Decoding Mycoplasma Nucleases: Biological Functions and Pathogenesis
Abstract
1. Introduction
2. Characteristic Domain of Mycoplasma Nuclease
2.1. TNASE_3 Domain
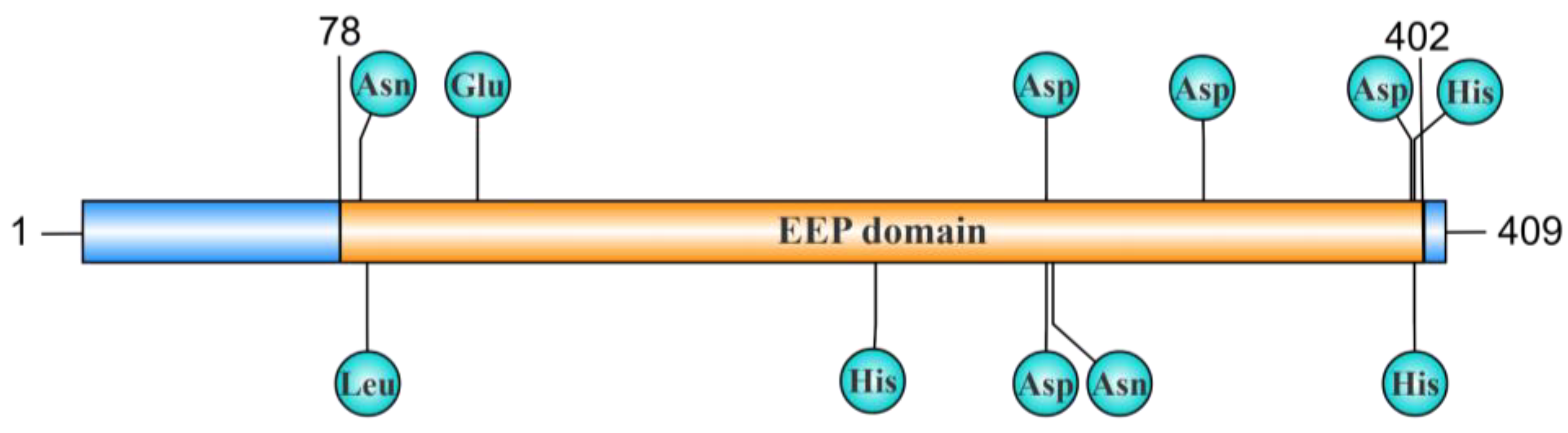
2.2. EEP Domain
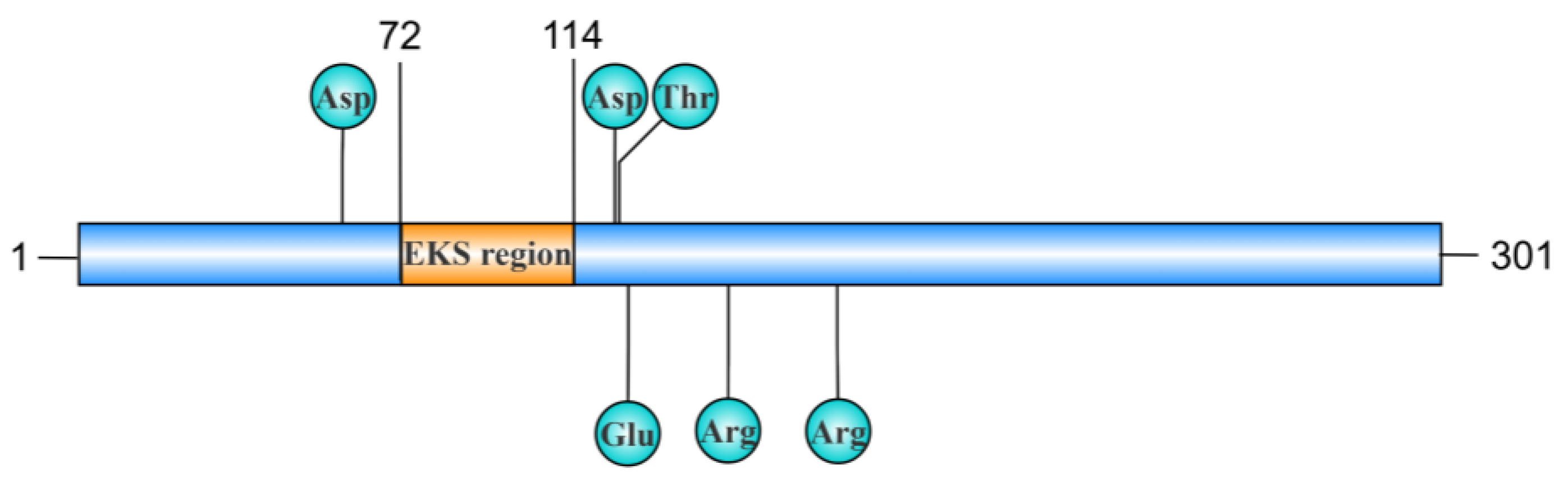
2.3. EKS Region
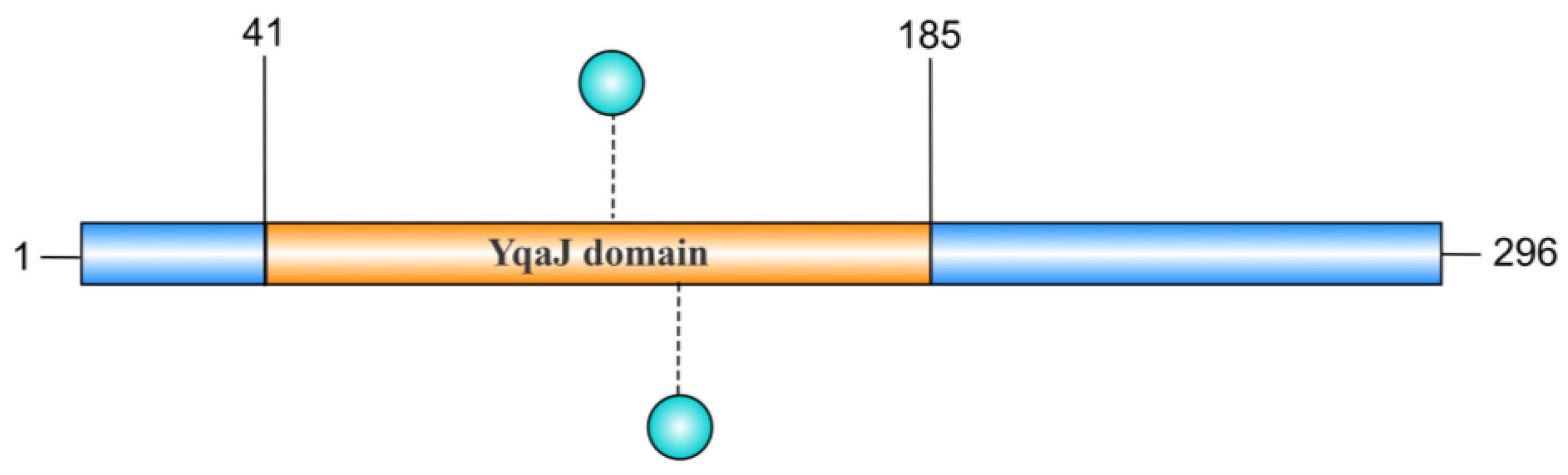
2.4. YqaJ Domain
3. Biochemical Characterization of Nuclease Activity
3.1. Metal Ions
3.2. Temperature
| Mycoplasma | Nuclease | Amino Acids | Cellular Location | Enzyme Activity | Required Divalent Cation | Substrate | Domain | Optimal Reaction Conditions | UniprotAccession Number | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| M. agalactiae | MAG_5040 | 390 | Membrane-associated | Endonuclease/exonuclease | Mg2+ | ssDNA, dsDNA, RNA, plasmid | TNASE_3 | Temperature: 37–45 °C, pH: 6–9 | A5IYU3 | [35] |
| M. bovis | MBOVPG45_0215 | 409 | Membrane-associated | Endonuclease/exonuclease | Ca2+, Mg2+ | dsDNA, plasmid | EEP | Temperature: 37–45 °C, pH: 6–9 | A0A454APR1 | [19] |
| M. bovis | MbovNase | 389 | Membrane-associated and secretory | Endonuclease | Ca2+ | dsDNA, RNA, plasmid | TNASE_3 | Temperature: 22–65 °C | Undefined | [20] |
| M. bovis | MbovP701 | 296 | Undefined | Exonuclease | Mg2+, Mn2+ | dsDNA, ssDNA, RNA, plasmid | YqaJ | Temperature: 43 °C, pH: 8.3 | Undefined | [25] |
| M. hominis | MHO_0730 | 321 | Membrane-associated | Endonuclease/exonuclease | Ca2+ | ssDNA, dsDNA, RNA, plasmid | TNASE_3 | Undefined | D1J7L0 | [32] |
| M. pneumoniae | Mpn133 | 301 | Membrane-associated | Endonuclease | Ca2+ | ssDNA, dsDNA, RNA, plasmid | TNASE_3EKS | Temperature: 37–49 °C, pH: 8.5 | P75265 | [18] |
| M. pneumoniae | Mpn491 | 474 | Secretory | DNAse | Mg2+ | DNA | EEP | Undefined | P75295 | [24] |
| M. genitalium | MG_186 | 250 | Membrane-associated | Endonuclease/exonuclease | Ca2+ | ssDNA, dsDNA, RNA, plasmid | TNASE_3 | Temperature: 37–55 °C, pH: 8.3 | P47432 | [17] |
| M. hyopneumoniae | Mhp379 | 310 | Membrane-associated | Endonuclease/exonuclease | Ca2+ | ssDNA, dsDNA, RNA, plasmid | TNASE_3 | Temperature: 37–45 °C, pH: 8.8 | Q600S5 | [13] |
| M. hyopneumoniae | Mhp597 | 377 | Secretory | Endonuclease/exonuclease | Ca2+, Mg2+ | ssDNA, dsDNA, RNA, plasmid | Undefined | Undefined | Q5ZZW0 | [36] |
| M. gallisepticum | MGA_0676 | 276 | Membrane-associated | Endonuclease/exonuclease | Ca2+ | ssDNA, dsDNA, RNA, plasmid | TNASE_3 | Temperature: 37–49 °C, pH: 8.3 | Q7NC48 | [14] |
| M. meleagridis | Mm19 | 646 | Membrane-associated | Endonuclease/exonuclease | Mg2+ | ssDNA, dsDNA, RNA, plasmid | Undefined | Undefined | A0A0U1YYX1 | [37] |
