Acetylated Deoxynivalenol Generates Differences of Gene Expression that Discriminate Trichothecene Toxicity
Abstract
:1. Introduction


2. Results and Discussion
2.1. Influences of Pleiotropic Drug Resistance Transporter Regulation

| Systematic Name | Gene Symbol | Localization | Description |
|---|---|---|---|
| YGL013C | PDR1 | cytoplasm, nucleus | Regulates pleiotropic drug resistance |
| YBL005W | PDR3 | cytoplasm, nucleus | Transcriptional activator of the pleiotropic drug resistance network |
| YDR011W | SNQ2 | plasma membrane | Plasma membrane ATP binding cassette (ABC) transporter |
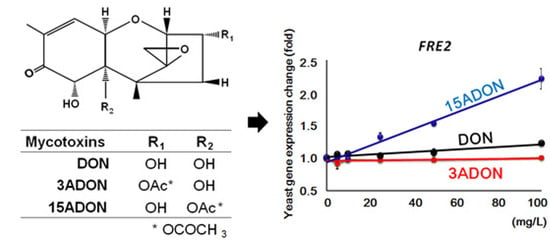
2.2. Stress-Responsive Redox Genes
| Systematic Name | Gene Symbol | Group * | Induction with | Localization | Description | |
|---|---|---|---|---|---|---|
| Low Level Iron | Low Level Copper | |||||
| YLR214W | FRE1 | II | + | ++ | plasma membrane | Ferric reductase and cupric reductase |
| YKL220C | FRE2 | I | ++ | + | Ferric reductase | |
| YOR381W | FRE3 | I | ++ | + | Ferric reductase | |
| YOL152W | FRE7 | II | - | ++ | Ferric reductase and cupric reductase | |
| YLR047C | FRE8 | III | - | - | Similarity to ferric/cupric reductases | |

2.3. Genes Specifically Responsive to 15ADON

| Systematic Name | Gene Symbol | Localization | Description |
|---|---|---|---|
| YBL016W | FUS3 | cytoplasm, nucleus | Mitogen-activated serine/threonine protein kinase |
| YJL153C | INO1 | cytoplasm | Inositol-3-phosphate synthase |
| YLR307C-A | - | unknown | Uncharacterized |
| YMR230W-A | - | unknown | Uncharacterized |
2.4. Glucose Transporter Genes Differentiate DON and its Derivatives

