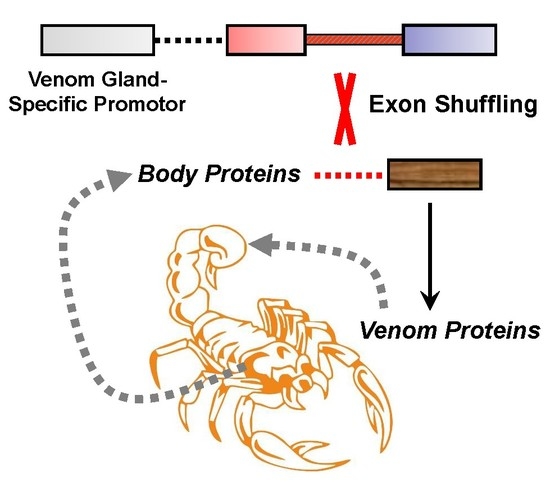
Exon Shuffling and Origin of Scorpion Venom Biodiversity
Abstract
:1. Introduction
2. Scorpion Venom Biodiversity
2.1. CSαβ-Type Peptides
2.2. ICK-Type Peptides
2.3. Alpha-Helical AMPs
2.4. Proteases and Protease Inhibitors
3. Exon-Intron Structures of Scorpion Venom Gland-Expressed Genes
3.1. Genes Encoding CSαβ Fold Peptides
3.2. Genes Encoding ICK Fold Peptides
3.3. Genes Encoding α-Helical AMPs
3.4. Genes Encoding Proteases and Protease Inhibitors
4. Origin of Scorpion Venom Diversity by Exon Shuffling
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhang, L.; Shi, W.; Zeng, X.C.; Ge, F.; Yang, M.; Nie, Y.; Bao, A.; Wu, S.; E, G. Unique diversity of the venom peptides from the scorpion Androctonus bicolor revealed by transcriptomic and proteomic analysis. J. Proteom. 2015, 128, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G. From genome to “venome”: Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005, 15, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, A.D.; Swain, M.T.; Hegarty, M.J.; Logan, D.W.; Mulley, J.F. Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins. Genome Biol. Evol. 2014, 6, 2088–2095. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Peigneur, S.; Gao, B.; Umetsu, Y.; Ohki, S.; Tytgat, J. Experimental conversion of a defensin into a neurotoxin: Implications for origin of toxic function. Mol. Biol. Evol. 2014, 31, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Gao, B.; Deng, M.; Yuan, Y.; Luo, L.; Peigneur, S.; Xiao, Y.; Liang, S.; Tytgat, J. Drosotoxin, a selective inhibitor of tetrodotoxin-resistant sodium channels. Biochem. Pharmacol. 2010, 80, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Umetsu, Y.; Gao, B.; Ohki, S.; Zhu, S. Mesomartoxin, a new Kv1.2-selective scorpion toxin interacting with the channel selectivity filter. Biochem. Pharmacol. 2015, 93, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Harvey, P.J.; Craik, D.J.; Ronjat, M.; De Waard, M.; Zhu, S. Functional evolution of scorpion venom peptides with an inhibitor cystine knot fold. Biosci. Rep. 2013, 33, e00047. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Xu, J.; Rodriguez Mdel, C.; Lanz-Mendoza, H.; Hernández-Rivas, R.; Du, W.; Zhu, S. Characterization of two linear cationic antimalarial peptides in the scorpion Mesobuthus eupeus. Biochimie 2010, 92, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Yongqing, T.; Wilmann, P.G.; Reeve, S.B.; Coetzer, T.H.; Smith, A.I.; Whisstock, J.C.; Pike, R.N.; Wijeyewickrema, L.C. The X-ray crystal structure of mannose-binding lectin-associated serine proteinase-3 reveals the structural basis for enzyme inactivity associated with the Carnevale, Mingarelli, Malpuech, and Michels (3MC) syndrome. J. Biol. Chem. 2013, 288, 22399–22407. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, F.; Feng, J.; Yang, W.; Zeng, D.; Zhao, R.; Cao, Z.; Liu, M.; Li, W.; Jiang, L.; et al. Genomic and structural characterization of Kunitz-type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins. PLoS ONE 2013, 8, e60201. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Gao, B.; Tytgat, J. Phylogenetic distribution, functional epitopes and evolution of the CSαβ superfamily. Cell. Mol. Life Sci. 2005, 62, 2257–2269. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Gao, B. Molecular characterization of a new scorpion venom lipolysis activating peptide: Evidence for disulfide bridge-mediated functional switch of peptides. FEBS Lett. 2006, 580, 6825–6836. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Yu, Y.; Wu, Y.; Hao, P.; Di, Z.; He, Y.; Chen, Z.; Yang, W.; Shen, Z.; He, X.; et al. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat. Commun. 2013, 4, 2602. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Hernández, V.; Jiménez-Vargas, J.M.; Gurrola, G.B.; Valdivia, H.H.; Possani, L.D. Scorpion venom components that affect ion-channels function. Toxicon 2013, 76, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez de la Vega, R.C.; Possani, L.D. Current views on scorpion toxins specific for K+-channels. Toxicon 2004, 43, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Possani, L.D.; Becerril, B.; Delepierre, M.; Tytgat, J. Scorpion toxins specific for Na+-channels. Eur. J. Biochem. 1999, 264, 287–300. [Google Scholar] [CrossRef] [PubMed]
