Single-Chain Variable Fragments: Targeting Snake Venom Phospholipase A2 and Serine Protease
Abstract
:1. Introduction
2. Results and Discussion
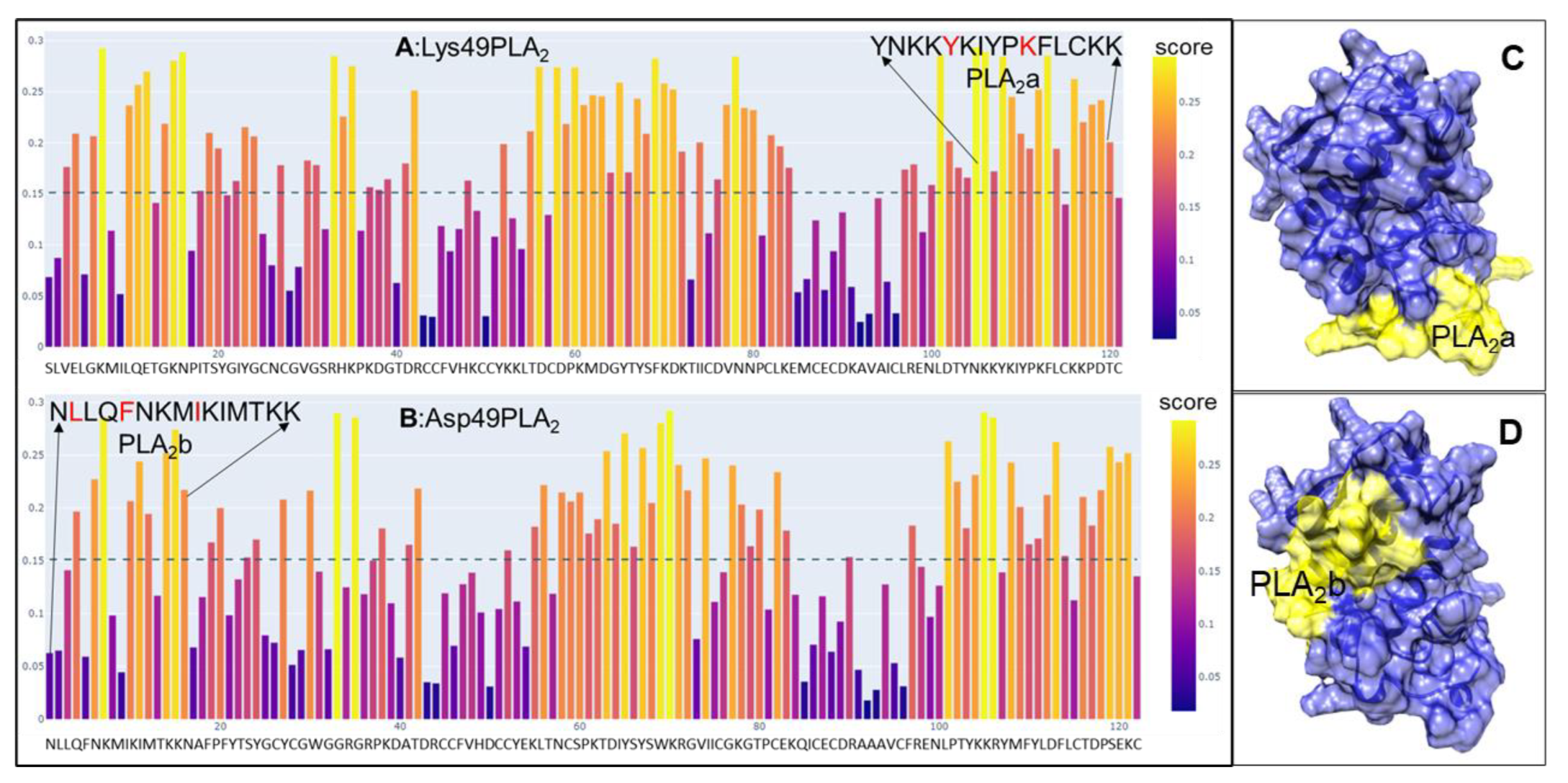
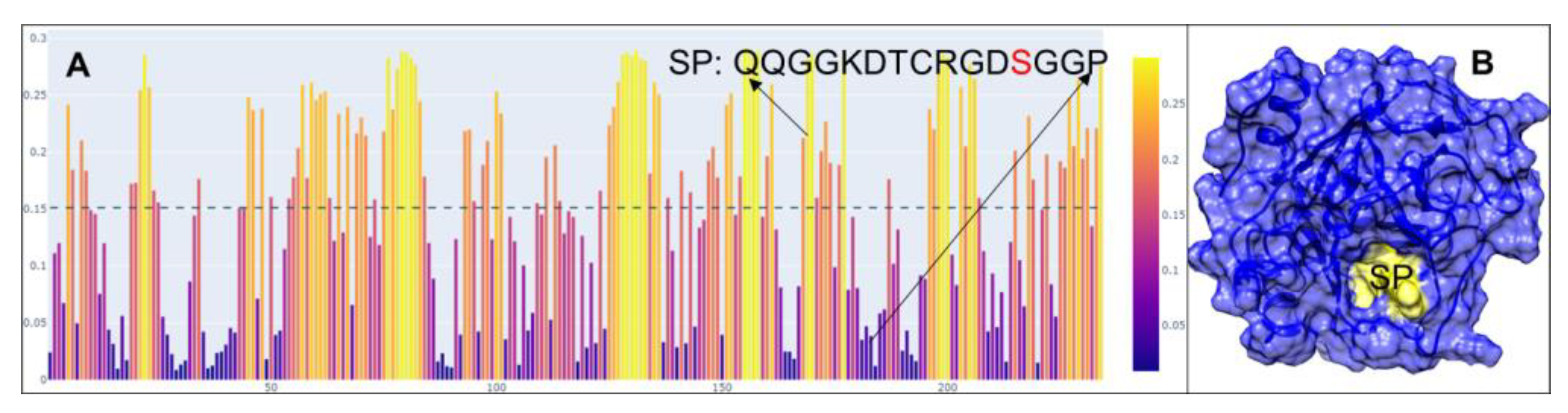
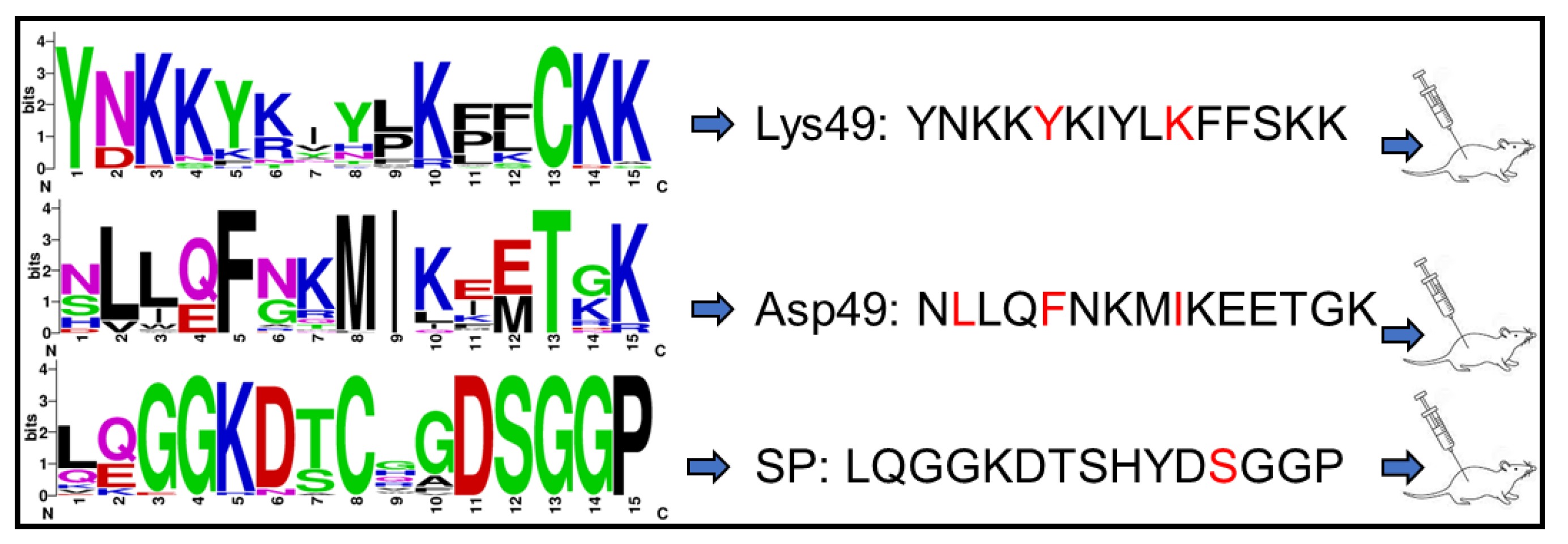
2.1. Peptide Design and Synthesis
2.2. Animal Immunization
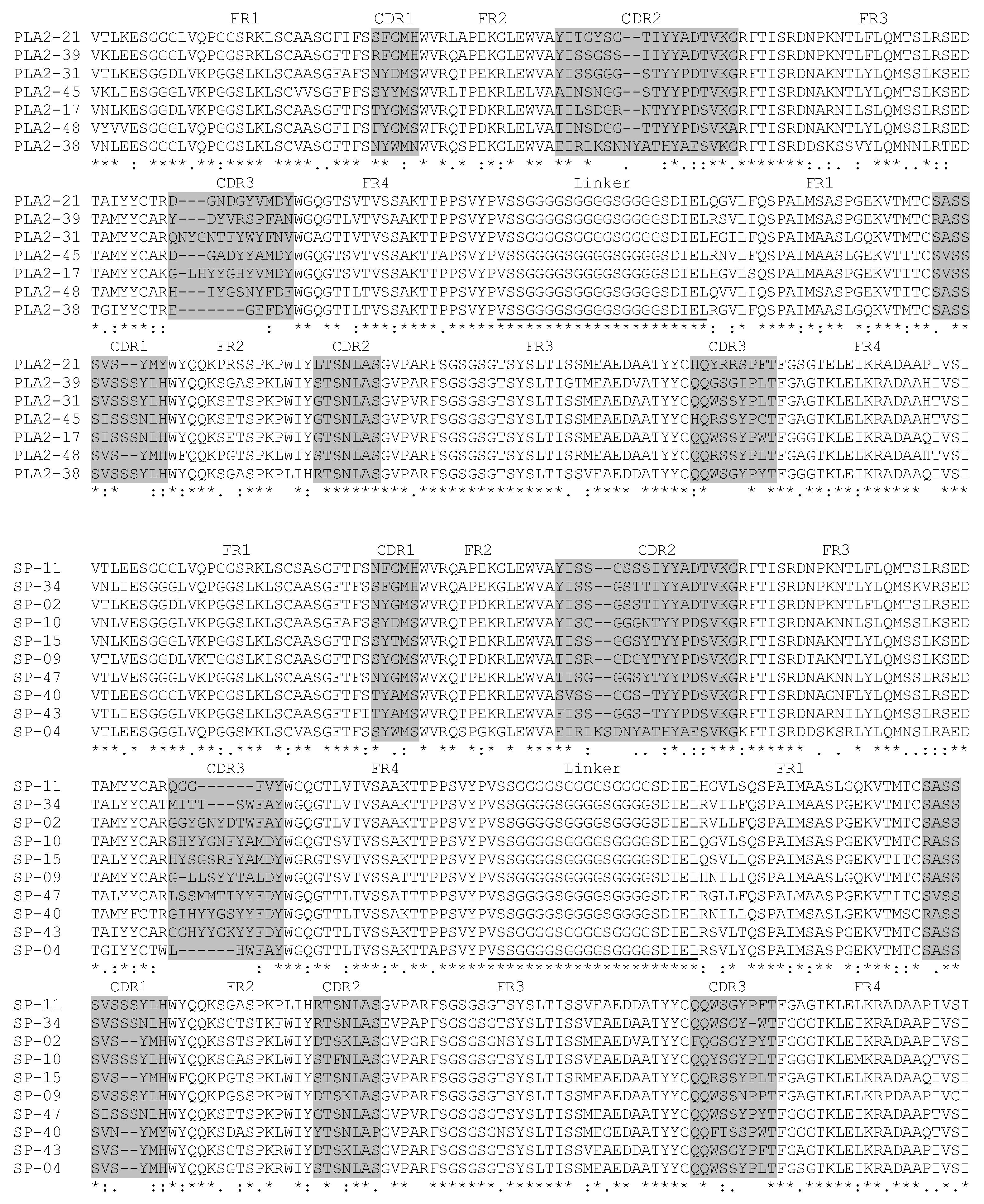
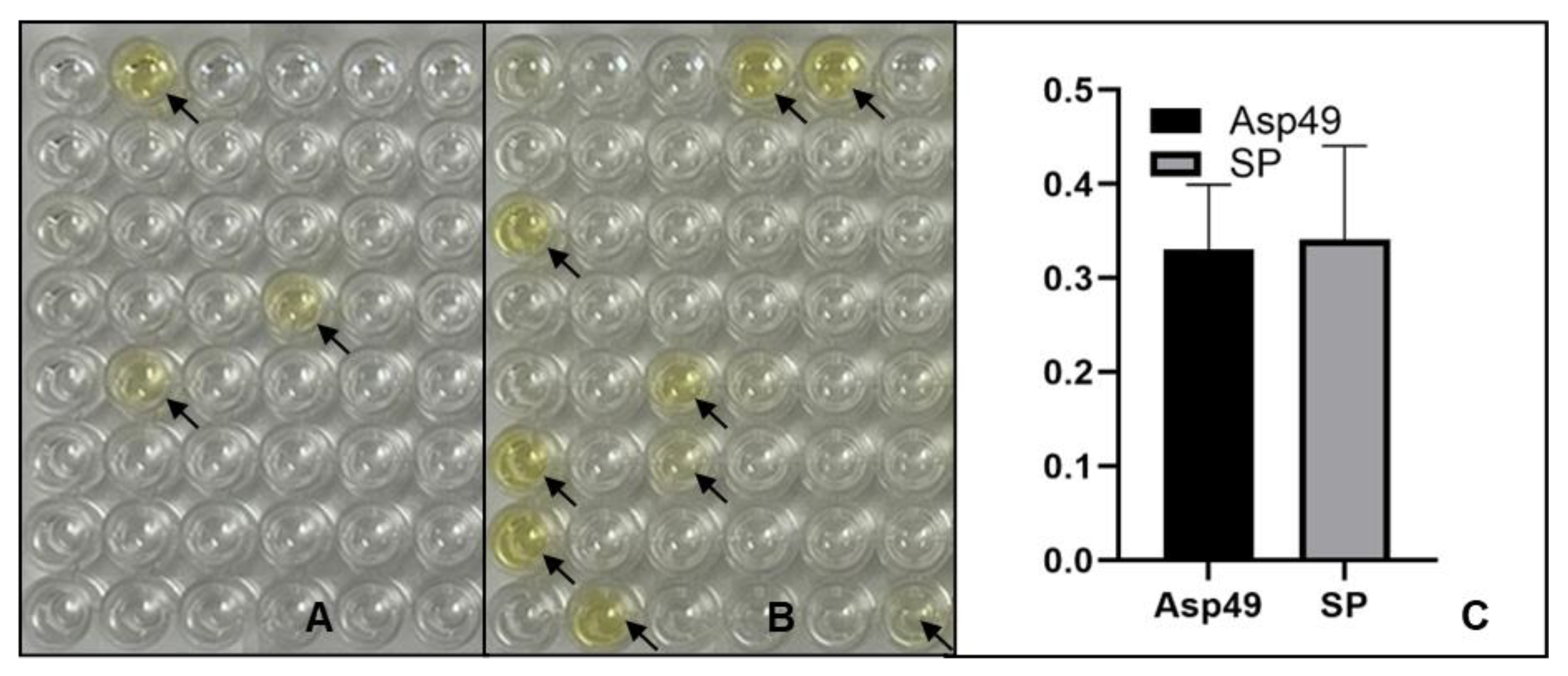
2.3. Characterization of scFv
3. Conclusions
4. Materials and Methods
4.1. Peptide Design
4.2. Animal Immunization
4.3. Enzyme-Linked Immunosorbent Assay
4.4. IgG Complementary DNA Generation
4.5. scFv Generation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chippaux, J.P. Snakebite envenomation turns again into a neglected tropical disease! J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 38. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J. Snakebite envenoming. Nat. Rev. Dis. Primers 2017, 3, 17063. [Google Scholar] [CrossRef]
- Sanz, L.; de Freitas-Lima, L.N.; Quesada-Bernat, S.; Graça-De-Souza, V.K.; Soares, A.M.; Calderón, L.d.A.; Calvete, J.J.; Caldeira, C.A. Comparative Venomics of Brazilian Coral Snakes: Micrurus frontalis, Micrurus spixii spixii, and Micrurus surinamensis. Toxicon 2019, 166, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Lomonte, B.; Saviola, A.J.; Bonilla, F.; Sasa, M.; Williams, D.J.; Undheim, E.A.B.; Sunagar, K.; Jackson, T.N.W. Mutual enlightenment: A toolbox of concepts and methods for integrating evolutionary and clinical toxinology via snake venomics and contextual stance. Toxicon X 2021, 9–10, 100070. [Google Scholar] [CrossRef]
