Characterization of the Phosphotransferase from Bacillus subtilis 1101 That Is Responsible for the Biotransformation of Zearalenone
Abstract
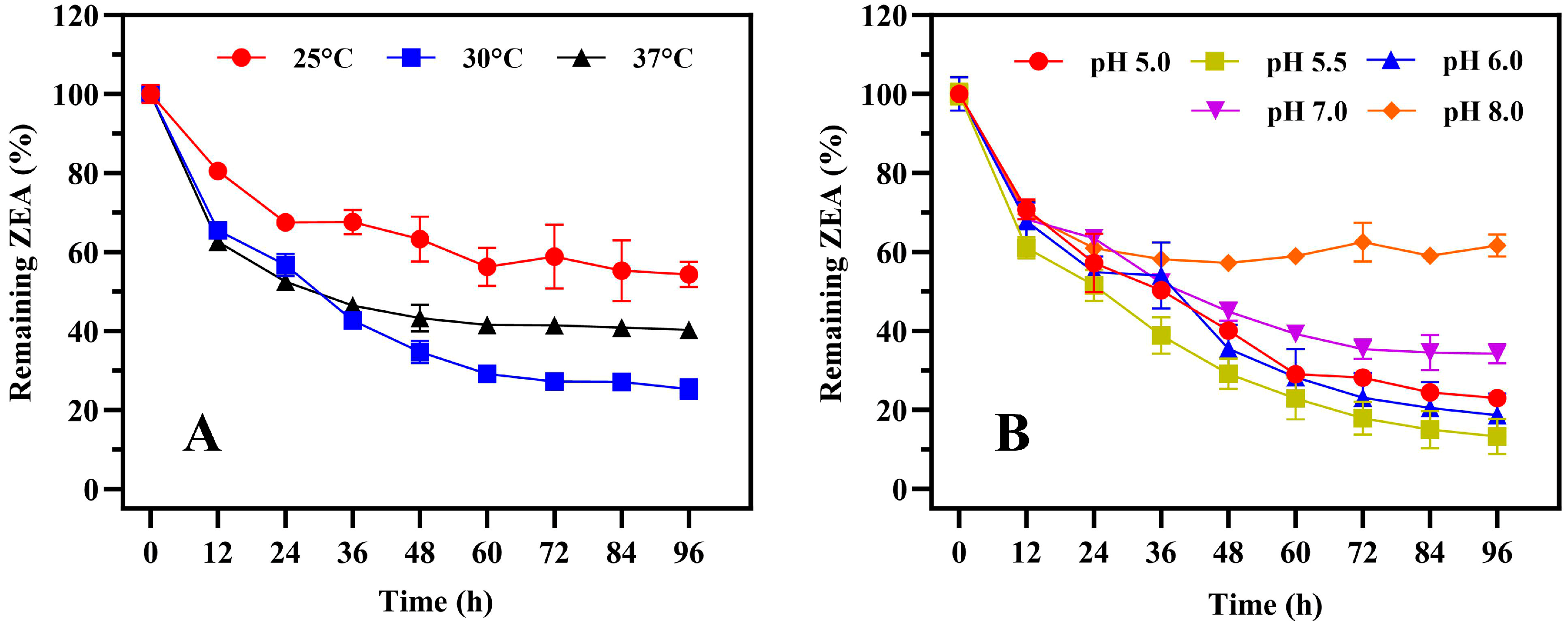
1. Introduction
2. Results
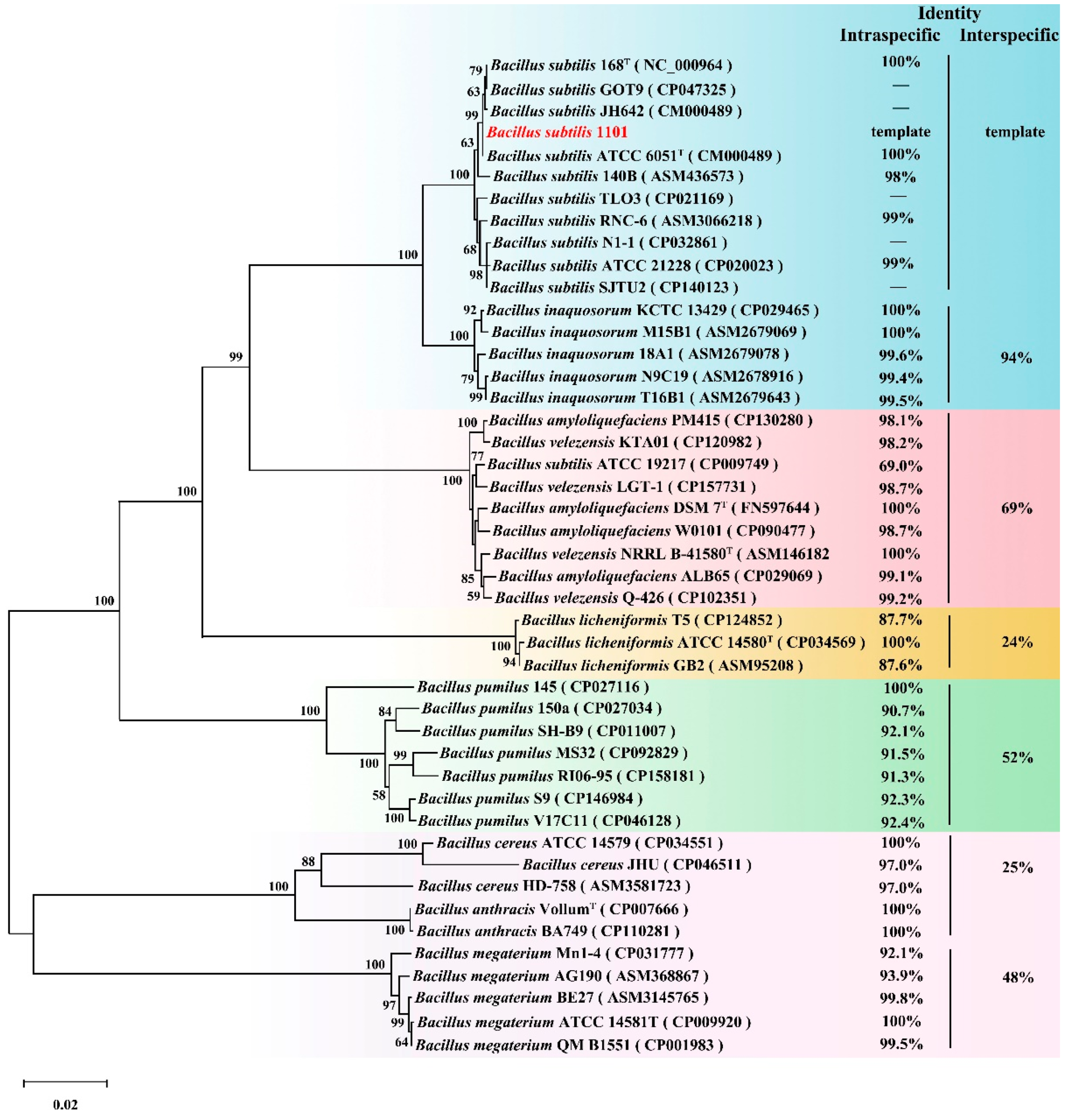
2.1. Isolation, Identification, and Characteristics of Strain 1101
2.2. Expression and Purification of Recombinant ZPH1101
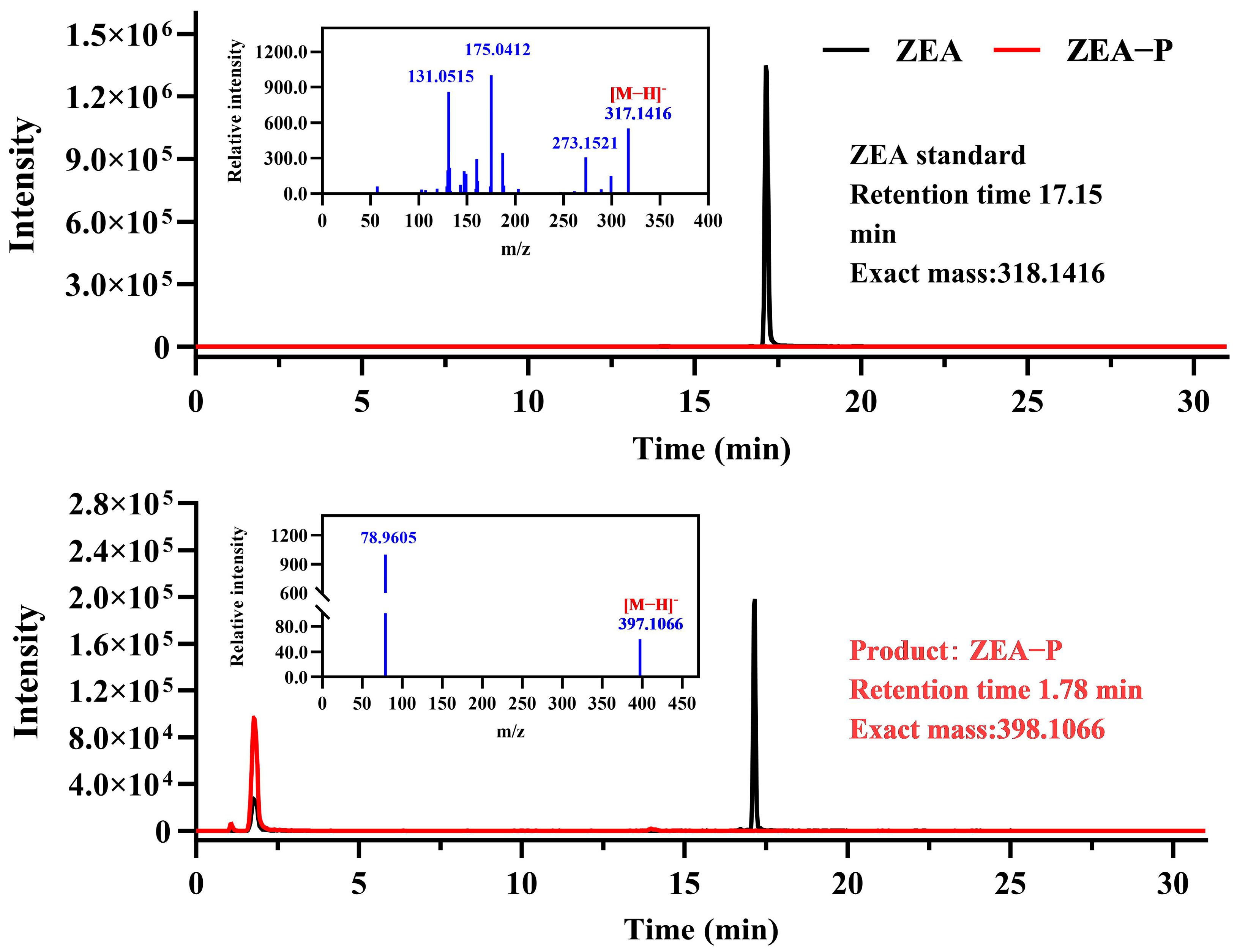
2.3. Biotransformation of ZEA by Recombinant ZPH1101
2.4. Characterization of Recombinant ZPH1101
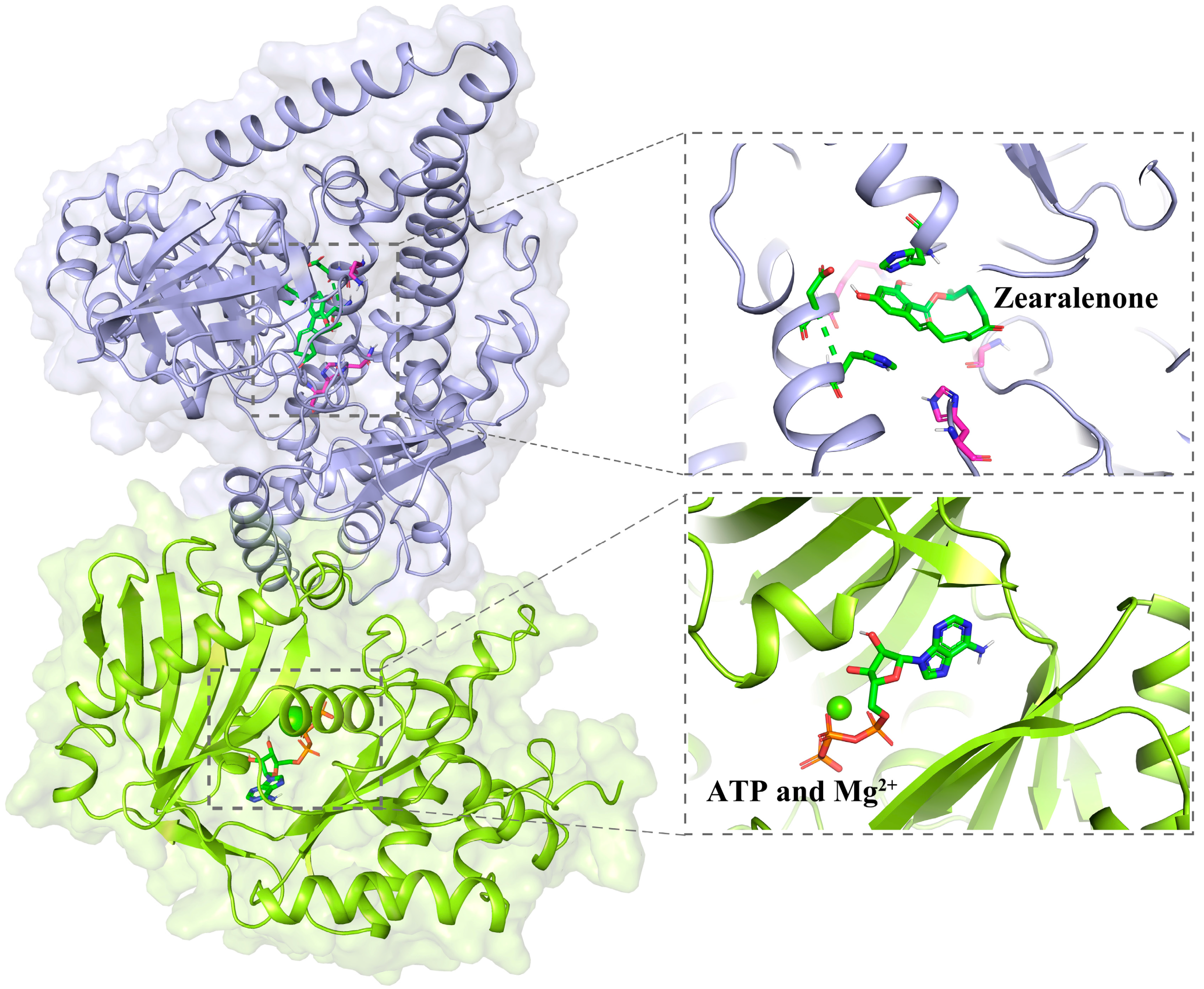
2.5. Sequence and Structure Analysis of ZPH1101
2.6. Characteristics of Knockout Mutant
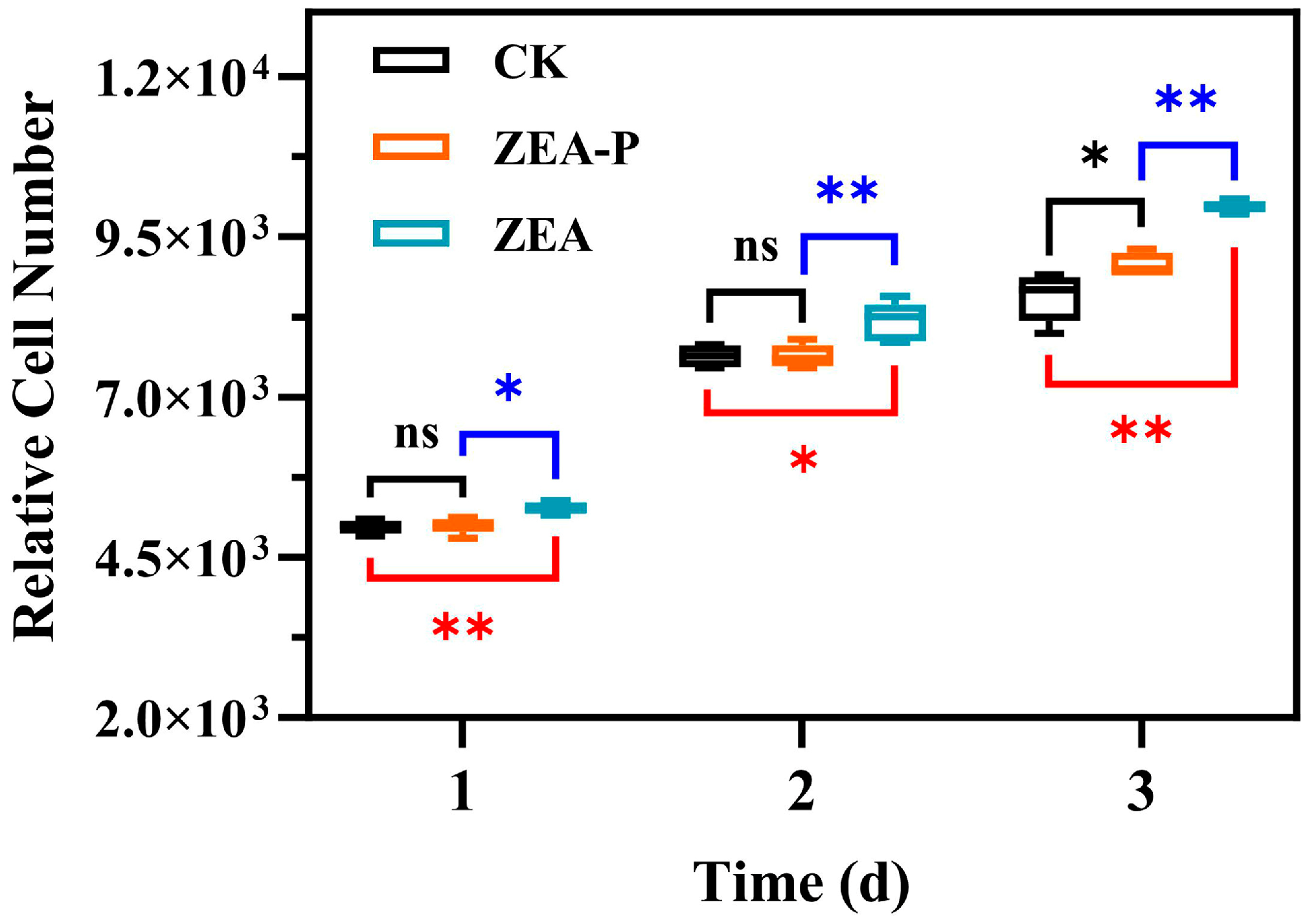
2.7. Estrogenic Activity of ZEA-P
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Samples, Chemicals, Strains, and Plasmid
5.2. Isolation and Identification of Strain 1101
5.3. Growth and Biotransformation Characteristics of Strain 1101
5.3.1. Determination of Optimal Growth Temperature and pH
5.3.2. Determination of Optimal Biotransformation Temperature and pH
5.4. Genome Sequencing and Annotation
5.5. Gene Cloning and Expression
5.6. Knockout of the Gene zph1101
5.7. Enzyme Activity Assay
5.8. Sequence Analysis and Molecular Docking
5.9. Detection of ZEA and Its Biotransformation Products
5.10. Determination of the Estrogenic Toxicity of ZEA and Its Biotransformation Products
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cai, P.; Feng, N.; Zou, H.; Gu, J.; Liu, X.; Liu, Z.; Yuan, Y.; Bian, J. Zearalenone damages the male reproductive system of rats by destroying testicular focal adhesion. Environ. Toxicol. 