| M. pulmonis | MnuA | 470 | Membrane-associated | Undefined | Undefined | DNA | Undefined | Undefined | Q50321 | [38] |
| M. penetrans | P40 | Undefined | Membrane-associated | Endonuclease/exonuclease | Ca2+, Mg2+ | ssDNA, dsDNA, RNA, plasmid | Undefined | Temperature: 37 °C, pH: 7–8 | Undefined | [34] |
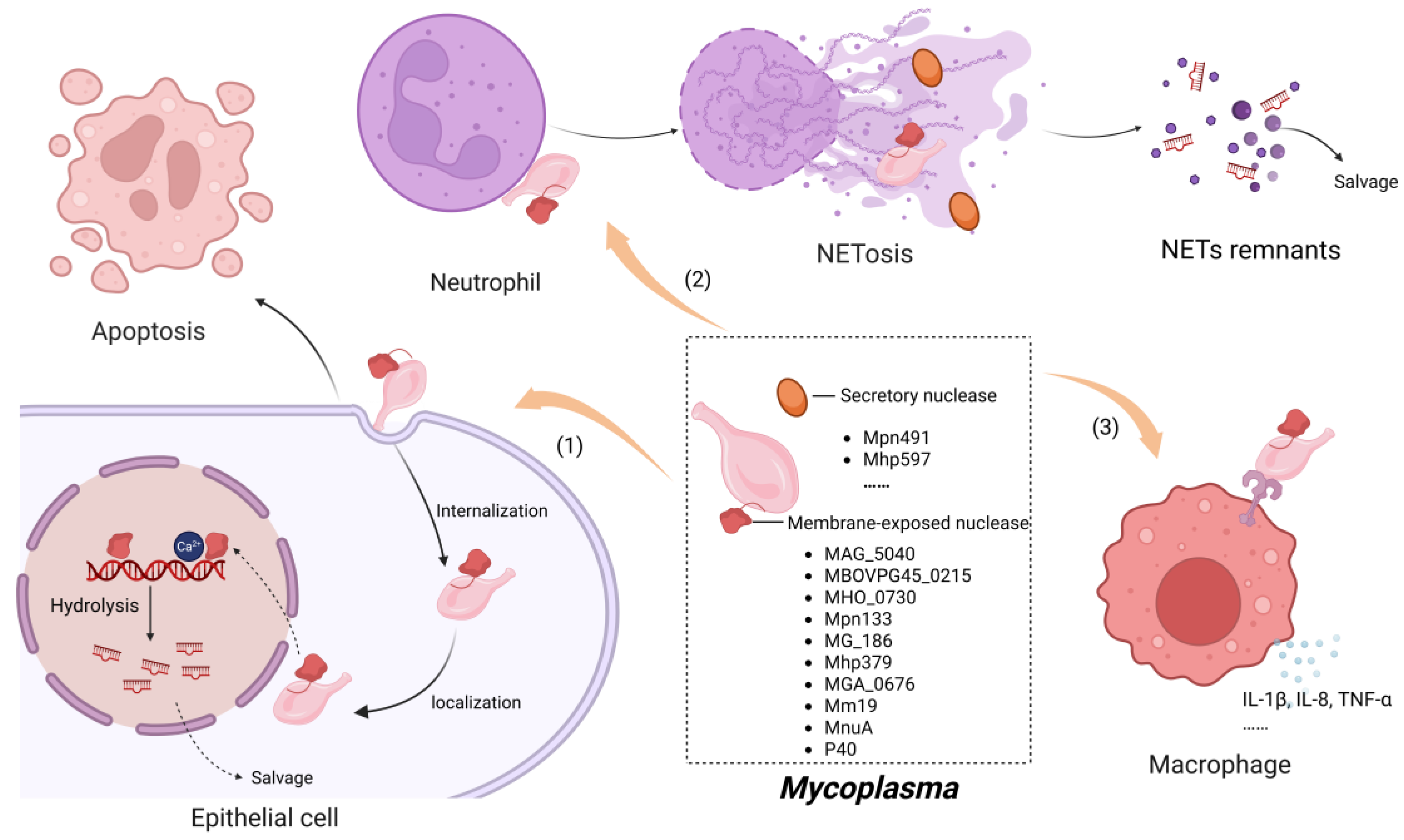
4. Mechanisms Underlying Nuclease Activity in Mycoplasma Pathogenesis
4.1. Induction of Host Cell Apoptosis
4.2. Modulation of Inflammation-Related Molecule Expression by Nucleases
4.3. The Evasion of Mycoplasmas from Neutrophil Extracellular Traps Facilitated by Nucleases
4.3.1. Formation of NETs
4.3.2. Degradation of NETs by Mycoplasma Nuclease
5. Prospects and Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baseman, J.B.; Tully, J.G. Mycoplasmas: Sophisticated, reemerging, and burdened by their notoriety. Emerg. Infect. Dis. 1997, 3, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.M.; Gocayne, J.D.; White, O.; Adams, M.D.; Clayton, R.A.; Fleischmann, R.D.; Bult, C.J.; Kerlavage, A.R.; Sutton, G.; Kelley, J.M.; et al. The minimal gene complement of Mycoplasma genitalium. Science 1995, 270, 397–403. [Google Scholar] [CrossRef]
- Garcia Gonzalez, J.; Hernandez, F.J. Nuclease activity: An exploitable biomarker in bacterial infections. Expert. Rev. Mol. Diagn. 2022, 22, 265–294. [Google Scholar] [CrossRef]
- Razin, S.; Knight, B.C.J.G. The Effects of Ribonucleic Acid and Deoxyribonucleic Acid on the Growth of Mycoplasma. J. Gen. Microbiol. 1960, 22, 504–519. [Google Scholar] [CrossRef]
- Araki, T. Über enzymatische Zersetzung der Nucleinsäure. Biol. Chem. 1903, 38, 84–97. [Google Scholar] [CrossRef]
- Iwanoff, L. Über die fermentative Zersetzung der Thymonucleinsäure durch Schimmelpilze. Biol. Chem. 1903, 39, 31–43. [Google Scholar] [CrossRef]
- Razin, S.; Knyszynski, A.; Lifshitz, Y. Nucleases of Mycoplasma. J. Gen. Microbiol. 1964, 36, 323–332. [Google Scholar] [CrossRef]
- Minion, F.C.; Jarvill-Taylor, K.J.; Billings, D.E.; Tigges, E. Membrane-associated nuclease activities in mycoplasmas. J. Bacteriol. 1993, 175, 7842–7847. [Google Scholar] [CrossRef]
- Hosie, A.H.; Allaway, D.; Jones, M.A.; Walshaw, D.L.; Johnston, A.W.; Poole, P.S. Solute-binding protein-dependent ABC transporters are responsible for solute efflux in addition to solute uptake. Mol. Microbiol. 2001, 40, 1449–1459. [Google Scholar] [CrossRef]
