Transcriptomics as an Early Warning of Domoic Acid Exposure in Pacific Razor Clams (Siliqua patula)
Abstract
:1. Introduction
2. Results
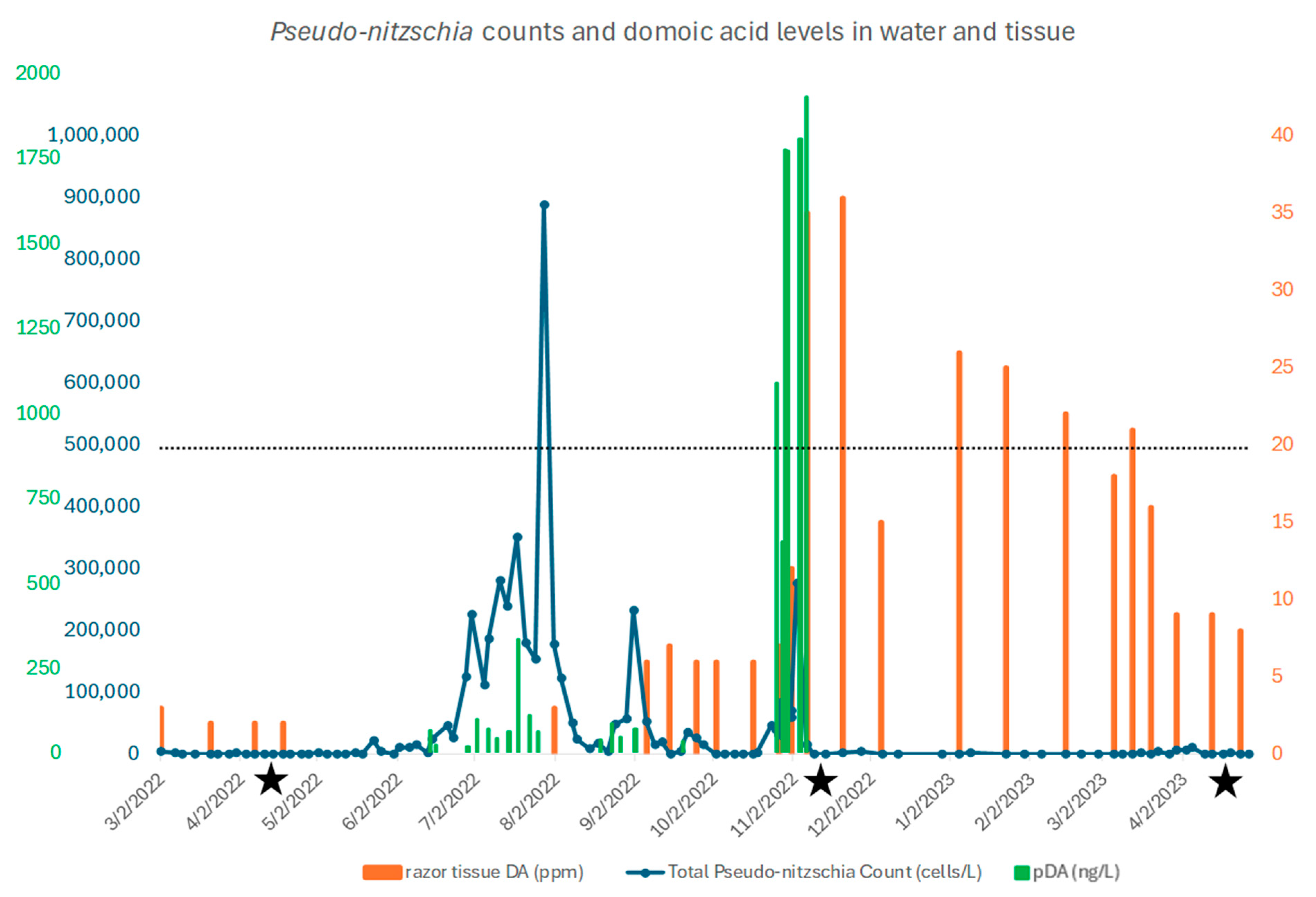
2.1. Pseudo-Nitzschia and Domoic Acid Levels
2.2. Differential Expression and Functional Annotation
2.3. Enriched GO Terms
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Harmful Algal Bloom Monitoring
5.2. Cell Counts and Particulate Domoic Acid
5.3. Domoic Acid Tissue Testing
5.4. Sample Collection
5.5. RNA Extraction
5.6. RNAseq
5.7. Differential Expression Analysis
5.8. Functional Annotation
5.9. GO Enrichment
5.10. Superclusters
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berdalet, E.; Fleming, L.E.; Gowen, R.; Davidson, K.; Hess, P.; Backer, L.C.; Moore, S.K.; Hoagland, P.; Enevoldsen, H. Marine harmful algal blooms, human health and wellbeing: Challenges and opportunities in the 21st century. J. Mar. Biol. Assoc. United Kingd. 2016, 96, 61–91. [Google Scholar] [CrossRef] [PubMed]
- Gobler, C.J.; Doherty, O.M.; Hattenrath-Lehmann, T.K.; Griffith, A.W.; Kang, Y.; Litaker, R.W. Ocean warming since 1982 has expanded the niche of toxic algal blooms in the North Atlantic and North Pacific oceans. Proc. Natl. Acad. Sci. USA 2017, 114, 4975–4980. [Google Scholar] [CrossRef] [PubMed]
- McCabe, R.M.; Hickey, B.M.; Kudela, R.M.; Lefebvre, K.A.; Adams, N.G.; Bill, B.D.; Gulland, F.M.D.; Thomson, R.E.; Cochlan, W.P.; Trainer, V.L. An unprecedented coastwide toxic algal bloom linked to anomalous ocean conditions. Geophys. Res. Lett. 2016, 43, 10366–10376. [Google Scholar] [CrossRef]
- Anderson, D.M.; Fensin, E.; Gobler, C.J.; Hoeglund, A.E.; Hubbard, K.A.; Kulis, D.M.; Landsberg, J.H.; Lefebvre, K.A.; Provoost, P.; Richlen, M.L.; et al. Marine harmful algal blooms (HABs) in the United States: History, current status and future trends. Harmful Algae 2021, 102, 101975. [Google Scholar] [CrossRef]
- Zahir, M.; Su, Y.; Shahzad, M.I.; Ayub, G.; Rehman, S.U.; Ijaz, J. A review on monitoring, forecasting, and early warning of harmful algal bloom. Aquaculture 2024, 593, 741351. [Google Scholar] [CrossRef]
- Wells, M.L.; Karlson, B.; Wulff, A.; Kudela, R.; Trick, C.; Asnaghi, V.; Berdalet, E.; Cochlan, W.; Davidson, K.; De Rijcke, M.; et al. Future HAB science: Directions and challenges in a changing climate. Harmful Algae 2020, 91, 101632. [Google Scholar] [CrossRef]
- Du, M.; Li, Z.; Wang, J.; Wang, F.; Zan, S.; Gu, C. Anaerobic biotransformation mechanism of marine toxin domoic acid. J. Hazard. Mater. 2022, 421, 126798. [Google Scholar] [CrossRef]
- Beyer, J.; Green, N.W.; Brooks, S.; Allan, I.J.; Ruus, A.; Gomes, T.; Bråte, I.L.N.; Schøyen, M. Blue mussels (Mytilus edulis spp.) as sentinel organisms in coastal pollution monitoring: A review. Mar. Environ. Res. 2017, 130, 338–365. [Google Scholar] [CrossRef]
- Bowen, L.; Counihan, K.L.; Ballachey, B.; Coletti, H.; Hollmen, T.; Pister, B.; Wilson, T.L. Monitoring nearshore ecosystem health using Pacific razor clams (Siliqua patula) as an indicator species. PeerJ 2020, 8, e8761. [Google Scholar] [CrossRef]
- Coletti, H.A.; Bowen, L.; Ballachey, B.E.; Wilson, T.L.; Waters, S.; Booz, M.; Counihan, K.L.; Hollmen, T.E.; Pister, B. Gene Expression Profiles in Two Razor Clam Populations: Discerning Drivers of Population Status. Life 2021, 11, 1288. [Google Scholar] [CrossRef]
- Jaramillo, M.; Joens, J.A.; O’Shea, K.E. Fundamental studies of the singlet oxygen reactions with the potent marine toxin domoic acid. Environ. Sci. Technol. 2020, 54, 6073–6081. [Google Scholar] [CrossRef] [PubMed]
- Jennings, E.D.; Parker, M.S.; Simenstad, C.A. Domoic acid depuration by intertidal bivalves fed on toxin-producing Pseudo-nitzschia multiseries. Toxicon X 2020, 6, 100027. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, A.C.; VanDola, F.M.; Gulland, F.M.; Rowles, T.K.; Schwacke, L.H. Production and toxicity of the marine biotoxin domoic acid and its effects on wildlife: A review. Hum. Ecol. Risk Assess. 2008, 14, 544–567. [Google Scholar] [CrossRef]
