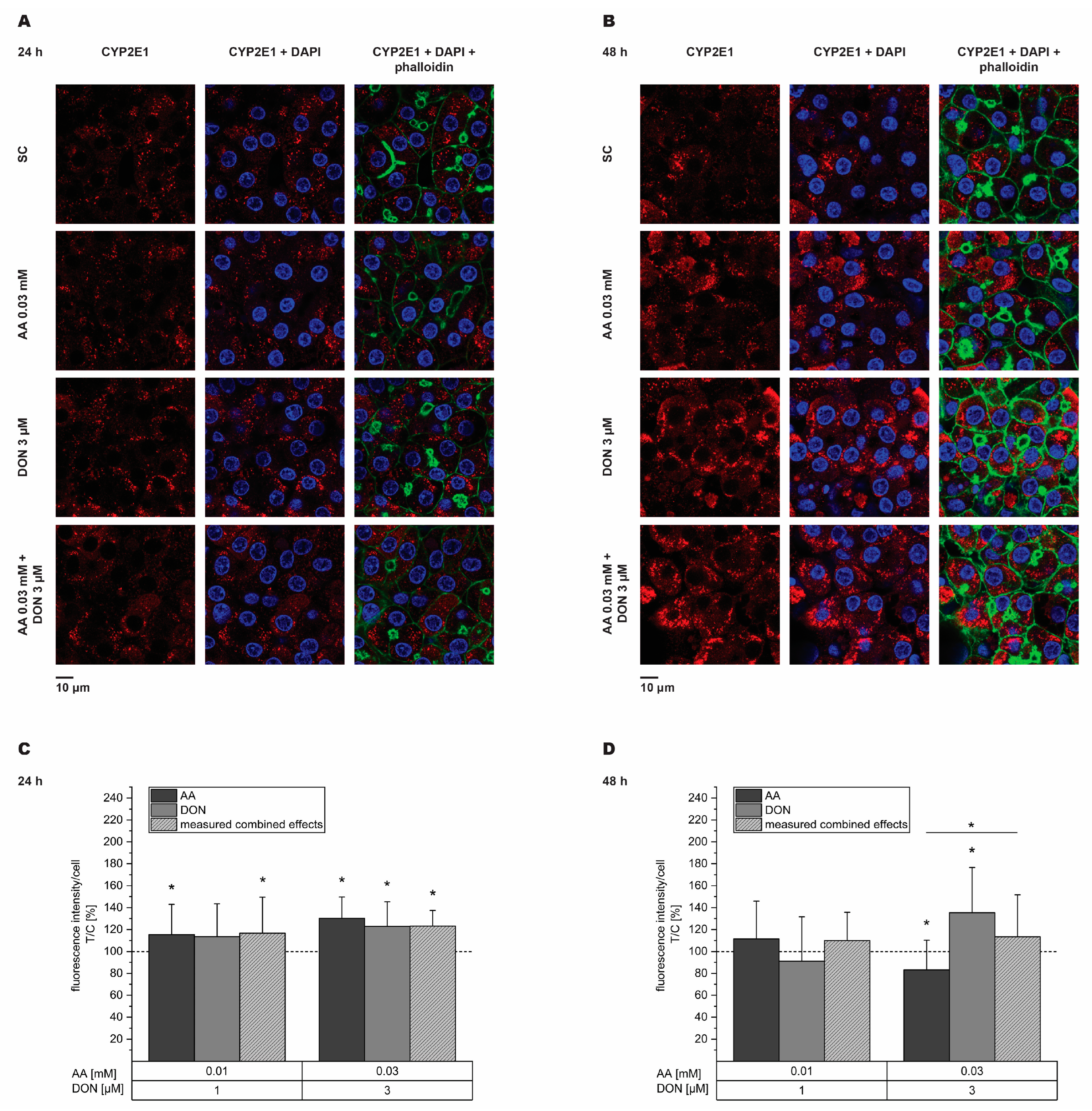
2.4. Impairment of CYP2E1 and CYP3A4 Expression
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Materials
5.2. Cell Culture
5.3. Dosage Information
5.4. Neutral Red Uptake Assay
5.5. Quantitative Real-Time PCR
5.6. CYP1A Activity
5.7. Immunofluorescence Staining
5.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, M.-C.; Madec, S.; Coton, E.; Hymery, N. Natural Co-Occurrence of Mycotoxins in Foods and Feeds and their in vitro Combined Toxicological Effects. Toxins 2016, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Heys, K.A.; Shore, R.F.; Pereira, M.G.; Jones, K.C.; Martin, F.L. Risk assessment of environmental mixture effects. RSC Adv. 2016, 6, 47844–47857. [Google Scholar] [CrossRef]
- WHO|JECFA. Safety Evaluation of Certain Contaminants in Food—Acrylamide. WHO Food Additives Series 63, FAO JECFA Monographs 8. Available online: https://apps.who.int/food-additives-contaminants-jecfa-database/Home/Chemical/5198 (accessed on 17 July 2024).