- Masukagami, Y.; Tivendale, K.A.; Mardani, K.; Ben-Barak, I.; Markham, P.F.; Browning, G.F. The Mycoplasma gallisepticum virulence factor lipoprotein MslA is a novel polynucleotide binding protein. Infect. Immun. 2013, 81, 3220–3226. [Google Scholar] [CrossRef]
- Balish, M.F.; Henrich, B.; Kretzmer, F.; Deenen, R.; Köhrer, K. Validation of a novel Mho microarray for a comprehensive characterisation of the Mycoplasma hominis action in HeLa cell infection. PLoS ONE 2017, 12, e0181383. [Google Scholar] [CrossRef]
- Diedershagen, M.; Overbeck, S.; Arlt, S.; Plumakers, B.; Lintges, M.; Rink, L. Mycoplasma arthritidis-derived superantigen (MAM) displays DNase activity. FEMS Immunol. Med. Microbiol. 2007, 49, 266–271. [Google Scholar] [CrossRef]
- Schmidt, J.A.; Browning, G.F.; Markham, P.F. Mycoplasma hyopneumoniae mhp379 is a Ca2+-dependent, sugar-nonspecific exonuclease exposed on the cell surface. J. Bacteriol. 2007, 189, 3414–3424. [Google Scholar] [CrossRef]
- Xu, J.; Teng, D.; Jiang, F.; Zhang, Y.; El-Ashram, S.A.; Wang, H.; Sun, Z.; He, J.; Shen, J.; Wu, W.; et al. Mycoplasma gallisepticum MGA_0676 is a membrane-associated cytotoxic nuclease with a staphylococcal nuclease region essential for nuclear translocation and apoptosis induction in chicken cells. Appl. Microbiol. Biotechnol. 2015, 99, 1859–1871. [Google Scholar] [CrossRef]
- Rechnitzer, H.; Brzuszkiewicz, E.; Strittmatter, A.; Liesegang, H.; Lysnyansky, I.; Daniel, R.; Gottschalk, G.; Rottem, S. Genomic features and insights into the biology of Mycoplasma fermentans. Microbiology 2011, 157, 760–773. [Google Scholar] [CrossRef]
- Tang, J.; Zhou, R.; Shi, X.; Kang, M.; Wang, H.; Chen, H. Two thermostable nucleases coexisted in Staphylococcus aureus: Evidence from mutagenesis and in vitro expression. FEMS Microbiol. Lett. 2008, 284, 176–183. [Google Scholar] [CrossRef]
- Li, L.; Krishnan, M.; Baseman, J.B.; Kannan, T.R. Molecular cloning, expression, and characterization of a Ca2+-dependent, membrane-associated nuclease of Mycoplasma genitalium. J. Bacteriol. 2010, 192, 4876–4884. [Google Scholar] [CrossRef]
- Somarajan, S.R.; Kannan, T.R.; Baseman, J.B. Mycoplasma pneumoniae Mpn133 is a cytotoxic nuclease with a glutamic acid-, lysine- and serine-rich region essential for binding and internalization but not enzymatic activity. Cell Microbiol. 2010, 12, 1821–1831. [Google Scholar] [CrossRef]