2023, 38, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Zhi, Y.; Xu, H.; Fang, H.; Jia, X. Zearalenone causes embryotoxicity and induces oxidative stress and apoptosis in differentiated human embryonic stem cells. Toxicol. Vitro 2019, 54, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Ren, Z.; Gao, S.; Chen, Y.; Yang, Y.; Yang, D.; Deng, J.; Zuo, Z.; Wang, Y.; Shen, L. Individual and combined effects of deoxynivalenol and zearalenone on mouse kidney. Environ. Toxicol. Pharmacol. 2015, 40, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Ryu, D.; Jackson, L.S.; Bullerman, L.B. Effects of processing on zearalenone. Adv. Exp. Med. Biol. 2002, 504, 205–216. [Google Scholar]
- Bullerman, L.B.; Bianchini, A. Stability of mycotoxins during food processing. Int. J. Food Microbiol. 2007, 119, 140–146. [Google Scholar] [CrossRef]
- Scudamore, K.A.; Guy, R.C.; Kelleher, B.; MacDonald, S.J. Fate of Fusarium mycotoxins in maize flour and grits during extrusion cooking. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2008, 25, 1374–1384. [Google Scholar] [CrossRef]
- He, J.; Zhou, T. Patented techniques for detoxification of mycotoxins in feeds and food matrices. Recent Pat. Food Nutr. Agric. 2010, 2, 96–104. [Google Scholar] [CrossRef]
- Adegoke, T.V.; Yang, B.; Xing, F.; Tian, X.; Wang, G.; Tai, B.; Si, P.; Hussain, S.; Jahan, I. Microbial enzymes involved in the biotransformation of major mycotoxins. J. Agric. Food Chem. 2023, 71, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, B.; Li, X.; Dong, L.; Javed, M.T.; Xu, L.; Saleemi, M.K.; Li, G.; Jin, B.; Cui, H.; Ali, A.; et al. Microbial and enzymatic battle with food contaminant zearalenone (ZEN). Appl. Microbiol. Biotechnol. 2022, 106, 4353–4365. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Jiang, L.; Huang, H. Improvement of the enzymatic detoxification activity towards mycotoxins through structure-based engineering. Biotechnol. Adv. 2022, 56, 107927. [Google Scholar] [CrossRef] [PubMed]