- Scientific Opinion on acrylamide in food. EFSA J. 2015, 13, 4104. [CrossRef]
- Wang, Z.; Wu, Q.; Kuča, K.; Dohnal, V.; Tian, Z. Deoxynivalenol: Signaling pathways and human exposure risk assessment—An update. Arch. Toxicol. 2014, 88, 1915–1928. [Google Scholar] [CrossRef]
- Munkvold, G.P. Fusarium Species and Their Associated Mycotoxins. Methods Mol. Biol. 2017, 1542, 51–106. [Google Scholar] [CrossRef] [PubMed]
- Thielecke, F.; Nugent, A.P. Contaminants in Grain-A Major Risk for Whole Grain Safety? Nutrients 2018, 10, 1213. [Google Scholar] [CrossRef]
- Bogdanova, E.; Rozentale, I.; Pugajeva, I.; Emecheta, E.E.; Bartkevics, V. Occurrence and risk assessment of myco-toxins, acrylamide, and furan in Latvian beer. Food Addit. Contam. Part B Surveill. 2018, 11, 126–137. [Google Scholar] [CrossRef]
- Balbo, C.; Woźniak, Ł. Dietary exposure and risk characterisation of multiple chemical contaminants in rye-wheat bread marketed in Poland. EFSA J. 2022, 20, e200911. [Google Scholar] [CrossRef]
- Tareke, E.; Rydberg, P.; Karlsson, P.; Eriksson, S.; Törnqvist, M. Analysis of acrylamide, a carcinogen formed in heated foodstuffs. J. Agric. Food Chem. 2002, 50, 4998–5006. [Google Scholar] [CrossRef]
- Koszucka, A.; Nowak, A. Thermal processing food-related toxicants: A review. Crit. Rev. Food Sci. Nutr. 2019, 59, 3579–3596. [Google Scholar] [CrossRef]
- Semla, M.; Goc, Z.; Martiniaková, M.; Omelka, R.; Formicki, G. Acrylamide: A common food toxin related to physiological functions and health. Physiol. Res. 2017, 66, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Taeymans, D.; Wood, J.; Ashby, P.; Blank, I.; Studer, A.; Stadler, R.H.; Gondé, P.; van Eijck, P.; Lalljie, S.; Lingnert, H.; et al. A review of acrylamide: An industry perspective on research, analysis, formation, and control. Crit. Rev. Food Sci. Nutr. 2004, 44, 323–347. [Google Scholar] [CrossRef] [PubMed]
- De Borba, V.S.; Lemos, A.C.; Cerqueira, M.B.R.; Badiale-Furlong, E. Pasta cooking influence on in vitro bioaccessibility of type B trichothecenes, acrylamide and hydroxymethylfurfural. Food Res. Int. 2023, 169, 112863. [Google Scholar] [CrossRef] [PubMed]
- Kraus, D.; Rokitta, D.; Fuhr, U.; Tomalik-Scharte, D. The role of human cytochrome P450 enzymes in metabolism of acrylamide in vitro. Toxicol. Mech. Methods 2013, 23, 346–351. [Google Scholar] [CrossRef]
- Ghanayem, B.I.; McDaniel, L.P.; Churchwell, M.I.; Twaddle, N.C.; Snyder, R.; Fennell, T.R.; Doerge, D.R. Role of CYP2E1 in the epoxidation of acrylamide to glycidamide and formation of DNA and hemoglobin adducts. Toxicol. Sci. 2005, 88, 311–318. [Google Scholar] [CrossRef]
- Sen, A.; Ozgun, O.; Arinç, E.; Arslan, S. Diverse action of acrylamide on cytochrome P450 and glutathione S-transferase isozyme activities, mRNA levels and protein levels in human hepatocarcinoma cells. Cell Biol. Toxicol. 2012, 28, 175–186. [Google Scholar] [CrossRef]
- Mei, N.; Guo, L.; Tseng, J.; Dial, S.L.; Liao, W.; Manjanatha, M.G. Gene expression changes associated with xenobiotic metabolism pathways in mice exposed to acrylamide. Environ. Mol. Mutagen. 2008, 49, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Nixon, B.J.; Katen, A.L.; Stanger, S.J.; Schjenken, J.E.; Nixon, B.; Roman, S.D. Mouse spermatocytes express CYP2E1 and respond to acrylamide exposure. PLoS ONE 2014, 9, e94904. [Google Scholar] [CrossRef]