- Sharma, S.; Tivendale, K.A.; Markham, P.F.; Browning, G.F. Disruption of the membrane nuclease gene (MBOVPG45_0215) of Mycoplasma bovis greatly reduces cellular nuclease activity. J. Bacteriol. 2015, 197, 1549–1558. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, G.; Guo, Y.; Menghwar, H.; Chen, Y.; Chen, H.; Guo, A. Mycoplasma bovis MBOV_RS02825 Encodes a Secretory Nuclease Associated with Cytotoxicity. Int. J. Mol. Sci. 2016, 17, 628. [Google Scholar] [CrossRef]
- Wang, H.; Morita, M.; Yang, X.; Suzuki, T.; Yang, W.; Wang, J.; Ito, K.; Wang, Q.; Zhao, C.; Bartlam, M.; et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 2010, 29, 2566–2576. [Google Scholar] [CrossRef] [PubMed]
- Mian, I.S.; Worthey, E.A.; Salavati, R. Taking U out, with two nucleases? BMC Bioinform. 2006, 7, 305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, Y.; McMahon, A.; Driscoll, G.; Bullock, S.; Zhao, J.; Yan, S. Function and molecular mechanisms of APE2 in genome and epigenome integrity. Mutat. Res. Rev. Mutat. Res. 2021, 787, 108347. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Kida, Y.; Sakamoto, Y.; Kuwano, K. Mpn491, a secreted nuclease of Mycoplasma pneumoniae, plays a critical role in evading killing by neutrophil extracellular traps. Cell Microbiol. 2017, 19, e12666. [Google Scholar] [CrossRef]
- Hao, Z.; Lu, D.; Li, X.; Raheem, A.; Zhao, G.; Dawood, A.S.; Chen, Y.; Chen, X.; Hu, C.; Chen, J.; et al. Novel Nuclease MbovP701 with a Yqaj Domain Is Interrelated with the Growth of Mycoplasma bovis. Microorganisms 2024, 12, 2509. [Google Scholar] [CrossRef]
- Marti, T.M.; Fleck, O. DNA repair nucleases. Cell. Mol. Life Sci. (CMLS) 2004, 61, 336–354. [Google Scholar] [CrossRef]
- Yang, W.; Lee, J.Y.; Nowotny, M. Making and breaking nucleic acids: Two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 2006, 22, 5–13. [Google Scholar] [CrossRef]
- Minion, F.C.; Goguen, J.D. Identification and preliminary characterization of external membrane-bound nuclease activities in Mycoplasma pulmonis. Infect. Immun. 1986, 51, 352–354. [Google Scholar] [CrossRef]
- Bendjennat, M.; Blanchard, A.; Loutfi, M.; Montagnier, L.; Bahraoui, E. Purification and characterization of Mycoplasma penetrans Ca2+/Mg2+-dependent endonuclease. J. Bacteriol. 1997, 179, 2210–2220. [Google Scholar] [CrossRef][Green Version]
- Paddenberg, R.; Weber, A.; Wulf, S.; Mannherz, H.G. Mycoplasma nucleases able to induce internucleosomal DNA degradation in cultured cells possess many characteristics of eukaryotic apoptotic nucleases. Cell Death Differ. 1998, 5, 517–528. [Google Scholar] [CrossRef]