- Ekstrom, J.A.; Moore, S.K.; Klinger, T. Examining harmful algal blooms through a disaster risk management lens: A case study of the 2015 US West Coast domoic acid event. Harmful Algae 2020, 94, 101740. [Google Scholar] [CrossRef]
- Moore, S.K.; Cline, M.R.; Blair, K.; Klinger, T.; Varney, A.; Norman, K. An index of fisheries closures due to harmful algal blooms and a framework for identifying vulnerable fishing communities on the US West Coast. Mar. Policy 2019, 110, 103543. [Google Scholar] [CrossRef]
- Scholin, C.A.; Gulland, F.; Doucette, G.J.; Benson, S.; Busman, M.; Chavez, F.P.; Cordaro, J.; DeLong, R.; De Vogelaere, A.; Harvey, J.; et al. Mortality of sea lions along the central California coast linked to a toxic diatom bloom. Nature 2000, 403, 80–84. [Google Scholar] [CrossRef]
- Chadsey, M.; Trainer, V.L.; Leschine, T.M. Cooperation of science and management for harmful algal blooms: Domoic acid and the Washington coast razor clam fishery. Coast. Manag. 2012, 40, 33–54. [Google Scholar] [CrossRef]
- Petroff, R.L.; Williams, C.; Li, J.L.; MacDonald, J.W.; Bammler, T.K.; Richards, T.; English, C.N.; Baldessari, A.; Shum, S.; Jing, J.; et al. Prolonged, low-level exposure to the marine toxin, domoic acid, and measures of neurotoxicity in nonhuman primates. Environ. Health Perspect. 2022, 130, 097003. [Google Scholar] [CrossRef]
- Chi, C.; Zhang, C.; Liu, J.; Zheng, X. Effects of marine toxin domoic acid on innate immune responses in bay scallop Argopecten irradians. J. Mar. Sci. Eng. 2019, 7, 407. [Google Scholar] [CrossRef]
- Dizer, H.; Fischer, B.; Harabawy, A.S.A.; Hennion, M.C.; Hansen, P.D. Toxicity of domoic acid in the marine mussel Mytilus edulis. Aquat. Toxicol. 2001, 55, 149–156. [Google Scholar] [CrossRef]
- Ventoso, P.; Pazos, A.J.; Blanco, J.; Pérez-Parallé, M.L.; Triviño, J.C.; Sánchez, J.L. Transcriptional response in the digestive gland of the King Scallop (Pecten maximus) after the injection of domoic acid. Toxins 2021, 13, 339. [Google Scholar] [CrossRef] [PubMed]
- Bowen, L.; Knowles, S.; Lefebvre, K.; St Martin, M.; Murray, M.; Kloecker, K.; Monson, D.; Weitzman, B.; Ballachey, B.; Coletti, H.; et al. Divergent Gene Expression Profiles in Alaskan Sea Otters: An Indicator of Chronic Domoic Acid Exposure? Oceans 2022, 3, 401–418. [Google Scholar] [CrossRef]
- Acevedo-Whitehouse, K.; Duffus, A.L.J. Effects of Environmental Change on Wildlife Health. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 3429–3438. [Google Scholar] [CrossRef]
- Blanchong, J.A.; Robinson, S.J.; Samuel, M.D.; Foster, J.T. Application of genetics and genomics to wildlife epidemiology. J. Wildl. Manag. 2016, 80, 593–608. [Google Scholar] [CrossRef]
- Danovaro, R.; Carugati, L.; Berzano, M.; Cahill, A.E.; Carvalho, S.; Chenuil, A.; Corinaldesi, C.; Cristina, S.; David, R.; Dell’Anno, A.; et al. Implementing and innovating marine monitoring approaches for assessing marine environmental status. Front. Mar. Sci. 2016, 3, 213. [Google Scholar] [CrossRef]
- Bowen, L.; Miles, A.K.; Ballachey, B.; Waters, S.; Bodkin, J. Gene transcript profiling in sea otters post-Exxon Valdez oil spill: A tool for marine ecosystem health assessment. J. Mar. Sci. Eng. 2016, 4, 39. [Google Scholar] [CrossRef]
- Bowen, L.; von Biela, V.R.; McCormick, S.D.; Regish, A.M.; Waters, S.C.; Durbin-Johnson, B.; Britton, M.; Settles, M.L.; Donnelly, D.S.; Laske, S.M.; et al. Transcriptomic response to elevated water temperatures in adult migrating Yukon River Chinook salmon (Oncorhynchus tshawytscha). Conserv. Physiol. 2020, 8, coaa084. [Google Scholar] [CrossRef]