- Cao, J.; Liu, Y.; Jia, L.; Jiang, L.-P.; Geng, C.-Y.; Yao, X.-F.; Kong, Y.; Jiang, B.-N.; Zhong, L.-F. Curcumin attenuates acrylamide-induced cytotoxicity and genotoxicity in HepG2 cells by ROS scavenging. J. Agric. Food Chem. 2008, 56, 12059–12063. [Google Scholar] [CrossRef]
- Chen, W.; Feng, L.; Shen, Y.; Su, H.; Li, Y.; Zhuang, J.; Zhang, L.; Zheng, X. Myricitrin inhibits acrylamide-mediated cytotoxicity in human Caco-2 cells by preventing oxidative stress. Biomed. Res. Int. 2013, 2013, 724183. [Google Scholar] [CrossRef]
- Kacar, S.; Vejselova, D.; Kutlu, H.M.; Sahinturk, V. Acrylamide-derived cytotoxic, anti-proliferative, and apoptotic effects on A549 cells. Hum. Exp. Toxicol. 2018, 37, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-H.; Tsou, T.-C.; Chiu, I.-M.; Chou, C.-C. Proliferation inhibition, DNA damage, and cell-cycle arrest of human astrocytoma cells after acrylamide exposure. Chem. Res. Toxicol. 2010, 23, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Srivastava, S.; Dewangan, J.; Divakar, A.; Kumar Rath, S. Global occurrence of deoxynivalenol in food commodities and exposure risk assessment in humans in the last decade: A survey. Crit. Rev. Food Sci. Nutr. 2020, 60, 1346–1374. [Google Scholar] [CrossRef] [PubMed]
- Stadler, D.; Lambertini, F.; Woelflingseder, L.; Schwartz-Zimmermann, H.; Marko, D.; Suman, M.; Berthiller, F.; Krska, R. The Influence of Processing Parameters on the Mitigation of Deoxynivalenol during Industrial Baking. Toxins 2019, 11, 317. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. Deoxynivalenol in food and feed: Occurrence and exposure. EFSA J. 2013, 11, 3379. [Google Scholar] [CrossRef]
- Payros, D.; Alassane-Kpembi, I.; Pierron, A.; Loiseau, N.; Pinton, P.; Oswald, I.P. Toxicology of deoxynivalenol and its acetylated and modified forms. Arch. Toxicol. 2016, 90, 2931–2957. [Google Scholar] [CrossRef]
- Warth, B.; Sulyok, M.; Berthiller, F.; Schuhmacher, R.; Krska, R. New insights into the human metabolism of the Fusarium mycotoxins deoxynivalenol and zearalenone. Toxicol. Lett. 2013, 220, 88–94. [Google Scholar] [CrossRef]
- EFSA CONTAM Panel (EFSA Panel on Contaminants in the Food Chain); Knutsen, H. K.; Alexander, J.; Barregård, L.; Bignami, M.; Brüschweiler, B.; Ceccatelli, S.; Cottrill, B.; Dinovi, M.; Grasl-Kraupp, B.; et al. Scientific Opinion on the risks to human and animal health related to the presence of deoxynivalenol and its acetylated and modified forms in food and feed. EFSA J. 2017, 15, e04718. [Google Scholar] [CrossRef]
- Ueno, Y. Mode of action of trichothecenes. Ann. Nutr. Aliment. 1977, 31, 885–900. [Google Scholar] [CrossRef]
- Bony, S.; Carcelen, M.; Olivier, L.; Devaux, A. Genotoxicity assessment of deoxynivalenol in the Caco-2 cell line model using the Comet assay. Toxicol. Lett. 2006, 166, 67–76. [Google Scholar] [CrossRef]