- Pollack, J.D.; Hoffmann, P.J. Properties of the nucleases of mollicutes. J. Bacteriol. 1982, 152, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Cacciotto, C.; Dessi, D.; Cubeddu, T.; Cocco, A.R.; Pisano, A.; Tore, G.; Fiori, P.L.; Rappelli, P.; Pittau, M.; Alberti, A. MHO_0730 as a Surface-Exposed Calcium-Dependent Nuclease of Mycoplasma hominis Promoting Neutrophil Extracellular Trap Formation and Escape. J. Infect. Dis. 2019, 220, 1999–2008. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, K.; Ward, W.S. A novel nuclease activity that is activated by Ca2+ chelated to EGTA. Syst. Biol. Reprod. Med. 2009, 55, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Bendjennat, M.; Blanchard, A.; Loutfi, M.; Montagnier, L.; Bahraoui, E. Role of Mycoplasma penetrans endonuclease P40 as a potential pathogenic determinant. Infect. Immun. 1999, 67, 4456–4462. [Google Scholar] [CrossRef]
- Cacciotto, C.; Addis, M.F.; Coradduzza, E.; Carcangiu, L.; Nuvoli, A.M.; Tore, G.; Dore, G.M.; Pagnozzi, D.; Uzzau, S.; Chessa, B.; et al. Mycoplasma agalactiae MAG_5040 is a Mg2+-dependent, sugar-nonspecific SNase recognised by the host humoral response during natural infection. PLoS ONE 2013, 8, e57775. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Y.; Li, X.; Zhou, W.; Li, X.; Jiang, F.; Wu, W. Mycoplasma hyopneumoniae Mhp597 is a cytotoxicity, inflammation and immunosuppression associated nuclease. Vet. Microbiol. 2019, 235, 53–62. [Google Scholar] [CrossRef]
- Yacoub, E.; Ben Abdelmoumen Mardassi, B. Mm19, a Mycoplasma meleagridis Major Surface Nuclease that Is Related to the RE_AlwI Superfamily of Endonucleases. PLoS ONE 2016, 11, e0152171. [Google Scholar] [CrossRef]
- Jarvill-Taylor, K.J.; VanDyk, C.; Minion, F.C. Cloning of mnuA, a membrane nuclease gene of Mycoplasma pulmonis, and analysis of its expression in Escherichia coli. J. Bacteriol. 1999, 181, 1853–1860. [Google Scholar] [CrossRef]
- Newton, K.; Strasser, A.; Kayagaki, N.; Dixit, V.M. Cell death. Cell 2024, 187, 235–256. [Google Scholar] [CrossRef]
- van Elsland, D.; Neefjes, J. Bacterial infections and cancer. EMBO Rep. 2018, 19, e46632. [Google Scholar] [CrossRef]
- Sokolova, I.A.; Vaughan, A.T.; Khodarev, N.N. Mycoplasma infection can sensitize host cells to apoptosis through contribution of apoptotic-like endonuclease(s). Immunol. Cell Biol. 1998, 76, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Gondaira, S.; Higuchi, H.; Nishi, K.; Iwano, H.; Nagahata, H. Mycoplasma bovis escapes bovine neutrophil extracellular traps. Vet. Microbiol. 2017, 199, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Perez-Casal, J. Pathogenesis and Virulence of Mycoplasma bovis. Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Kida, Y.; Kuwano, K. A dipalmitoylated lipoprotein from Mycoplasma pneumoniae activates NF-kappa B through TLR1, TLR2, and TLR6. J. Immunol. 2005, 175, 4641–4646. [Google Scholar] [CrossRef]