- Sunagar, K.; Khochare, S.; Senji Laxme, R.R.; Attarde, S.; Dam, P.; Suranse, V.; Khaire, A.; Martin, G.; Captain, A. A wolf in another wolf’s clothing: Post-genomic regulation dictates venom profiles of medically-important cryptic kraits in India. Toxins 2021, 13, 69. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Ledsgaard, L.; Laustsen, A.H.; Pus, U.; Wade, J.; Villar, P.; Boddum, K.; Slavny, P.; Masters, E.W.; Arias, A.S.; Oscoz, S.; et al. In vitro discovery of a human monoclonal antibody that neutralizes lethality of cobra snake venom. Mabs 2022, 14, 2085536. [Google Scholar] [CrossRef] [PubMed]
- Ledsgaard, L.; Wade, J.; Jenkins, T.P.; Boddum, K.; Oganesyan, I.; Harrison, J.A.; Villar, P.; Leah, R.A.; Zenobi, R.; Schoffelen, S.; et al. Discovery and optimization of a broadly- neutralizing human monoclonal antibody against long-chain α-neurotoxins from snakes. Nat. Commun. 2023, 14, 682. [Google Scholar] [CrossRef] [PubMed]
- Khalek, I.S.; Laxme, R.R.S.; Nauyen, Y.T.K.; Khochare, S.; Patel, R.N.; Woehl, J.; Smith, J.M.; Saye-Francisco, K.; Kim, Y.; Mindrebo, L.M.; et al. Synthetic development of a broadly neutralizing antibody against snake venom long-chain α-neurotoxins. Sci. Transl. Med. 2024, 16, 735. [Google Scholar] [CrossRef]
- Monnier, P.P.; Vigouroux, R.J.; Tassew, N.G. In vivo applications of single chain Fv (variable domain) (scFv) fragments. Antibodies 2013, 2, 193–208. [Google Scholar] [CrossRef]
- Sandomenico, A.; Sivaccumar, J.P.; Ruvo, M. Evolution of Escherichia coli Expression System in Producing Antibody Recombinant Fragments. Int. J. Mol. Sci. 2020, 21, 6324. [Google Scholar] [CrossRef] [PubMed]
- Yazaki, P.J.; Wu, A.M. Construction and characterization of minibodies for imaging and therapy of colorectal carcinomas. Methods Mol. Biol. 2003, 207, 351–364. [Google Scholar] [PubMed]