- Del Favero, G.; Woelflingseder, L.; Braun, D.; Puntscher, H.; Kütt, M.-L.; Dellafiora, L.; Warth, B.; Pahlke, G.; Dall’Asta, C.; Adam, G.; et al. Response of intestinal HT-29 cells to the trichothecene mycotoxin deoxynivalenol and its sulfated conjugates. Toxicol. Lett. 2018, 295, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Instanes, C.; Hetland, G. Deoxynivalenol (DON) is toxic to human colonic, lung and monocytic cell lines, but does not increase the IgE response in a mouse model for allergy. Toxicology 2004, 204, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Maresca, M.; Mahfoud, R.; Garmy, N.; Fantini, J. The mycotoxin deoxynivalenol affects nutrient absorption in human intestinal epithelial cells. J. Nutr. 2002, 132, 2723–2731. [Google Scholar] [CrossRef]
- Beisl, J.; Pahlke, G.; Abeln, H.; Ehling-Schulz, M.; Del Favero, G.; Varga, E.; Warth, B.; Sulyok, M.; Abia, W.; Ezekiel, C.N.; et al. Combinatory effects of cereulide and deoxynivalenol on in vitro cell viability and inflammation of human Caco-2 cells. Arch. Toxicol. 2020, 94, 833–844. [Google Scholar] [CrossRef]
- Peng, Z.; Chen, L.; Nüssler, A.K.; Liu, L.; Yang, W. Current sights for mechanisms of deoxynivalenol-induced hepatotoxicity and prospective views for future scientific research: A mini review. J. Appl. Toxicol. 2017, 37, 518–529. [Google Scholar] [CrossRef]
- Königs, M.; Schwerdt, G.; Gekle, M.; Humpf, H.-U. Effects of the mycotoxin deoxynivalenol on human primary hepatocytes. Mol. Nutr. Food Res. 2008, 52, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.E.; Lee, W.; Jeong, J.Y.; Lee, Y.; Lee, H.-J.; Kim, M.S.; Kim, D.-W.; Yu, D.; Cho, A.; Oh, Y.K.; et al. Effects of deoxynivalenol- and zearalenone-contaminated feed on the gene expression profiles in the kidneys of piglets. Asian-Australas. J. Anim. Sci. 2018, 31, 138–148. [Google Scholar] [CrossRef]
- Gajęcka, M.; Dąbrowski, M.; Otrocka-Domagała, I.; Brzuzan, P.; Rykaczewska, A.; Cieplińska, K.; Barasińska, M.; Gajęcki, M.T.; Zielonka, Ł. Correlations between exposure to deoxynivalenol and zearalenone and the immunohistochemical expression of estrogen receptors in the intestinal epithelium and the mRNA expression of selected colonic enzymes in pre-pubertal gilts. Toxicon 2020, 173, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]
- Bliss, C.I. The Toxicity of Poisons Applied Jointly. Ann. Appl. Biol. 1939, 26, 585–615. [Google Scholar] [CrossRef]
- Webb, J.L. Effect of more than one inhibitor. Enzym. Metab. Inhib. 1963, 1, 488–512. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Klarić, M.S.; Rašić, D.; Peraica, M. Deleterious effects of mycotoxin combinations involving ochratoxin A. Toxins 2013, 5, 1965–1987. [Google Scholar] [CrossRef]
- Knebel, C.; Neeb, J.; Zahn, E.; Schmidt, F.; Carazo, A.; Holas, O.; Pavek, P.; Püschel, G.P.; Zanger, U.M.; Süssmuth, R.; et al. Unexpected Effects of Propiconazole, Tebuconazole, and Their Mixture on the Receptors CAR and PXR in Human Liver Cells. Toxicol. Sci. 2018, 163, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Knebel, C.; Kebben, J.; Eberini, I.; Palazzolo, L.; Hammer, H.S.; Süssmuth, R.D.; Heise, T.; Hessel-Pras, S.; Lampen, A.; Braeuning, A.; et al. Propiconazole is an activator of AHR and causes concentration additive effects with an established AHR ligand. Arch. Toxicol. 2018, 92, 3471–3486. [Google Scholar] [CrossRef]