- Li, R.; Zheng, W.; Xiao, Y.; Yu, X.; Sheng, J.; Zhang, H.; Chen, C.; Ma, Z.; Wang, Y. Mycoplasma hyopneumoniae nuclease Mhp597 negatively regulates TBK1-IRF3-IFN-I pathway by targeting vimentin to facilitate infection. Int. J. Biol. Macromol. 2025, 306, 141351. [Google Scholar] [CrossRef]
- Fousert, E.; Toes, R.; Desai, J. Neutrophil Extracellular Traps (NETs) Take the Central Stage in Driving Autoimmune Responses. Cells 2020, 9, 915. [Google Scholar] [CrossRef]
- Baz, A.A.; Hao, H.; Lan, S.; Li, Z.; Liu, S.; Chen, S.; Chu, Y. Neutrophil extracellular traps in bacterial infections and evasion strategies. Front. Immunol. 2024, 15, 1357967. [Google Scholar] [CrossRef]
- Liew, P.X.; Kubes, P. The Neutrophil’s Role During Health and Disease. Physiol. Rev. 2019, 99, 1223–1248. [Google Scholar] [CrossRef]
- Nathan, C. Neutrophils and immunity: Challenges and opportunities. Nat. Rev. Immunol. 2006, 6, 173–182. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Kaplan, M.J.; Radic, M. Neutrophil extracellular traps: Double-edged swords of innate immunity. J. Immunol. 2012, 189, 2689–2695. [Google Scholar] [CrossRef] [PubMed]
- Doring, Y.; Libby, P.; Soehnlein, O. Neutrophil Extracellular Traps Participate in Cardiovascular Diseases: Recent Experimental and Clinical Insights. Circ. Res. 2020, 126, 1228–1241. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; Weinrauch, Y.; Brinkmann, V.; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007, 176, 231–241. [Google Scholar] [CrossRef]
- Cacciotto, C.; Cubeddu, T.; Addis, M.F.; Anfossi, A.G.; Tedde, V.; Tore, G.; Carta, T.; Rocca, S.; Chessa, B.; Pittau, M.; et al. Mycoplasma lipoproteins are major determinants of neutrophil extracellular trap formation. Cell Microbiol. 2016, 18, 1751–1762. [Google Scholar] [CrossRef]
- Schenk, M.; Belisle, J.T.; Modlin, R.L. TLR2 looks at lipoproteins. Immunity 2009, 31, 847–849. [Google Scholar] [CrossRef]
- Munoz-Caro, T.; Mena Huertas, S.J.; Conejeros, I.; Alarcon, P.; Hidalgo, M.A.; Burgos, R.A.; Hermosilla, C.; Taubert, A. Eimeria bovis-triggered neutrophil extracellular trap formation is CD11b-, ERK 1/2-, p38 MAP kinase- and SOCE-dependent. Vet. Res. 2015, 46, 23. [Google Scholar] [CrossRef]