- Takahashi-Ando, N.; Kimura, M.; Kakeya, H.; Osada, H.; Yamaguchi, I. A novel lactonohydrolase responsible for the detoxification of zearalenone: Enzyme purification and gene cloning. Biochem. J. 2002, 365, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, G.; Hou, M.; Du, S.; Han, J.; Yu, Y.; Gao, H.; He, D.; Shi, J.; Lee, Y.W.; et al. New hydrolase from Aeromicrobium sp. HA for the biodegradation of zearalenone: Identification, mechanism, and application. J. Agric. Food Chem. 2023, 71, 2411–2420. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Du, S.; Qiu, H.; Wu, Y.; Hong, Q.; Wang, G.; Mohamed, S.R.; Lee, Y.W.; Xu, J. A hydrolase produced by Rhodococcus erythropolis HQ is responsible for the detoxification of zearalenone. Toxins 2023, 15, 688. [Google Scholar] [CrossRef]
- Yu, Y.; Qiu, L.; Wu, H.; Tang, Y.; Yu, Y.; Li, X.; Liu, D. Degradation of zearalenone by the extracellular extracts of Acinetobacter sp. SM04 liquid cultures. Biodegradation 2011, 22, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, H.; Tang, Y.; Qiu, L. Cloning, expression of a peroxiredoxin gene from Acinetobacter sp. SM04 and characterization of its recombinant protein for zearalenone detoxification. Microbiol. Res. 2012, 167, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Hu, Y.; He, J.; Wu, L.; Liao, F.; Luo, B.; He, Y.; Zuo, Z.; Ren, Z.; Zhong, Z.; et al. Zearalenone degradation by two Pseudomonas strains from soil. Mycotoxin Res. 2014, 30, 191–196. [Google Scholar] [CrossRef]
- Tan, H.; Zhang, Z.; Hu, Y.; Wu, L.; Liao, F.; He, J.; Luo, B.; He, Y.; Zuo, Z.; Ren, Z.; et al. Isolation and characterization of Pseudomonas otitidis TH-N1 capable of degrading zearalenone. Food Control 2015, 47, 285–290. [Google Scholar] [CrossRef]