- Hartman, G.D.; Kuduk, S.D.; Espiritu, C.; Lam, A.M. P450s under Restriction (PURE) Screen Using HepaRG and Primary Human Hepatocytes for Discovery of Novel HBV Antivirals. ACS Med. Chem. Lett. 2020, 11, 1919–1927. [Google Scholar] [CrossRef]
- Andersson, T.B.; Kanebratt, K.P.; Kenna, J.G. The HepaRG cell line: A unique in vitro tool for understanding drug metabolism and toxicology in human. Expert Opin. Drug Metab. Toxicol. 2012, 8, 909–920. [Google Scholar] [CrossRef]
- Lasch, A.; Lichtenstein, D.; Marx-Stoelting, P.; Braeuning, A.; Alarcan, J. Mixture effects of chemicals: The difficulty to choose appropriate mathematical models for appropriate conclusions. Environ. Pollut. 2020, 260, 113953. [Google Scholar] [CrossRef] [PubMed]
- Le Hegarat, L.; Dumont, J.; Josse, R.; Huet, S.; Lanceleur, R.; Mourot, A.; Poul, J.-M.; Guguen-Guillouzo, C.; Guillouzo, A.; Fessard, V. Assessment of the genotoxic potential of indirect chemical mutagens in HepaRG cells by the comet and the cytokinesis-block micronucleus assays. Mutagenesis 2010, 25, 555–560. [Google Scholar] [CrossRef]
- Chen, X.; Murdoch, R.; Shafer, D.J.; Ajuwon, K.M.; Applegate, T.J. Cytotoxicity of various chemicals and mycotoxins in fresh primary duck embryonic fibroblasts: A comparison to HepG2 cells. J. Appl. Toxicol. 2016, 36, 1437–1445. [Google Scholar] [CrossRef]
- Juan-García, A.; Taroncher, M.; Font, G.; Ruiz, M.-J. Micronucleus induction and cell cycle alterations produced by deoxynivalenol and its acetylated derivatives in individual and combined exposure on HepG2 cells. Food Chem. Toxicol. 2018, 118, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Juan-García, A.; Juan, C.; Tolosa, J.; Ruiz, M.J. Effects of deoxynivalenol, 3-acetyl-deoxynivalenol and 15-acetyl-deoxynivalenol on parameters associated with oxidative stress in HepG2 cells. Mycotoxin Res. 2019, 35, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Eisenbrand, G. Revisiting the evidence for genotoxicity of acrylamide (AA), key to risk assessment of dietary AA exposure. Arch. Toxicol. 2020, 94, 2939–2950. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Liu, S.; Zhao, C.; Fan, L.; Hu, H.; Yin, S. The combination of T-2 toxin and acrylamide synergistically induces hepatotoxicity and nephrotoxicity via the activation of oxidative stress and the mitochondrial pathway. Toxicon 2021, 189, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Van de Walle, J.; Romier, B.; Larondelle, Y.; Schneider, Y.-J. Influence of deoxynivalenol on NF-kappaB activation and IL-8 secretion in human intestinal Caco-2 cells. Toxicol. Lett. 2008, 177, 205–214. [Google Scholar] [CrossRef]
- Kim, T.H.; Shin, S.; Kim, K.B.; Seo, W.S.; Shin, J.C.; Choi, J.H.; Weon, K.-Y.; Joo, S.H.; Jeong, S.W.; Shin, B.S. Determination of acrylamide and glycidamide in various biological matrices by liquid chromatography-tandem mass spectrometry and its application to a pharmacokinetic study. Talanta 2015, 131, 46–54. [Google Scholar] [CrossRef]