- Knowles, S.M.; Wu, A.M. Advances in immuno-positron emission tomography: Antibodies for molecular imaging in oncology. J. Clin. Oncol. 2012, 30, 3884–3892. [Google Scholar] [CrossRef] [PubMed]
- Holliger, P.; Hudson, P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005, 23, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.L.; Reichert, J.M. Development trends for therapeutic antibody fragments. Nat. Biotechnol. 2009, 27, 331–337. [Google Scholar] [CrossRef]
- Yokota, T.; Milenic, D.E.; Whitlow, M.; Schlom, J. Rapid tumor penetration of a single-chain Fv and comparison with other immunoglobulin forms. Cancer Res. 1992, 52, 3402–3408. [Google Scholar] [PubMed]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. scFv antibody: Principles and clinical application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef] [PubMed]
- Bates, A.; Power, C.A. David vs. Goliath: The structure, function, and clinical prospects of antibody fragments. Antibodies 2019, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.A.; Lamar, W.W. The Venomous Reptiles of the Western Hemisphere; Comstock Publishing Associates: Ithaca, NY, USA, 2004. [Google Scholar]
- Calvete, J.J.; Fasoli, E.; Sanz, L.; Boschetti, E.; Righetti, P.G. Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res. 2009, 8, 3055–3067. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Lopez, I.; Kowalski, P. Toxin transcripts in Crotalus atrox venom and in silico structure of toxins. J. Venom Res. 2020, 10, 18–22. [Google Scholar] [PubMed]
- Kini, R.M.; Evans, H.J. Structure-function relationships of phospholipases. J. Biol. Chem. 1987, 262, 14402–14407. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Kudo, I. Phospholipase A2. J. Biochem 2002, 131, 285–292. [Google Scholar] [CrossRef]
- Vega, C.; Jia, Y. Tarantula toxin transcripts and toxin structure prediction by AlphaFold. Chem. Pharm. Res. 2024, 6, 2. [Google Scholar] [CrossRef]