- De la Vega, R.C.R.; Possani, L.D. Overview of scorpion toxins specific for Na+ channels and related peptides: Biodiversity, structure-function relationships and evolution. Toxicon 2005, 46, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Tytgat, J.; Chandy, K.G.; Garcia, M.L.; Gutman, G.A.; Martin-Eauclaire, M.F.; van der Walt, J.J.; Possani, L.D. A unified nomenclature for short-chain peptides isolated from scorpion venoms: Alpha-KTx molecular subfamilies. Trends Pharmacol. Sci. 1999, 20, 444–447. [Google Scholar] [CrossRef]
- Legros, C.; Céard, B.; Bougis, P.E.; Martin-Eauclaire, M.F. Evidence for a new class of scorpion toxins active against K+ channels. FEBS Lett. 1998, 431, 375–380. [Google Scholar] [CrossRef]
- Gurevitz, M.; Froy, O.; Zilberberg, N.; Turkov, M.; Strugatsky, D.; Gershburg, E.; Lee, D.; Adams, M.E.; Tugarinov, V.; Anglister, J.; et al. Sodium channel modifiers from scorpion venom: Structure-activity relationship, mode of action and application. Toxicon 1998, 36, 1671–1682. [Google Scholar] [CrossRef]
- Zhu, S.; Bosmans, F.; Tytgat, J. Adaptive evolution of scorpion sodium channel toxins. J. Mol. Evol. 2004, 58, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Cestèle, S.; Catterall, W.A. Molecular mechanisms of neurotoxin action on voltage-gated sodium channels. Biochimie 2000, 82, 883–892. [Google Scholar] [CrossRef]
- Catterall, W.A. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron 2000, 26, 13–25. [Google Scholar] [CrossRef]
- Yount, N.Y.; Yeaman, M.R. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 7363–7368. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, B.; Zhu, S. A single-point mutation enhances dual functionality of a scorpion toxin. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2016, 179, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Diego-García, E.; Caliskan, F.; Tytgat, J. The Mediterranean scorpion Mesobuthus gibbosus (Scorpiones, Buthidae): Transcriptome analysis and organization of the genome encoding chlorotoxin-like peptides. BMC Genom. 2014, 15, 295. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.M.; Gao, B.; Zhu, S.Y. Origin of neurotoxins from defensins. Acta Physiol. Sin. 2015, 67, 239–247. [Google Scholar]
- Lukács, B.; Sztretye, M.; Almássy, J.; Sárközi, S.; Dienes, B.; Mabrouk, K.; Simut, C.; Szabó, L.; Szentesi, P.; de Waard, M.; et al. Charged surface area of maurocalcine determines its interaction with the skeletal ryanodine receptor. Biophys. J. 2008, 95, 3497–3509. [Google Scholar] [CrossRef] [PubMed]
- Gurrola, G.B.; Capes, E.M.; Zamudio, F.Z.; Possani, L.D.; Valdivia, H.H. Imperatoxin A, a cell-penetrating peptide from scorpion venom, as a probe of Ca2+ release channels/ryanodine receptors. Pharmaceuticals 2010, 3, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Torres-Larios, A.; Gurrola, G.B.; Zamudio, F.Z.; Possani, L.D. Hadrurin, a new antimicrobial peptide from the venom of the scorpion Hadrurus aztecus. Eur. J. Biochem. 2000, 267, 5023–5031. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Sherman, P.; Luo, L.; Bowie, J.; Zhu, S. Structural and functional characterization of two genetically related meucin peptides highlights evolutionary divergence and convergence in antimicrobial peptides. FASEB J. 2009, 23, 1230–1245. [Google Scholar] [CrossRef] [PubMed]
- Corzo, G.; Escoubas, P.; Villegas, E.; Barnham, K.J.; He, W.; Norton, R.S.; Nakajima, T. Characterization of unique amphipathic antimicrobial peptides from venom of the scorpion Pandinus imperator. Biochem. J. 2001, 359, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Ma, Y.; Dai, C.; Zhao, R.; Li, S.; Wu, Y.; Cao, Z.; Li, W. Imcroporin, a new cationic antimicrobial peptide from the venom of the scorpion Isometrus maculates. Antimicrob. Agents Chemother. 2009, 53, 3472–3477. [Google Scholar] [CrossRef] [PubMed]