- Doerge, D.R.; Young, J.F.; McDaniel, L.P.; Twaddle, N.C.; Churchwell, M.I. Toxicokinetics of acrylamide and glycidamide in Fischer 344 rats. Toxicol. Appl. Pharmacol. 2005, 208, 199–209. [Google Scholar] [CrossRef]
- Doerge, D.R.; Young, J.F.; McDaniel, L.P.; Twaddle, N.C.; Churchwell, M.I. Toxicokinetics of acrylamide and glycidamide in B6C3F1 mice. Toxicol. Appl. Pharmacol. 2005, 202, 258–267. [Google Scholar] [CrossRef]
- Zödl, B.; Schmid, D.; Wassler, G.; Gundacker, C.; Leibetseder, V.; Thalhammer, T.; Ekmekcioglu, C. Intestinal transport and metabolism of acrylamide. Toxicology 2007, 232, 99–108. [Google Scholar] [CrossRef]
- Schabacker, J.; Schwend, T.; Wink, M. Reduction of acrylamide uptake by dietary proteins in a Caco-2 gut model. J. Agric. Food Chem. 2004, 52, 4021–4025. [Google Scholar] [CrossRef]
- Meky, F.A.; Turner, P.C.; Ashcroft, A.E.; Miller, J.D.; Qiao, Y.-L.; Roth, M.J.; Wild, C.P. Development of a urinary biomarker of human exposure to deoxynivalenol. Food Chem. Toxicol. 2003, 41, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J.; Islam, Z.; Amuzie, C.J. Immunochemical assessment of deoxynivalenol tissue distribution following oral exposure in the mouse. Toxicol. Lett. 2008, 178, 83–87. [Google Scholar] [CrossRef]
- Wan, D.; Huang, L.; Pan, Y.; Wu, Q.; Chen, D.; Tao, Y.; Wang, X.; Liu, Z.; Li, J.; Wang, L.; et al. Metabolism, distribution, and excretion of deoxynivalenol with combined techniques of radiotracing, high-performance liquid chromatography ion trap time-of-flight mass spectrometry, and online radiometric detection. J. Agric. Food Chem. 2014, 62, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Beisl, J.; Varga, E.; Braun, D.; Warth, B.; Ehling-Schulz, M.; Del Favero, G.; Marko, D. Assessing Mixture Effects of Cereulide and Deoxynivalenol on Intestinal Barrier Integrity and Uptake in Differentiated Human Caco-2 Cells. Toxins 2021, 13, 189. [Google Scholar] [CrossRef] [PubMed]
- Sergent, T.; Parys, M.; Garsou, S.; Pussemier, L.; Schneider, Y.-J.; Larondelle, Y. Deoxynivalenol transport across human intestinal Caco-2 cells and its effects on cellular metabolism at realistic intestinal concentrations. Toxicol. Lett. 2006, 164, 167–176. [Google Scholar] [CrossRef]
- Ling, K.-H.; Wan, M.L.Y.; El-Nezami, H.; Wang, M. Protective Capacity of Resveratrol, a Natural Polyphenolic Compound, against Deoxynivalenol-Induced Intestinal Barrier Dysfunction and Bacterial Translocation. Chem. Res. Toxicol. 2016, 29, 823–833. [Google Scholar] [CrossRef]
- Pyo, M.C.; Shin, H.S.; Jeon, G.Y.; Lee, K.-W. Synergistic Interaction of Ochratoxin A and Acrylamide Toxins in Human Kidney and Liver Cells. Biol. Pharm. Bull. 2020, 43, 1346–1355. [Google Scholar] [CrossRef]
- Ivanova, L.; Denisov, I.G.; Grinkova, Y.V.; Sligar, S.G.; Fæste, C.K. Biotransformation of the Mycotoxin Enniatin B1 by CYP P450 3A4 and Potential for Drug-Drug Interactions. Metabolites 2019, 9, 158. [Google Scholar] [CrossRef]
- Manikandan, P.; Nagini, S. Cytochrome P450 Structure, Function and Clinical Significance: A Review. Curr. Drug Targets 2018, 19, 38–54. [Google Scholar] [CrossRef]