- Serrano, S.M.T. The long road of research on snake venom serine proteinases. Toxicon 2013, 62, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ekici, Ö.D.; Paetzel, M.; Dalbey, R.E. Unconventional serine proteinases: Variations on the catalytic ser/His/Asp triad configuration. Protein Sci. 2008, 17, 2023–2037. [Google Scholar] [CrossRef]
- Neurath, H. Evolution of proteolytic enzymes. Science 1984, 224, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Marsh, N.; Williams, V. Practical application of snake venom toxins in haemostasis. Toxicon 2005, 45, 1171–1181. [Google Scholar] [CrossRef]
- Madrigal, M.; Alape-Girón, A.; Barboza-Arguedas, E.; Aguilar-Ulloa, W.; Flores-Díaz, M. Identification of B cell recognized linear epitopes in a snake venom serine proteinase from the central American bushmaster Lachesis stenophrys. Toxicon 2017, 140, 72–82. [Google Scholar] [CrossRef]
- Clifford, J.N.; Høie, M.H.; Deleuran, S.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-3.0: Improved B-cell epitope prediction using protein language models. Protein Sci. 2022, 31, e4497. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B. Identification of linear B-cell epitopes on myotoxin II, a Lys49 phospholipase A2 homologue from Bothrops asper snake venom. Toxicon 2012, 60, 782–790. [Google Scholar] [CrossRef]
- Chioato, L.; Aragão, E.A.; Ferreira, T.L.; de Medeiros, A.I.; Faccioli, L.H.; Ward, R.J. Mapping of the structural determinants of artificial and biological membrane damaging activities of Lys49 phospholipase A2 by scanning alanine mutagenesis. Biochim. Biophys. Acta 2007, 1768, 1247–1257. [Google Scholar] [CrossRef]