- Song, J.A.; Choi, C.Y.; Park, H.S. Exposure to domoic acid causes oxidative stress in bay scallops Argopecten irradians. Fish. Sci. 2020, 86, 701–709. [Google Scholar] [CrossRef]
- Ventoso, P.; Pazos, A.J.; Pérez-Parallé, M.L.; Blanco, J.; Triviño, J.C.; Sánchez, J.L. RNA-Seq transcriptome profiling of the queen scallop (Aequipecten opercularis) digestive gland after exposure to domoic acid-producing Pseudo-nitzschia. Toxins 2019, 11, 97. [Google Scholar] [CrossRef]
- Mafra, L.L., Jr.; Bricelj, V.M.; Fennel, K. Domoic acid uptake and elimination kinetics in oysters and mussels in relation to body size and anatomical distribution of toxin. Aquat. Toxicol. 2010, 100, 17–29. [Google Scholar] [CrossRef]
- Blanco, J.; Arévalo, F.; Salgado, C.; Moroño, Á. Depuration of mussels (Mytilus galloprovincialis) contaminated with domoic acid. Aquat. Living Resour. 2002, 15, 53–60. [Google Scholar] [CrossRef]
- Mafra, L.L., Jr.; Bricelj, V.M.; Ouellette, C.; Léger, C.; Bates, S.S. Mechanisms contributing to low domoic acid uptake by oysters feeding on Pseudo-nitzschia cells. I. Filtration and pseudofeces production. Aquat. Biol. 2009, 6, 201–212. [Google Scholar] [CrossRef]
- Jones, T.O.; Whyte, J.N.C.; Townsend, L.D.; Ginther, N.G.; Iwama, G.K. Effects of domoic acid on haemolymph pH, PCO2 and PO2 in the Pacific oyster, Crassostrea gigas and the California mussel, Mytilus californianus. Aquat. Toxicol. 1995, 31, 43–55. [Google Scholar] [CrossRef]
- Jones, T.O.; Whyte, J.N.; Ginther, N.G.; Townsend, L.D.; Iwama, G.K. Haemocyte changes in the Pacific oyster, Crassostrea gigas, caused by exposure to domoic acid in the diatom Pseudonitzschia pungens f. multiseries. Toxicon 1995, 33, 347–353. [Google Scholar] [CrossRef]
- Liu, H.; Kelly, M.S.; Campbell, D.A.; Dong, S.L.; Zhu, J.X.; Wang, S.F. Exposure to domoic acid affects larval development of king scallop Pecten maximus (Linnaeus, 1758). Aquat. Toxicol. 2007, 81, 152–158. [Google Scholar] [CrossRef]
- Liu, H.; Kelly, M.S.; Campbell, D.A.; Fang, J.; Zhu, J. Accumulation of domoic acid and its effect on juvenile king scallop Pecten maximus (Linnaeus, 1758). Aquaculture 2008, 284, 224–230. [Google Scholar] [CrossRef]
- Hégaret, H.; Da Silva, P.M.; Wikfors, G.H.; Haberkorn, H.; Shumway, S.E.; Soudant, P. In vitro interactions between several species of harmful algae and haemocytes of bivalve molluscs. Cell Biol. Toxicol. 2011, 27, 249–266. [Google Scholar] [CrossRef]
- Pazos, A.J.; Ventoso, P.; Martínez-Escauriaza, R.; Pérez-Parallé, M.L.; Blanco, J.; Triviño, J.C.; Sánchez, J.L. Transcriptional response after exposure to domoic acid-producing Pseudo-nitzschia in the digestive gland of the mussel Mytilus galloprovincialis. Toxicon 2017, 140, 60–71. [Google Scholar] [CrossRef]
- Zafra, F.; Ibáñez, I.; Bartolomé-Martín, D.; Piniella, D.; Arribas-Blázquez, M.; Giménez, C. Glycine transporters and its coupling with NMDA receptors. Adv. Neurobiol. 2017, 16, 55–83. [Google Scholar]
- Morrow, J.A.; Collie, I.T.; Dunbar, D.R.; Walker, G.B.; Shahid, M.; Hill, D.R. Molecular cloning and functional expression of the human glycine transporter GlyT2 and chromosomal localisation of the gene in the human genome. EBS Lett. 1998, 439, 334–340. [Google Scholar] [CrossRef]
- Sun, X.; Wu, Y.; Wang, Y.; Xue, Q.; Cheng, X.; Zhang, G.; Zhu, Y.; Wang, X. β-1, 4-galactosyltransferase-I activates proliferation and participates in intercellular contacts of lymphocytes. Hum. Immunol. 2014, 75, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Graham, A.L.; Shuker, D.M.; Pollitt, L.C.; Auld, S.K.; Wilson, A.J.; Little, T.J. Fitness consequences of immune responses: Strengthening the empirical framework for ecoimmunology. Funct. Ecol. 2010, 25, 5–17. [Google Scholar] [CrossRef]