- Papayannopoulos, V.; Metzler, K.D.; Hakkim, A.; Zychlinsky, A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 2010, 191, 677–691. [Google Scholar] [CrossRef]
- Bjornsdottir, H.; Welin, A.; Michaelsson, E.; Osla, V.; Berg, S.; Christenson, K.; Sundqvist, M.; Dahlgren, C.; Karlsson, A.; Bylund, J. Neutrophil NET formation is regulated from the inside by myeloperoxidase-processed reactive oxygen species. Free Radic. Biol. Med. 2015, 89, 1024–1035. [Google Scholar] [CrossRef]
- de Bont, C.M.; Koopman, W.J.H.; Boelens, W.C.; Pruijn, G.J.M. Stimulus-dependent chromatin dynamics, citrullination, calcium signalling and ROS production during NET formation. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1621–1629. [Google Scholar] [CrossRef]
- Mitiku, F.; Hartley, C.A.; Sansom, F.M.; Coombe, J.E.; Mansell, P.D.; Beggs, D.S.; Browning, G.F. The major membrane nuclease MnuA degrades neutrophil extracellular traps induced by Mycoplasma bovis. Vet. Microbiol. 2018, 218, 13–19. [Google Scholar] [CrossRef]
- Berends, E.T.; Horswill, A.R.; Haste, N.M.; Monestier, M.; Nizet, V.; von Kockritz-Blickwede, M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2010, 2, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, S.; Stojkov, D.; Germic, N.; Simon, D.; Wang, X.; Benarafa, C.; Simon, H.U. Untangling “NETosis” from NETs. Eur. J. Immunol. 2019, 49, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Henthorn, C.R.; Chris Minion, F.; Sahin, O. Utilization of macrophage extracellular trap nucleotides by Mycoplasma hyopneumoniae. Microbiology (Reading) 2018, 164, 1394–1404. [Google Scholar] [CrossRef]
- Tan, C.; Aziz, M.; Wang, P. The vitals of NETs. J. Leukoc. Biol. 2021, 110, 797–808. [Google Scholar] [CrossRef]
- Denning, N.L.; Aziz, M.; Gurien, S.D.; Wang, P. DAMPs and NETs in Sepsis. Front. Immunol. 2019, 10, 2536. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Shao, J.; Cao, B.; Zhao, R.; Li, H.; Gao, W.; Chen, P.; Jin, L.; Cao, L.; Ji, S.; et al. The Significance of Neutrophil Extracellular Traps in Colorectal Cancer and Beyond: From Bench to Bedside. Front. Oncol. 2022, 12, 848594. [Google Scholar] [CrossRef]
- Konwar, B.; Mullick, P.; Das, G.; Ramesh, A. Anthraquinone-Based Ligands as MNase Inhibitors: Insights from Inhibition Studies and Generation of a Payload Nanocarrier for Potential Anti-MRSA Therapy. ChemMedChem 2023, 18, e202200711. [Google Scholar] [CrossRef]