- Fernandes, C.A.H.; Borges, R.J.; Lomonte, B.; Fontes, M.R. A structure-based proposal for a comprehensive myotoxic mechanism of phospholipase A2-like proteins from viperid snake venoms. Biochem. Biophys. Acta 2014, 12, 2265–2276. [Google Scholar] [CrossRef]
- de Castro, K.L.P.; Lopes-de-Souza, L.; de Oliveira, D.; Machado-de-Ávila, R.A.; Paiva, A.L.B.; de Freitas, C.F.; Ho, P.L.; Chávez-Olórtegui, C.; Guerra, C. A combined strategy to improve the development of a coral antivenom against Micrurus spp. Front. Immunol. 2019, 10, 2422. [Google Scholar] [CrossRef] [PubMed]
- Castro, K.L.; Duarte, C.G.; Ramos, H.R.; de Avila, R.A.M.; Schneider, F.S.; Oliveira, D.; Freitas, C.F.; Kalapothakis, E.; Ho, P.L.; Chávez-Olortegui, C. Identification and characterization of B-cell epitopes of 3FTx and PLA2 toxins from Micrurus corallinus snake venom. Toxicon 2015, 93, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.S.; Georgieva, D.; Genov, N.; Murakami, M.T.; Sinha, M.; Kumar, R.P.; Kaur, P.; Kumar, S.; Dey, S.; Sharma, S.; et al. Enzymatic toxins from snake venom: Structural characterization and mechanism of catalysis. FEBS J. 2011, 278, 4544–4576. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Means, N. Molecular farming of pharmaceutical proteins. Transgenic Res. 2000, 9, 279–299. [Google Scholar] [CrossRef] [PubMed]
- Kanter, G.; Yang, J.; Voloshin, A.; Levy, S.; Swartz, J.R.; Levy, R. Cell-free production of scFv fusion proteins: An efficient approach for personalized lymphoma vaccines. Blood 2007, 109, 3393–3399. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.H.; Kong, S.; Young, N.J.; Chuo, S.-W.; Shiah, J.V.; Connelly, E.J.; Rohweder, P.J.; Born, A.; Manglik, A.; Grandis, J.R.; et al. Rare antibody phage isolation and discrimination (RAPID) biopanning enables identification of high-affinity antibodies against challenging targets. Commun. Biol. 2023, 6, 1036. [Google Scholar] [CrossRef] [PubMed]