- Ndossi, D.G.; Frizzell, C.; Tremoen, N.H.; Fæste, C.K.; Verhaegen, S.; Dahl, E.; Eriksen, G.S.; Sørlie, M.; Connolly, L.; Ropstad, E. An in vitro investigation of endocrine disrupting effects of trichothecenes deoxynivalenol (DON), T-2 and HT-2 toxins. Toxicol. Lett. 2012, 214, 268–278. [Google Scholar] [CrossRef]
- Mann, A.; Pelz, T.; Rennert, K.; Mosig, A.; Decker, M.; Lupp, A. Evaluation of HepaRG cells for the assessment of indirect drug-induced hepatotoxicity using INH as a model substance. Hum. Cell 2017, 30, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, C.; Pelkonen, O.; Andersson, T.B.; Strickland, J.; Wilk-Zasadna, I.; Asturiol, D.; Cole, T.; Liska, R.; Worth, A.; Müller-Vieira, U.; et al. Validation of in vitro methods for human cytochrome P450 enzyme induction: Outcome of a multi-laboratory study. Toxicol. In Vitro 2019, 60, 212–228. [Google Scholar] [CrossRef] [PubMed]
- Abass, K.; Lämsä, V.; Reponen, P.; Küblbeck, J.; Honkakoski, P.; Mattila, S.; Pelkonen, O.; Hakkola, J. Characteriza-tion of human cytochrome P450 induction by pesticides. Toxicology 2012, 294, 17–26. [Google Scholar] [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef]
- Hoekstra, R.; Nibourg, G.A.A.; van der Hoeven, T.V.; Ackermans, M.T.; Hakvoort, T.B.M.; van Gulik, T.M.; Lamers, W.H.; Elferink, R.P.O.; Chamuleau, R.A.F.M. The HepaRG cell line is suitable for bioartificial liver application. Int. J. Biochem. Cell Biol. 2011, 43, 1483–1489. [Google Scholar] [CrossRef]
- Lamy, E.; Völkel, Y.; Roos, P.H.; Kassie, F.; Mersch-Sundermann, V. Ethanol enhanced the genotoxicity of acrylamide in human, metabolically competent HepG2 cells by CYP2E1 induction and glutathione depletion. Int. J. Hyg. Environ. Health 2008, 211, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, M.; Groothuis, G.M.M.; de Kanter, R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2006, 2, 875–894. [Google Scholar] [CrossRef]
- Repetto, G.; Del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Beisl, J.; Pahlke, G.; Ehling-Schulz, M.; Del Favero, G.; Marko, D. Cereulide and Deoxynivalenol Increase LC3 Protein Levels in HepG2 Liver Cells. Toxins 2022, 14, 151. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beisl, J.; Jochum, K.; Chen, Y.; Varga, E.; Marko, D. Combinatory Effects of Acrylamide and Deoxynivalenol on In Vitro Cell Viability and Cytochrome P450 Enzymes of Human HepaRG Cells. Toxins 2024, 16, 389. https://doi.org/10.3390/toxins16090389
Beisl J, Jochum K, Chen Y, Varga E, Marko D. Combinatory Effects of Acrylamide and Deoxynivalenol on In Vitro Cell Viability and Cytochrome P450 Enzymes of Human HepaRG Cells. Toxins. 2024; 16(9):389. https://doi.org/10.3390/toxins16090389
Chicago/Turabian StyleBeisl, Julia, Kristina Jochum, Yi Chen, Elisabeth Varga, and Doris Marko. 2024. "Combinatory Effects of Acrylamide and Deoxynivalenol on In Vitro Cell Viability and Cytochrome P450 Enzymes of Human HepaRG Cells" Toxins 16, no. 9: 389. https://doi.org/10.3390/toxins16090389
APA StyleBeisl, J., Jochum, K., Chen, Y., Varga, E., & Marko, D. (2024). Combinatory Effects of Acrylamide and Deoxynivalenol on In Vitro Cell Viability and Cytochrome P450 Enzymes of Human HepaRG Cells. Toxins, 16(9), 389. https://doi.org/10.3390/toxins16090389