- Martin, L.B.; Hopkins, W.A.; Mydlarz, L.D.; Rohr, J.R. The effects of anthropogenic global changes on immune functions and disease resistance. Ann. N.Y. Acad. Sci. 2010, 1195, 129–148. [Google Scholar] [CrossRef]
- Hidayat, A.S.; Lefebvre, K.A.; MacDonald, J.; Bammler, T.; Aluru, N. Symptomatic and asymptomatic domoic acid exposure in zebrafish (Danio rerio) revealed distinct non-overlapping gene expression patterns in the brain. Aquat. Toxicol. 2022, 252, 106310. [Google Scholar] [CrossRef]
- Neely, B.A.; Soper, J.L.; Gulland, F.M.; Bell, P.D.; Kindy, M.; Arthur, J.M.; Janech, M.G. Proteomic analysis of cerebrospinal fluid in California sea lions (Zalophus californianus) with domoic acid toxicosis identifies proteins associated with neurodegeneration. Proteomics 2015, 15, 4051–4063. [Google Scholar] [CrossRef]
- Vranyac-Tramoundanas, A.; Harrison, J.C.; Sawant, P.M.; Kerr, D.S.; Sammut, I.A. Ischemic cardiomyopathy following seizure induction by domoic acid. Am. J. Pathol. 2011, 179, 141–154. [Google Scholar] [CrossRef]
- Pulido, O.M. Domoic Acid Toxicologic Pathology: A Review. Mar. Drugs 2008, 6, 180–219. [Google Scholar] [CrossRef]
- Niu, D.; Wang, L.; Sun, F.; Liu, Z.; Li, J. Development of molecular resources for an intertidal clam, Sinonovacula constricta, using 454 transcriptome sequencing. PLoS ONE 2013, 8, e67456. [Google Scholar] [CrossRef]
- Wang, Z.; Shao, Y.; Li, C.; Zhang, W.; Duan, X.; Zhao, X.; Qiu, Q.; Jin, C. RNA-seq analysis revealed ROS-mediated related genes involved in cadmium detoxification in the razor clam Sinonovacula constricta. Fish Shellfish Immunol. 2016, 57, 350–361. [Google Scholar] [CrossRef]
- Peng, C.; Zhao, X.; Liu, S.; Shi, W.; Han, Y.; Guo, C.; Jiang, J.; Wan, H.; Shen, T.; Liu, G. Effects of anthropogenic sound on digging behavior, metabolism, Ca2+/Mg2+ ATPase activity, and metabolism-related gene expression of the bivalve Sinonovacula constricta. Sci. Rep. 2016, 6, 24266. [Google Scholar] [CrossRef]
- Niu, D.; Xie, S.; Bai, Z.; Wang, L.; Jin, K.; Li, J. Identification, expression, and responses to bacterial challenge of the cathepsin C gene from the razor clam Sinonovacula constricta. Dev. Comp. Immunol. 2014, 46, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Wang, F.; Zhao, H.; Wang, Z.; Xie, S.; Li, J. Identification, expression, and innate immune responses of two insulin-like peptide genes in the razor clam Sinonovacula constricta. Fish Shellfish Immunol. 2016, 51, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, C.L.; Wekell, J.C.; Gauglitz, E.J., Jr.; Bamett, H.J. Salt clean-up procedure for the determination of domoic acid by HPLC. Nat. Toxins 1994, 2, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Trainer, V.L.; Cochlan, W.P.; Erickson, A.; Bill, B.D.; Cox, F.H.; Borchert, J.A.; Lefebvre, K.A. Recent domoic acid closures of shellfish harvest areas in Washington State inland waterways. Harmful Algae 2007, 6, 449–459. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- The R Project for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 20 August 2024).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple hypothesis testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G.; et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g: Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- Thomas, P.D. The gene ontology and the meaning of biological function. Methods Mol. Biol. 2017, 1446, 15–24. [Google Scholar]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]

| (a) Top 20 down-regulated genes in bloom compared with pre-bloom razor clams | ||||
| Transcript_ID | Description | logFC | adj.P.Val | Representative GO ID and Function |
| a_HQ_transcript/79367 | Sodium- and chloride-dependent glycine transporter 2 | −4.83 | 1.85 × 10−07 | GO:0001504 neurotransmitter uptake |