- Murtaza, B.; Wang, L.; Li, X.; Ali, A.; Haq, S.U.; Ji-bin, L.; Xu, Y. Novel Lactobacillus consortium for effective zearalenone adsorption and biodegradation. Int. Biodeterior. Biodegrad. 2024, 194, 105889. [Google Scholar] [CrossRef]
- Mwabulili, F.; Xie, Y.; Sun, S.; Ma, W.; Li, Q.; Yang, Y.; Jia, H.; Li, X. Thermo-Alkali-Tolerant recombinant laccase from Bacillus swezeyi and its degradation potential against zearalenone and aflatoxin B(1). J. Agric. Food Chem. 2024, 72, 13371–13381. [Google Scholar] [CrossRef]
- Li, Y.; Chen, S.; Yu, Z.; Yao, J.; Jia, Y.; Liao, C.; Chen, J.; Wei, Y.; Guo, R.; He, L.; et al. A novel Bacillus velezensis for efficient degradation of zearalenone. Foods 2024, 13, 530. [Google Scholar] [CrossRef]
- Cho, K.J.; Kang, J.S.; Cho, W.T.; Lee, C.H.; Ha, J.K.; Song, K.B. In vitro degradation of zearalenone by Bacillus subtilis. Biotechnol. Lett. 2010, 32, 1921–1924. [Google Scholar] [CrossRef]
- Tinyiro, S.E.; Wokadala, C.; Xu, D.; Yao, W. Adsorption and degradation of zearalenone by Bacillus strains. Folia Microbiol. 2011, 56, 321–327. [Google Scholar] [CrossRef]
- Hsu, T.C.; Yi, P.J.; Lee, T.Y.; Liu, J.R. Probiotic characteristics and zearalenone-removal ability of a Bacillus licheniformis strain. PLoS ONE 2018, 13, e0194866. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, Y.; Liu, Y.; Ma, Q.; Ji, C.; Zhao, L. Detoxification of the mycoestrogen zearalenone by Bacillus licheniformis spore CotA laccase and application of immobilized laccase in contaminated corn meal. LWT-Food Sci. Technol. 2022, 163, 113548. [Google Scholar] [CrossRef]