- Keyser, P.D.; Kalichuk, V.; Zögg, T.; Wohlkönig, A.; Schenck, S.; Brunner, J.; Pardon, E.; Steyaert, J. A biosensor-based phage display selection method for automated, high-throughput Nanobody discovery. Biosens. Bioelectron. 2024, 271, 116951. [Google Scholar] [CrossRef] [PubMed]
- Ledsgaad, L.; Ljungars, A.; Rimbault, C.; Sørensen, C.V.; Tulika, T.; Wade, J.; Wouters, Y.; McCafferty, J.; Laustsen, A.H. Advances in antibody phage display technology. Drug Discov. Today 2022, 27, 2151–2169. [Google Scholar] [CrossRef]
- Palma, M. Epitopes and mimotopes identification using phage display for vaccine development against infectious pathogens. Vaccines 2023, 11, 1176. [Google Scholar] [CrossRef]
- Woloschak, G.E.; Krco, C.J. Regulation of kappa/lambda immunoglobulin light chain expression in normal murine lymphocytes. Mol. Immunol. 1987, 24, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Kabat, E.A.; Wu, T.; Gottesman, K.S.; Perry, H.M.; Foeller, C. Sequences of Proteins of Immunological Interest, 5th ed.; US Department of Health and Human Services: Washington, DC, USA; Diane Publishing Co.: Collingdale, PA, USA, 1991. [Google Scholar]
- Wilson, I.A.; Stanfield, R.L. Antibody-antigen interactions: New structures and new conformational changes. Curr. Opin. Struct. Biol. 1994, 4, 857–867. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Slutzki, M.; Barak, Y.; Reshef, D.; Schueler-Furman, O.; Lamed, R.; Bayer, E.A. Indirect ELISA-based approach for comparative measurement of high-affinity cohesin-dockerin interactions. J. Mol. Recognit. 2012, 25, 616–622. [Google Scholar] [CrossRef]