| a_HQ_transcript/131580 | Spectrin alpha chain, non-erythrocytic 1; | −4.73 | 5.88 × 10−07 | GO:0030036 actin cytoskeleton organization |
| a_HQ_transcript/122962 | Uncharacterized | −4.51 | 4.56 × 10−06 | Uncharacterized |
| a_HQ_transcript/79902 | Beta-1,4-galactosyltransferase galt-1 | −4.34 | 4.56 × 10−06 | GO:0070085 glycosylation |
| a_HQ_transcript/75321 | Myosin heavy chain, striated muscle | −3.52 | 4.56 × 10−06 | GO:0005524 ATP binding |
| a_HQ_transcript/141107 | Fatty acid-binding protein homolog 6 | −4.25 | 0.00 | GO:0015908 fatty acid transport |
| a_HQ_transcript/133860 | Tubulin alpha-1A chain | −3.89 | 0.00 | GO:1902065 response to L-glutamate |
| a_HQ_transcript/145194 | Full = Septin-7 | −5.52 | 0.00 | GO:0030154 cell differentiation55 |
| a_HQ_transcript/34261 | Nuclear receptor ROR-alpha | −2.56 | 0.00 | GO:0071356 cellular response to tumor necrosis factor |
| a_HQ_transcript/48432 | Complement C3 | −6.27 | 0.00 | GO:0006954 inflammatory response |
| a_HQ_transcript/123862 | Uncharacterized | −3.88 | 0.00 | Uncharacterized |
| a_HQ_transcript/60848 | Metalloprotease mig-17 | −3.65 | 0.00 | GO:0016540 protein autoprocessing |
| a_HQ_transcript/108352 | Ankyrin repeat and EF-hand domain-containing protein 1 | −3.03 | 0.00 | Uncharacterized |
| a_HQ_transcript/98226 | Nuclear receptor ROR-alpha | −2.44 | 0.00 | GO:0071356 cellular response to tumor necrosis factor |
| a_HQ_transcript/49343 | Polyubiquitin-A | −3.91 | 0.00 | GO:0019941 modification-dependent protein catabolic process |
| a_HQ_transcript/33295 | Collagen alpha-2(I) chain | −4.12 | 0.00 | GO:0007179 transforming growth factor beta receptor signaling pathway |
| a_HQ_transcript/76594 | Glutamine synthetase | −5.21 | 0.00 | GO:0006536 glutamate metabolic process |
| a_HQ_transcript/66242 | Glutamine synthetase | −4.47 | 0.00 | GO:0006536 glutamate metabolic process |
| a_HQ_transcript/102242 | Uncharacterized | −2.17 | 0.00 | Uncharacterized |
| a_HQ_transcript/7159 | Uncharacterized | −1.70 | 0.00 | Uncharacterized |
| (b) Top 20 up-regulated genes in bloom compared with pre-bloom razor clams | ||||
| Transcript_ID | Description | logFC | adj.P.Val | Representative GO ID and Function |
| a_HQ_transcript/118139 | Uncharacterized | 6.80 | 0.00 | Uncharacterized |
| a_HQ_transcript/53710 | Uncharacterized | 3.02 | 0.00 | Uncharacterized |
| a_HQ_transcript/138234 | Adapter molecule | 5.01 | 0.00 | GO:0043087 regulation of GTPase activity |
| a_HQ_transcript/114723 | Uncharacterized | 3.02 | 0.00 | Uncharacterized |
| a_HQ_transcript/135178 | Uncharacterized | 5.39 | 0.00 | Uncharacterized |
| a_HQ_transcript/41636 | Uncharacterized | 3.52 | 0.00 | Uncharacterized |
| a_HQ_transcript/152158 | Uncharacterized | 1.42 | 0.01 | Uncharacterized |
| a_HQ_transcript/70915 | Dynein beta chain, ciliary | 3.24 | 0.01 | GO:0007018 microtubule-based movement |
| a_HQ_transcript/51541 | Opioid growth factor receptor-like protein 1 | 3.85 | 0.01 | GO:0140625 opioid growth factor receptor activity |
| a_HQ_transcript/105284 | Isoamyl acetate-hydrolyzing esterase 1 homolog | 3.92 | 0.01 | GO:0016042 lipid catabolic process |
| a_HQ_transcript/101470 | Lysosomal-associated transmembrane protein 4A | 4.15 | 0.01 | GO:0005765 lysosomal membrane |
| a_HQ_transcript/79577 | Uncharacterized | 4.41 | 0.01 | Uncharacterized |
| a_HQ_transcript/149006 | Uncharacterized | 4.76 | 0.01 | Uncharacterized |
| a_HQ_transcript/130597 | Uncharacterized | 2.93 | 0.01 | Uncharacterized |
| a_HQ_transcript/95361 | Uncharacterized | 3.71 | 0.01 | Uncharacterized |
| a_HQ_transcript/110997 | Uncharacterized | 5.25 | 0.02 | Uncharacterized |
| a_HQ_transcript/62554 | Heat shock 70 kDa protein 12A | 1.76 | 0.02 | GO:0005524 ATP binding |
| a_HQ_transcript/84948 | Coiled-coil domain-containing protein 81 | 4.20 | 0.02 | GO:0005737 cytoplasm |