- Wang, G.; Yu, M.; Dong, F.; Shi, J.; Xu, J. Esterase activity inspired selection and characterization of zearalenone degrading bacteria Bacillus pumilus ES-21. Food Control 2017, 77, 57–64. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, C.; Yang, J.; Tang, Y. Conversion of zearalenone to β-zearalenol and zearalenone-14,16-diglucoside by Candida parapsilosis ATCC 7330. Food Control 2022, 131, 108429. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, J.; Yu, Y.; Tang, Y. A novel glycosyltransferase from Bacillus subtilis achieves zearalenone detoxification by diglycosylation modification. Food Funct. 2024, 15, 6042–6053. [Google Scholar] [CrossRef]
- Sun, Z.; You, Y.; Xu, H.; You, Y.; He, W.; Wang, Z.; Li, A.; Xia, Y. Food-Grade expression of two laccases in Pichia pastoris and study on their enzymatic degradation characteristics for mycotoxins. J. Agric. Food Chem. 2024, 72, 9365–9375. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.B.; Zheng, H.C.; Xu, J.Y.; Zhao, X.Y.; Shu, W.J.; Li, X.M.; Song, H.; Ma, Y.H. New biotransformation mode of zearalenone identified in Bacillus subtilis Y816 revealing a novel ZEN conjugate. J. Agric. Food Chem. 2021, 69, 7409–7419. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Drouin, P.; Lepp, D.; Li, X.-Z.; Zhu, H.; Castex, M.; Zhou, T. A novel microbial zearalenone transformation through phosphorylation. Toxins 2021, 13, 294. [Google Scholar] [CrossRef]
- Wang, L.T.; Lee, F.L.; Tai, C.J.; Kasai, H. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA-DNA hybridization in the Bacillus subtilis group. Int. J. Syst. Evol. Microbiol. 2007, 57, 1846–1850. [Google Scholar] [CrossRef] [PubMed]
- Narindrasorasak, S.; Bridger, W.A. Phosphoenolypyruvate synthetase of Escherichia coli: Molecular weight, subunit composition, and identification of phosphohistidine in phosphoenzyme intermediate. J. Biol. Chem. 1977, 252, 3121–3127. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Zhou, Q.; Sun, X.; Li, L.; Zhou, B.; Zeng, F.; Zhao, Y.; Shen, W.; Sun, Z. Effect of low-dose zearalenone exposure on reproductive capacity of male mice. Toxicol. Appl. Pharmacol. 2017, 333, 60–67. [Google Scholar] [CrossRef]