| Systematic Name | Gene Symbol | Localization | Description |
|---|---|---|---|
| YKL038W | RGT1 | nucleus | Glucose-responsive transcription factor |
| YDL138W | RGT2 | plasma membrane | Plasma membrane glucose sensor |
| YDL194W | SNF3 | plasma membrane | Plasma membrane low glucose sensor |
| YHR094C | HXT1 | plasma membrane | Low-affinity glucose transporter |
| YMR011W | HXT2 | plasma membrane | High-affinity glucose transporter |
| YHR092C | HXT4 | plasma membrane | High-affinity glucose transporter |
3. Conclusions
4. Experimental Section
4.1. Chemicals
4.2. Cell Strain and Growth Conditions
4.3. RNA and cDNA Preparation, Semi-Quantitative PCR
| Name | Direction | Forward or Reverse |
|---|---|---|
| 5′ 3′ | ||
| ACT1 | ATTGCCGAAAGAATGCAAAAGG | F |
| CGCACAAAAGCAGAGATTAGAAACA | R | |
| FRE1 | GCTCGGAAATAAAACTCTCAGAA | F |
| ATTATTAACAAGGGGCCTTACCA | R | |
| FRE2 | CGACCAAATGTTAAGGAACTTCTAC | F |
| AAATGATCACCAGCATTGATACTCT | R | |
| FRE3 | TAAATGGATCGTTAGCTGTGGTT | F |
| TTGTGATAGGTAAAATAGTGAGGAAA | R | |
| FRE7 | GCTTCATTTGTTCAGGTTCTGAC | F |
| TGCTTGAATGATATTTCACATGG | R | |
| FRE8 | ACGGATTCTATTCAATGTTGCAG | F |
| TCAAATACGTGAATTTTCCAAGC | R | |
| FUS3 | ATCCGTATTTGCAAACATACCAC | F |
| TGTATACATTGTTCTTCGGGTTGATA | R | |
| HXT1 | CGTCTTTTTCTTCGTTCCAGA | F |
| GAGCTTGTTTAGTTTATTTCCTGCT | R | |
| HXT2 | GGGTATGTCTTCATGGGCTGT | F |
| TATAATCTCTTATTCCTCGGAAACTC | R | |
| HXT4 | TTCTTCGTTCCAGAAACTAAAGG | F |
| CCGAACATCTTCTTGTAAAATGG | R | |
| INO1 | AAGATGCTGGCAAATTCGAG | F |
| TGAGATTACAACAATCTCTCTTCG | R | |
| PDR1 | ACAATATTAACAACAACAACAGTAACAA | F |
| GGAAGGAAGTTTTTGAGAACTTTTA | R | |
| PDR3 | CAACAGACAAAAAGACAACATTCTG | F |
| CCATTTACTATGGTTATGCTCTGCT | R | |
| RGT1 | CCTCCGCGAGTCATCAGT | F |
| ACCTGTCAATACCAGCCTAACTC | R | |
| RGT2 | CAATAACAACACTGAACGAAATGG | F |
| GGGGAAGTGTATTGGCTGTG | R | |
| SNF3 | GAACGAATGGCGCAGTTT | F |
| CAAATCATTATTTTCATTTACAGGTTG | R | |
| SNQ2 | GCTACTTGTGGAGAAATTTTGGA | F |
| GCAGATGAATGCACAAAATGTTA | R | |
| YLR307C-A | GGGATGCTAGTACTACTCAATGTCG | F |
| TTAATTCAAATTATACTTTTACGTGCTC | R | |
| YMR230W-A | GGATGTGTTACGATGCAGACA | F |
| ATATGGCGCGTTCTTGAAGG | R |
Acknowledgments
Author Contributions
Conflicts of Interest
Nomenclature
References
- Pestka, J.J.; Zhou, H.R.; Moon, Y.; Chung, Y.J. Cellular and molecular mechanisms for immune modulation by deoxynivalenol and other trichothecenes: unraveling a paradox. Toxicol. Lett. 2004, 153, 61–73. [Google Scholar] [PubMed]
- Pestka, J.J.; Uzarski, R.L.; Islam, Z. Induction of apoptosis and cytokine production in the Jurkathuman T cells by deoxynivalenol: Role of mitogen-activated protein kinases and comparison to other 8-ketotrichothecenes. Toxicology 2005, 206, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Iwahashi, Y. Comprehensive gene expression analysis of type B trichothecenes. J. Agric. Food Chem. 2012, 60, 9519–9527. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, J.E.; Bin-Umer, M.A.; Tortora, A.; Mendez, N.; McCormick, S.; Tumer, N.E. A genome-wide screen in Saccharomyces cerevisiae reveals a critical role for the mitochondria in the toxicity of a trichothecene mycotoxin. Proc. Natl. Acad. Sci. USA 2009, 106, 21883–21888. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, A.E.; McCormick, S.P.; Appell, M. Structure-activity relationships of trichothecene toxins in an Arabidopsis thaliana leaf assay. J. Agric. Food Chem. 2007, 55, 6487–6492. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J. Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 2010, 84, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Ward, T.J.; Clear, R.M.; Rooney, A.P.; O'Donnell, K.; Gaba, D.; Patrick, S.; Starkey, D.E.; Gilbert, J.; Geiser, D.M.; Nowicki, T.W. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 2008, 45, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Alexander, N.J.; McCormick, S.P.; Ziegenhorn, S.L. Phytotoxicity of selected trichothecenes using Chlamydomonas reinhardtii as a model systemt. Nat. Toxins 1999, 7, 265–269. [Google Scholar] [CrossRef]
- Pestka, J.J. Deoxynivalenol: Toxicity, mechanisms and animal health risks. Anim. Feed Sci. Technol. 2007, 137, 283–298. [Google Scholar] [CrossRef]
- Alassane-Kpembi, I.; Kolf-Clauw, M.; Gauthier, T.; Abrami, R.; Abiola, F.A.; Oswald, I.P.; Puel, O. New insights into mycotoxin mixtures: The toxicity of low doses of Type B trichothecenes on intestinal epithelial cells is synergistic. Toxicol. Appl. Pharmacol. 2013, 272, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Iwahashi, Y. Phytotoxicity evaluation of type B trichothecenes using a Chlamydomonas reinhardtii model system. Toxins 2014, 6, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Sundstøl Eriksen, G.; Pettersson, H.; Lundh, T. Comparative cytotoxicity of deoxynivalenol, nivalenol, their acetylated derivatives and de-epoxy metabolites. Food Chem. Toxicol. 2004, 42, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.W.; Duncan, G.; Richardson, A.J. The human fecal microbiota metabolizes deoxynivalenol and deoxynivalenol-3-glucoside and may be responsible for urinary deepoxy-deoxynivalenol. Appl. Environ. Microbiol. 2013, 79, 1821–1825. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA). Deoxynivalenol in Food And feed: Occurrence and Exposure. In EFSA Journal; EFSA: Parma, Italy, 2013; Volume 11, pp. 1–56. [Google Scholar]