- Sharma, P.; Garg, N.; Sharma, A.; Capalash, N.; Singh, R. Nucleases of bacterial pathogens as virulence factors, therapeutic targets and diagnostic markers. Int. J. Med. Microbiol. 2019, 309, 151354. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Wolfs, J.M.; Kolaczyk, T.; Roberts, A.K.; Hu, S.X.; Edgell, D.R. Monomeric site-specific nucleases for genome editing. Proc. Natl. Acad. Sci. USA 2012, 109, 8061–8066. [Google Scholar] [CrossRef]
- Liu, R.M.; Liang, L.L.; Freed, E.; Chang, H.; Oh, E.; Liu, Z.Y.; Garst, A.; Eckert, C.A.; Gill, R.T. Synthetic chimeric nucleases function for efficient genome editing. Nat. Commun. 2019, 10, 5524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, X.; Lv, J.; Huang, H.; Wang, J.; Zhuo, M.; Tan, Z.; Huang, G.; Liu, J.; Liu, Y.; et al. Engineered circular guide RNAs boost CRISPR/Cas12a- and CRISPR/Cas13d-based DNA and RNA editing. Genome Biology 2023, 24, 145. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.J.; Gupta, A.; Bednarski, C.; Gehrig-Giannini, S.; Richter, F.; Pitzler, C.; Gamalinda, M.; Galonska, C.; Takeuchi, R.; Wang, K.; et al. Improved CRISPR genome editing using small highly active and specific engineered RNA-guided nucleases. Nat. Commun. 2021, 12, 4219. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Liu, G.; Tang, Z.; Xiang, S.; Yang, L.; Huang, L.; He, Y.; Fan, T.; Liu, S.; Zheng, X.; et al. Efficient plant genome engineering using a probiotic sourced CRISPR-Cas9 system. Nat. Commun. 2023, 14, 6102. [Google Scholar] [CrossRef]
- Zhang, G.; Han, L.; Li, Z.; Chen, Y.; Li, Q.; Wang, S.; Shi, H. Screening of immunogenic proteins and evaluation of vaccine candidates against Mycoplasma synoviae. NPJ Vaccines 2023, 8, 121. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, X.; Huang, Y.; Li, X.; Xu, H.; Liu, C.; Li, C.; Zeng, Q.; Luo, H.; Ye, Z.; He, J.; et al. Decoding Mycoplasma Nucleases: Biological Functions and Pathogenesis. Toxins 2025, 17, 215. https://doi.org/10.3390/toxins17050215
Yi X, Huang Y, Li X, Xu H, Liu C, Li C, Zeng Q, Luo H, Ye Z, He J, et al. Decoding Mycoplasma Nucleases: Biological Functions and Pathogenesis. Toxins. 2025; 17(5):215. https://doi.org/10.3390/toxins17050215
Chicago/Turabian StyleYi, Xinchao, Ying Huang, Xinru Li, Hao Xu, Chang Liu, Chao Li, Qianrui Zeng, Haodang Luo, Zufeng Ye, Jun He, and et al. 2025. "Decoding Mycoplasma Nucleases: Biological Functions and Pathogenesis" Toxins 17, no. 5: 215. https://doi.org/10.3390/toxins17050215
APA StyleYi, X., Huang, Y., Li, X., Xu, H., Liu, C., Li, C., Zeng, Q., Luo, H., Ye, Z., He, J., & You, X. (2025). Decoding Mycoplasma Nucleases: Biological Functions and Pathogenesis. Toxins, 17(5), 215. https://doi.org/10.3390/toxins17050215