- Wang, J.; Tian, H.; Liu, H.; Wen, J.; Huang, R.; Zou, K.; Hou, L.; Li, P. Low dose of zearalenone inhibited the proliferation of porcine prospermatogonia and transformed the physiology through cytokine-cytokine receptor interaction. Theriogenology 2023, 211, 49–55. [Google Scholar] [CrossRef]
- Catteuw, A.; Broekaert, N.; De Baere, S.; Lauwers, M.; Gasthuys, E.; Huybrechts, B.; Callebaut, A.; Ivanova, L.; Uhlig, S.; De Boevre, M.; et al. Insights into in vivo absolute oral bioavailability, biotransformation, and toxicokinetics of zearalenone, alpha-zearalenol, beta-zearalenol, zearalenone-14-glucoside, and zearalenone-14-sulfate in pigs. J. Agric. Food Chem. 2019, 67, 3448–3458. [Google Scholar] [CrossRef]
- Gajecka, M.; Zielonka, L.; Babuchowski, A.; Gajecki, M.T. Exposure to Low Zearalenone Doses and Changes in the Homeostasis and Concentrations of Endogenous Hormones in Selected Steroid-Sensitive Tissues in Pre-Pubertal Gilts. Toxins 2022, 14, 790. [Google Scholar] [CrossRef] [PubMed]
- Asaduzzaman, M.; Pavlov, I.; St-Jean, G.; Zhu, Y.; Castex, M.; Chorfi, Y.; Del Castillo, J.R.E.; Zhou, T.; Alassane-Kpembi, I. Phosphorylation of zearalenone retains its toxicity. J. Agric. Food Chem. 2024, 72, 26491–26503. [Google Scholar] [CrossRef] [PubMed]
- Binder, S.B.; Schwartz-Zimmermann, H.E.; Varga, E.; Bichl, G.; Michlmayr, H.; Adam, G.; Berthiller, F. Metabolism of zearalenone and its major modified forms in pigs. Toxins 2017, 9, 56. [Google Scholar] [CrossRef]
- Qi, X.; Lin, W.; Ma, M.; Wang, C.; He, Y.; He, N.; Gao, J.; Zhou, H.; Xiao, Y.; Wang, Y.; et al. Structural basis of rifampin inactivation by rifampin phosphotransferase. Proc. Natl. Acad. Sci. USA 2016, 113, 3803–3808. [Google Scholar] [CrossRef]