- Leonard, P.J.; Rathod, P.K.; Golin, J. Loss of function mutation in the yeast multiple drug resistance gene PDR5 causes a reduction in chloramphenicol efflux. Antimicrob. Agents Chemother. 1994, 38, 2492–2494. [Google Scholar] [CrossRef] [PubMed]
- Mitterbauer, R.; Adam, G. Saccharomyces cerevisae and Arabidopsis thaliana: Useful model systems for the identification of molecular mechanisms involved in resistance of plants to toxins. Eur. J. Plant Pathol. 2002, 108, 699–703. [Google Scholar] [CrossRef]
- Wehrschütz-Sigl, E.; Jungwirth, H.; Bergler, H.; Högenauer, G. The transporters Pdr5p and Snq2p mediate diazaborine resistance and are under the control of the gain-of-function allele PDR1–12. Eur. J. Biochem. 2004, 271, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Sirisattha, S.; Mori, K.; Iwahashi, Y. Mycotoxin toxicity in Saccharomyces cerevisiae differs depending on gene mutations. Food Sci. Technol. Res. 2009, 6, 453–458. [Google Scholar] [CrossRef]
- Suzuki, T.; Iwahashi, Y. Low toxicity of deoxynivalenol-3-glucoside in microbial cells. Toxins 2015, 7, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Mitterbauer, R.; Weindorfer, H.; Safaie, N.; Krska, R.; Lemmens, M.; Ruckenbauer, P.; Kuchler, K.; Adam, G. A sensitive and inexpensive yeast bioassay for the mycotoxin zearalenone and other compounds with estrogenic activity. Appl. Environ. Microbiol. 2003, 69, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Jungwirth, H.; Kuchler, K. Yeast ABC transporters—A tale of sex, stress, drugs and aging. FEBS Lett. 2006, 580, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Georgatsou, E.; Alexandraki, D. Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast 1999, 15, 573–584. [Google Scholar] [CrossRef]
- Georgatsou, E.; Alexandraki, D. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994, 14, 3065–3073. [Google Scholar] [CrossRef] [PubMed]
- Shatwell, K.P.; Dancis, A.; Cross, A.R.; Klausner, R.D.; Segal, A.W. The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J. Biol. Chem. 1996, 271, 14240–14244. [Google Scholar] [PubMed]
- Hajji, K.; Clotet, J.; Ariño, J. Disruption and phenotypic analysis of seven ORFs from the left arm of chromosome XV of Saccharomyces cerevisiae. Yeast 1999, 15, 435–441. [Google Scholar] [CrossRef]
- Suzuki, T.; Iwahashi, Y. Gene expression profile of MAP kinase PTC1 mutant exposed to Aflatoxin B1: Dysfunctions of gene expression in glucose utilization and sphingolipid metabolism. Chem. Biol. Informat. J. 2009, 9, 94–107. [Google Scholar] [CrossRef]
- Soriano, J.M.; Gonzalez, L.; Catala, A.I. Mechanism of action of sphingolipids and their metabolites in the toxicity of fumonisin B1. Prog. Lipid Res. 2005, 44, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Cherkasova, V.; Lyons, D.M.; Elion, E.A. Fus3p and Kss1p control G1 arrest in Saccharomyces cerevisiae through a balance of distinct arrest and proliferative functions that operate in parallel with Far1p. Genetics 1999, 151, 989–1004. [Google Scholar] [PubMed]
- Ozcan, S.; Johnston, M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 1995, 15, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, S.; Johnston, M. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 1999, 63, 554–569. [Google Scholar] [PubMed]
- Kim, J.H.; Roy, A.; Jouandot, D.; Cho, K.H. The glucose signaling network in yeast. Biochim. Biophys. Acta 2013, 1830, 5204–5210. [Google Scholar] [CrossRef] [PubMed]
- Evaluations of the Joint FAO/WHO Expert Committee on Food Additives (JECFA). Available online: http://apps.who.int/food-additives-contaminants-jecfa-database/chemical.aspx?chemID=2947 (accessed on 25 November 2015).
- Saccharomyces Genome Database. Available online: http://www.yeastgenome.org/ (accessed on 15 September 2015).
- Iwahashi, H.; Odani, M.; Ishidou, E.; Kitagawa, E. Adaptation of Saccharomyces cerevisiae to high hydrostatic pressure causing growth inhibition. FEBS Lett. 2005, 579, 2847–2852. [Google Scholar] [CrossRef] [PubMed]
- Primer3. Available online: http://frodo.wi.mit.edu/ (accessed on 6 April 2015).
- Gene Expression Omnibus Database. Available online: http://www.ncbi.nlm.nih.gov/geo/ (accessed on 12 October 2015).
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, T.; Iwahashi, Y. Acetylated Deoxynivalenol Generates Differences of Gene Expression that Discriminate Trichothecene Toxicity. Toxins 2016, 8, 42. https://doi.org/10.3390/toxins8020042
Suzuki T, Iwahashi Y. Acetylated Deoxynivalenol Generates Differences of Gene Expression that Discriminate Trichothecene Toxicity. Toxins. 2016; 8(2):42. https://doi.org/10.3390/toxins8020042
Chicago/Turabian StyleSuzuki, Tadahiro, and Yumiko Iwahashi. 2016. "Acetylated Deoxynivalenol Generates Differences of Gene Expression that Discriminate Trichothecene Toxicity" Toxins 8, no. 2: 42. https://doi.org/10.3390/toxins8020042
APA StyleSuzuki, T., & Iwahashi, Y. (2016). Acetylated Deoxynivalenol Generates Differences of Gene Expression that Discriminate Trichothecene Toxicity. Toxins, 8(2), 42. https://doi.org/10.3390/toxins8020042