| a_HQ_transcript/91398 | Transcription elongation regulator 1 | 5.34 | 0.02 | GO:0006397 mRNA processing |
| a_HQ_transcript/135047 | Uncharacterized | 4.42 | 0.02 | Uncharacterized |
| (a) Top 20 down-regulated genes in bloom compared with post-bloom razor clams | ||||
| Transcript_ID | Description | logFC | adj.P.Val | Representative GO ID and Function |
| a_HQ_transcript/72650 | a_HQ_transcript/72650 | −5.47 | 7.62 × 10−07 | GO:0098609 cell-cell adhesion |
| a_HQ_transcript/45368 | Cilia- and flagella- associated protein 210 | −7.24 | 3.35 × 10−06 | GO:0005879 axonemal microtubule |
| a_HQ_transcript/23807 | Steroid 17-alpha-hydroxylase | −4.50 | 3.35 × 10−06 | GO:0042446 hormone biosynthetic process |
| a_HQ_transcript/27296 | Uncharacterized | −7.17 | 8.17 × 10−06 | Uncharacterized |
| a_HQ_transcript/1070 | Uncharacterized | −6.16 | 8.17 × 10−06 | Uncharacterized |
| a_HQ_transcript/55144 | ATP-dependent translocase ABCB1 | −4.07 | 8.17 × 10−06 | GO:1905039 carboxylic acid transmembrane transport |
| a_HQ_transcript/95382 | Uncharacterized | −6.62 | 9.54 × 10−06 | Uncharacterized |
| a_HQ_transcript/66970 | ATP-dependent translocase ABCB1 | −2.94 | 9.54 × 10−06 | GO:1905039 carboxylic acid transmembrane transport |
| a_HQ_transcript/102242 | Uncharacterized | −3.02 | 9.54 × 10−06 | Uncharacterized |
| a_HQ_transcript/34261 | Nuclear receptor ROR-alpha | −4.89 | 9.54 × 10−06 | GO:0071456 cellular response to hypoxia |
| a_HQ_transcript/125280 | Uncharacterized | −4.91 | 2.74 × 10−05 | Uncharacterized |
| a_HQ_transcript/146424 | Uncharacterized | −2.57 | 2.74 × 10−05 | Uncharacterized |
| a_HQ_transcript/58336 | Nose resistant to fluoxetine protein 6 | −5.68 | 2.74 × 10−05 | GO:0006869 lipid transport |
| a_HQ_transcript/129123 | Uncharacterized | −9.53 | 2.74 × 10−05 | Uncharacterized |
| a_HQ_transcript/73314 | Uncharacterized | −3.72 | 5.25 × 10−05 | Uncharacterized |
| a_HQ_transcript/22218 | Steroid 17-alpha-hydroxylase | −4.21 | 5.58 × 10−05 | GO:0042446 hormone biosynthetic process |
| a_HQ_transcript/92691 | Adenylate kinase isoenzyme 1 | −3.66 | 5.62 × 10−05 | GO:0006172 ADP biosynthetic process |
| a_HQ_transcript/79902 | Beta-1,4-galactosyltransferase galt-1 | −7.11 | 6.74 × 10−05 | GO:0070085 glycosylation |
| a_HQ_transcript/35228 | Steroid 17-alpha-hydroxylase | −5.73 | 9.66 × 10−05 | GO:0006694 steroid biosynthetic process |
| a_HQ_transcript/95521 | S-crystallin SL11 | −5.60 | 0.0001407 | GO:0005212 structural constituent of eye lens |
| (b) Top 20 up-regulated genes in bloom compared with post-bloom razor clams | ||||
| Transcript_ID | Description | logFC | adj.P.Val | Representative GO ID and Function |
| a_HQ_transcript/75035 | Uncharacterized | 5.34 | 0.00 | Uncharacterized |
| a_HQ_transcript/135178 | Uncharacterized | 7.34 | 0.00 | Uncharacterized |
| a_HQ_transcript/36072 | Macrophage-expressed gene 1 protein | 3.18 | 0.01 | GO:0002250 adaptive immune response |
| a_HQ_transcript/53566 | ATP-dependent translocase ABCB1 | 4.10 | 0.01 | GO:1905039 carboxylic acid transmembrane transport |
| a_HQ_transcript/101470 | Lysosomal-associated transmembrane protein 4A | 4.16 | 0.01 | GO:0005765 lysosomal membrane |
| a_HQ_transcript/52617 | Dynein regulatory complex protein 10 | 2.03 | 0.01 | GO:0031514 motile cilium |
| a_HQ_transcript/146734 | Uncharacterized | 4.00 | 0.01 | Uncharacterized |
| a_HQ_transcript/130548 | Uncharacterized | 4.54 | 0.01 | Uncharacterized |
| a_HQ_transcript/47331 | Uncharacterized | 3.63 | 0.01 | Uncharacterized |
| a_HQ_transcript/131746 | Uncharacterized | 5.99 | 0.01 | Uncharacterized |
| a_HQ_transcript/15084 | Sodium-dependent phosphate transport protein 2B | 3.73 | 0.01 | GO:0006412 translation |
| a_HQ_transcript/17220 | NADPH--cytochrome P450 reductase | 4.47 | 0.01 | GO:0006805 xenobiotic metabolic process |