- Stogios, P.J.; Cox, G.; Spanogiannopoulos, P.; Pillon, M.C.; Waglechner, N.; Skarina, T.; Koteva, K.; Guarné, A.; Savchenko, A.; Wright, G.D. Rifampin phosphotransferase is an unusual antibiotic resistance kinase. Nat. Commun. 2016, 7, 11343. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Oesterling, R.M. Dual divalent cation requirement for activation of pyruvate kinase; essential roles of both enzyme- and nucleotide-bound metal ions. Biochem. J. 1976, 15, 2881–2887. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Shu, M.; Zhong, C.; Zhao, Y.; Bao, S.; Pan, H.; Wang, S.; Wu, G. Characterization of a broad-spectrum endolysin rLysJNwz and its utility against Salmonella in foods. Appl. Microbiol. Biotechnol. 2023, 107, 3229–3241. [Google Scholar] [CrossRef]
- Maguire, O.R.; Smokers, I.B.A.; Oosterom, B.G.; Zheliezniak, A.; Huck, W.T.S. A prebiotic precursor to life’s phosphate transfer system with an ATP analog and histidyl peptide organocatalysts. J. Am. Chem. Soc. 2024, 146, 7839–7849. [Google Scholar] [CrossRef] [PubMed]
- Jing, Z.; Liu, C.; Qi, R.; Ren, P. Many-body effect determines the selectivity for Ca2+ and Mg2+ in proteins. Proc. Natl. Acad. Sci. USA 2018, 115, e7495–e7501. [Google Scholar] [CrossRef] [PubMed]
- Fukui, K.; Yamamoto, T.; Murakawa, T.; Baba, S.; Kumasaka, T.; Yano, T. Catalytic mechanism of the zinc-dependent MutL endonuclease reaction. Life Sci. Alliance 2023, 6, e202302001. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Froger, A.; Hall, J.E. Transformation of plasmid DNA into E. coli using the heat shock method. J. Vis. Exp. 2007, 6, e253. [Google Scholar] [CrossRef]