- Wang, R.; Fang, S.; Wu, D.; Lian, J.; Fan, J.; Zhang, Y.; Wang, S.; Lin, W. Screening for a single-chain variable-fragment antibody that can effectively neutralize the cytotoxicity of the Vibrio parahaemolyticus thermolabile hemolysin. Appl. Environ. Microbiol. 2012, 78, 4967–4975. [Google Scholar] [CrossRef] [PubMed]
- Lang, Q.L.; Yu, L.; He, Q.L.; Ge, L.-P.; Yang, X. Construction and screening of a phage display library of single chain Fv antibody efficiently from mouse immunized with ovalbumin. China Biotechnol. 2018, 38, 25–31. [Google Scholar]


| Primer | Target | Sequences |
|---|---|---|
| VhF | VH | GTKAMKCTRRWRGAGTCTGG |
| VhR | VH | TCCACCTGAGGAGACTGGATAGACHGATGGGGSTGT |
| VkF | VL | TCGGACATCGAGCTCCRMRDTVTKCTSWHHCARWCW |
| VkR | VL | GATRGATACARTKKGTGCAGCATC |
| Linker | gtctcctcaggtggaggcggttcaggcggaggtggctctggcggtggcggatcggacatcgagctc | |
| scFv-EcoRI | scFv | GGTGAATTCGTKAMKCTRRWRGAGTCTGG |
| scFv-XhoI | scFv | GGTCTCGAGTCATTAGATRGATACART |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Y.; Garcia, A.; Reyes, E. Single-Chain Variable Fragments: Targeting Snake Venom Phospholipase A2 and Serine Protease. Toxins 2025, 17, 55. https://doi.org/10.3390/toxins17020055
Jia Y, Garcia A, Reyes E. Single-Chain Variable Fragments: Targeting Snake Venom Phospholipase A2 and Serine Protease. Toxins. 2025; 17(2):55. https://doi.org/10.3390/toxins17020055
Chicago/Turabian StyleJia, Ying, Ariane Garcia, and Elizabeth Reyes. 2025. "Single-Chain Variable Fragments: Targeting Snake Venom Phospholipase A2 and Serine Protease" Toxins 17, no. 2: 55. https://doi.org/10.3390/toxins17020055
APA StyleJia, Y., Garcia, A., & Reyes, E. (2025). Single-Chain Variable Fragments: Targeting Snake Venom Phospholipase A2 and Serine Protease. Toxins, 17(2), 55. https://doi.org/10.3390/toxins17020055