| a_HQ_transcript/49759 | Phospholipase A and acyltransferase 3 | 1.68 | 0.02 | GO:0046485 ether lipid metabolic process |
| a_HQ_transcript/8232 | Uncharacterized | 2.92 | 0.02 | Uncharacterized |
| a_HQ_transcript/57283 | Fibropellin-3 | 2.44 | 0.02 | GO:0005509 calcium ion binding |
| a_HQ_transcript/76697 | Skeletal aspartic acid-rich protein 1 | 5.11 | 0.02 | GO:0005576 extracellular region |
| a_HQ_transcript/22375 | Steroid 17-alpha-hydroxylase | 1.53 | 0.02 | GO:0042446 hormone biosynthetic process |
| a_HQ_transcript/52827 | Tektin-4 | 4.93 | 0.02 | GO:0030030 cell projection organization |
| a_HQ_transcript/107872 | Fascin | 3.86 | 0.02 | GO:0030036 actin cytoskeleton organization |
| a_HQ_transcript/109347 | Malate dehydrogenase | 3.24 | 0.03 | GO:0006108 malate metabolic process |
| (a) Top 20 down-regulated genes in pre-bloom compared with post-bloom razor clams | ||||
| Transcript_ID | Description | logFC | adj.P.Val | Representative GO ID and Function |
| a_HQ_transcript/72650 | protocadherin | −7.28 | 2.02 × 10−15 | GO:0098609 cell-cell adhesion |
| a_HQ_transcript/45368 | Cilia and flagella associate protein | −5.47 | 7.62 × 10−07 | GO:0005879 axonemal microtubule |
| a_HQ_transcript/23807 | Steroid 17-alpha-hydroxylase | −7.24 | 3.35 × 10−06 | GO:0042446 hormone biosynthetic process |
| a_HQ_transcript/27296 | Uncharacterized | −4.50 | 3.35 × 10−06 | Uncharacterized |
| a_HQ_transcript/1070 | Uncharacterized | −7.17 | 8.17 × 10−06 | Uncharacterized |
| a_HQ_transcript/55144 | ATP-dependent translocase ABCB1 | −6.16 | 8.17 × 10−06 | GO:1905039 carboxylic acid transmembrane transport |
| a_HQ_transcript/95382 | Uncharacterized | −4.07 | 8.17 × 10−06 | Uncharacterized |
| a_HQ_transcript/66970 | ATP-dependent translocase ABCB1 | −6.62 | 9.54 × 10−06 | GO:1905039 carboxylic acid transmembrane transport |
| a_HQ_transcript/125280 | Uncharacterized | −4.89 | 9.54 × 10−06 | Uncharacterized |
| a_HQ_transcript/34261 | Nuclear receptor ROR-alpha | −3.02 | 9.54 × 10−06 | GO:0071456 cellular response to hypoxia |
| a_HQ_transcript/102242 | Uncharacterized | −2.94 | 9.54 × 10−06 | Uncharacterized |
| a_HQ_transcript/73314 | Uncharacterized | −9.53 | 2.74 × 10−05 | Uncharacterized |
| a_HQ_transcript/129123 | Uncharacterized | −5.68 | 2.74 × 10−05 | Uncharacterized |
| a_HQ_transcript/146424 | Uncharacterized | −4.91 | 2.74 × 10−05 | Uncharacterized |
| a_HQ_transcript/58336 | Nose resistant to fluoxetine protein 6 | −2.57 | 2.74 × 10−05 | GO:0006869 lipid transport |
| a_HQ_transcript/22218 | Steroid 17-alpha-hydroxylase | −3.72 | 5.25 × 10−05 | GO:0042446 hormone biosynthetic process |
| a_HQ_transcript/92691 | Adenylate kinase isoenzyme 1 | −4.21 | 5.58 × 10−05 | GO:0046034 ATP metabolic process |
| a_HQ_transcript/79902 | Beta-1,4-galactosyltransferase galt-1 | −3.66 | 5.62 × 10−05 | GO:0070085 glycosylation |
| a_HQ_transcript/35228 | Steroid 17-alpha-hydroxylase | −7.11 | 6.74 × 10−05 | GO:0006694 steroid biosynthetic process |
| a_HQ_transcript/95521 | S-crystallin SL11 | −5.73 | 9.66 × 10−05 | GO:0005212 structural constituent of eye lens |
| (b) Top 20 up-regulated genes in pre-bloom compared with post-bloom razor clams | ||||
| Transcript_ID | Description | logFC | adj.P.Val | Representative GO ID and Function |
| a_HQ_transcript/135178 | Uncharacterized | 5.34 | 0.00 | Uncharacterized |
| a_HQ_transcript/107478 | Tubulin alpha-1A chain | 7.34 | 0.00 | GO:0071277 cellular response to calcium ion |
| a_HQ_transcript/70915 | Dynein beta chain | 3.18 | 0.01 | GO:0030030 cell projection organization |
| a_HQ_transcript/101470 | Lysosomal-associated transmembrane protein 4A | 4.10 | 0.01 | GO:0005765 lysosomal membrane |
| a_HQ_transcript/136081 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 | 4.16 | 0.01 | GO:0032981 mitochondrial respiratory chain complex I assembly |