- Miao, F.; Kompala, D.S. Overexpression of cloned genes using recombinant Escherichia coli regulated by a T7 promoter: I. Batch cultures and kinetic modeling. Biotechnol. Bioeng. 1992, 40, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Kompala, D.S. Overexpression of cloned genes using recombinant Escherichia coli regulated by a T7 promoter: II. Two-stage continuous cultures and model simulations. Biotechnol. Bioeng. 1993, 42, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Spizizen, J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 1958, 44, 1072–1078. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, w320–w324. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, w296–w303. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Zhou, Q.; Hu, J.; Shan, Y.; Liu, J.; Wang, G.; Lee, Y.-W.; Shi, J.; Xu, J. Characterization of the Phosphotransferase from Bacillus subtilis 1101 That Is Responsible for the Biotransformation of Zearalenone. Toxins 2025, 17, 21. https://doi.org/10.3390/toxins17010021
Wu Y, Zhou Q, Hu J, Shan Y, Liu J, Wang G, Lee Y-W, Shi J, Xu J. Characterization of the Phosphotransferase from Bacillus subtilis 1101 That Is Responsible for the Biotransformation of Zearalenone. Toxins. 2025; 17(1):21. https://doi.org/10.3390/toxins17010021
Chicago/Turabian StyleWu, Yuzhuo, Qiuyu Zhou, Junqiang Hu, Yunfan Shan, Jinyue Liu, Gang Wang, Yin-Won Lee, Jianrong Shi, and Jianhong Xu. 2025. "Characterization of the Phosphotransferase from Bacillus subtilis 1101 That Is Responsible for the Biotransformation of Zearalenone" Toxins 17, no. 1: 21. https://doi.org/10.3390/toxins17010021
APA StyleWu, Y., Zhou, Q., Hu, J., Shan, Y., Liu, J., Wang, G., Lee, Y.-W., Shi, J., & Xu, J. (2025). Characterization of the Phosphotransferase from Bacillus subtilis 1101 That Is Responsible for the Biotransformation of Zearalenone. Toxins, 17(1), 21. https://doi.org/10.3390/toxins17010021