| a_HQ_transcript/90430 | Uncharacterized | 2.03 | 0.01 | Uncharacterized |
| a_HQ_transcript/62043 | U2 small nuclear ribonucleoprotein A | 4.00 | 0.01 | GO:0000398 mRNA splicing, via spliceosome |
| a_HQ_transcript/140232 | Peptidyl-prolyl cis-trans isomerase | 4.54 | 0.01 | GO:0006457 protein folding |
| a_HQ_transcript/131746 | Uncharacterized | 3.63 | 0.01 | Uncharacterized |
| a_HQ_transcript/119340 | Syntaxin-12 | 5.99 | 0.01 | GO:0033344 cholesterol efflux |
| a_HQ_transcript/114019 | Uncharacterized | 3.73 | 0.01 | Uncharacterized |
| a_HQ_transcript/80242 | Soluble calcium-activated nucleotidase 1 | 4.47 | 0.01 | GO:0030166 proteoglycan biosynthetic process |
| a_HQ_transcript/60560 | Uncharacterized | 1.68 | 0.02 | Uncharacterized |
| a_HQ_transcript/57283 | Fibropellin-3 | 2.92 | 0.02 | GO:0005509 calcium ion binding |
| a_HQ_transcript/112742 | Poly [ADP-ribose] polymerase 2 | 2.44 | 0.02 | GO:0006284 base-excision repair |
| a_HQ_transcript/115926 | Uncharacterized | 5.11 | 0.02 | Uncharacterized |
| a_HQ_transcript/135559 | Uncharacterized | 1.53 | 0.02 | Uncharacterized |
| a_HQ_transcript/107872 | Fascin | 4.93 | 0.02 | GO:0030036 actin cytoskeleton organization |
| a_HQ_transcript/73908 | Uncharacterized | 3.86 | 0.02 | Uncharacterized |
| a_HQ_transcript/101113 | N-alpha-acetyltransferase 40 | 3.24 | 0.03 | GO:0006629 lipid metabolic process |
| Supercluster | Bloom vs. Pre-Bloom | Bloom vs. Post-Bloom | Pre-Bloom vs. Post-Bloom |
|---|---|---|---|
| Basement membrane assembly | X | ||
| Binding of sperm to zona pellucida | X | ||
| Cell adhesion | X | ||
| Cellular organofluorine metabolic process | X | ||
| Cerebellar cortex morphogenesis | X | X | |
| Cilium assembly | X | ||
| Cilium movement involved in cell motility | X | X | |
| Cilium-dependent cell motility | X | ||
| Collagen fibril organization | X | ||
| Complement activation, classical pathway | X | ||
| Defense response to Gram-positive bacterium | X | X | |
| Epithelial cilium movement involved in extracellular fluid movement | X | ||
| Gonad morphogenesis | X | ||
| Homophilic cell adhesion via plasma membrane adhesion molecules | X | X | X |
| Immune effector process | X | ||
| Locomotion | X | ||
| Maintenance of location in cell | X | ||
| Mitochondrial transmembrane transport | X | ||
| Negative regulation of necroptotic process | X | ||
| Nucleoside triphosphate biosynthetic process | X | X | |
| Positive regulation of anion channel activity | X | ||
| Positive regulation of epithelial cell differentiation | X | ||
| Protein galactosylation | X | X | |
| Retinoic acid catabolic process | X | ||
| Viral genome replication | X |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bowen, L.; Waters, S.; Ballachey, B.; Coletti, H.; Forster, Z.; Li, J.; Jenner, B. Transcriptomics as an Early Warning of Domoic Acid Exposure in Pacific Razor Clams (Siliqua patula). Toxins 2025, 17, 194. https://doi.org/10.3390/toxins17040194
Bowen L, Waters S, Ballachey B, Coletti H, Forster Z, Li J, Jenner B. Transcriptomics as an Early Warning of Domoic Acid Exposure in Pacific Razor Clams (Siliqua patula). Toxins. 2025; 17(4):194. https://doi.org/10.3390/toxins17040194
Chicago/Turabian StyleBowen, Lizabeth, Shannon Waters, Brenda Ballachey, Heather Coletti, Zachary Forster, Jie Li, and Bradley Jenner. 2025. "Transcriptomics as an Early Warning of Domoic Acid Exposure in Pacific Razor Clams (Siliqua patula)" Toxins 17, no. 4: 194. https://doi.org/10.3390/toxins17040194
APA StyleBowen, L., Waters, S., Ballachey, B., Coletti, H., Forster, Z., Li, J., & Jenner, B. (2025). Transcriptomics as an Early Warning of Domoic Acid Exposure in Pacific Razor Clams (Siliqua patula). Toxins, 17(4), 194. https://doi.org/10.3390/toxins17040194