- Remijsen, Q.; Verdonck, F.; Willems, J. Parabutoporin, a cationic amphipathic peptide from scorpion venom: Much more than an antibiotic. Toxicon 2010, 55, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.C.; Zhou, L.; Shi, W.; Luo, X.; Zhang, L.; Nie, Y.; Wang, J.; Wu, S.; Cao, B.; Cao, H. Three new antimicrobial peptides from the scorpion Pandinus imperator. Peptides 2013, 45, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Hwang, P.M.; Vogel, H.J. Structure-function relationships of antimicrobial peptides. Biochem. Cell Biol. 1998, 76, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Chhatwal, G.S.; Habermann, E. Neurotoxins, protease inhibitors and histamine releasers in the venom of the Indian red scorpion (Buthus tamulus): Isolation and partial characterization. Toxicon 1981, 19, 807–823. [Google Scholar] [CrossRef]
- Schwartz, E.F.; Diego-Garcia, E.; de la Vega, R.C.R.; Possani, L.D. Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones). BMC Genom. 2007, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Hu, Y.T.; Yang, W.S.; He, Y.W.; Feng, J.; Wang, B.; Zhao, R.M.; Ding, J.P.; Cao, Z.J.; Li, W.X.; et al. Hg1, novel peptide inhibitor specific for Kv1.3 channels from first scorpion Kunitz-type potassium channel toxin family. J. Biol. Chem. 2012, 287, 13813–13821. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cao, Z.; Li, W.; Wu, Y. Cloning and characterization of a novel Kunitz-type inhibitor from scorpion with unique cysteine framework. Toxicon 2013, 72, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, B.; Hu, J.; Yang, W.; Cao, Z.; Zhuo, R.; Li, W.; Wu, Y. SjAPI, the first functionally characterized Ascaris-type protease inhibitor from animal venoms. PLoS ONE 2013, 8, e57529. [Google Scholar] [CrossRef] [PubMed]
- Grasberger, B.L.; Clore, G.M.; Gronenborn, A.M. High-resolution structure of Ascaris trypsin inhibitor in solution: Direct evidence for a pH-induced conformational transition in the reactive site. Structure 1994, 2, 669–678. [Google Scholar] [CrossRef]
- Patthy, L. Introns: Phase Compatibility. eLS 2008. [Google Scholar] [CrossRef]
- Tordai, H.; Patthy, L. Insertion of spliceosomal introns in proto-splice sites: The case of secretory signal peptides. FEBS Lett. 2004, 575, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Vibranovski, M.D.; Sakabe, N.J.; de Souza, S.J. A possible role of exon-shuffling in the evolution of signal peptides of human proteins. FEBS Lett. 2006, 580, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Xie, Y.; Dai, C.; Zhu, S.; Yin, S.; Wu, Y.; Li, W. Cloning and characterization of a novel calcium channel toxin-like gene BmCa1 from Chinese scorpion Mesobuthus martensii Karsch. Peptides 2006, 27, 1235–1240. [Google Scholar]
- Zhu, S.; Darbon, H.; Dyason, K.; Verdonck, F.; Tytgat, J. Evolutionary origin of inhibitor cystine knot peptides. FASEB J. 2003, 17, 1765–1767. [Google Scholar] [CrossRef] [PubMed]
- Froy, O.; Sagiv, T.; Poreh, M.; Urbach, D.; Zilberberg, N.; Gurevitz, M. Dynamic diversification from a putative common ancestor of scorpion toxins affecting sodium, potassium, and chloride channels. J. Mol. Evol. 1999, 48, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Froy, O.; Gurevitz, M. Arthropod defensins illuminate the divergence of scorpion neurotoxins. J. Pept. Sci. 2004, 10, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Nie, Y.; Zeng, X.C.; Cao, H.; Zhang, L.; Zhou, L.; Yang, Y.; Luo, X.; Liu, Y. Genomic and functional characterization of three new venom peptides from the scorpion Heterometrus spinifer. Peptides 2014, 53, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; de Souza, S.J.; Rosenberg, C.; Gilbert, W. Exon shuffling and the origin of the mitochondrial targeting function in plant cytochrome c1 precursor. Proc. Natl. Acad. Sci. USA 1996, 93, 7727–7731. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Tytgat, J. The scorpine family of defensins: Gene structure, alternative polyadenylation and fold recognition. Cell. Mol. Life Sci. 2004, 61, 1751–1763. [Google Scholar] [CrossRef] [PubMed]
- Froy, O.; Gurevitz, M. Arthropod and mollusk defensins—Evolution by exon-shuffling. Trends Genet. 2003, 19, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Kaessmann, H.; Zöllner, S.; Nekrutenko, A.; Li, W.H. Signatures of domain shuffling in the human genome. Genome Res. 2002, 12, 1642–1650. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Undheim, E.A.; Chan, A.H.; Koludarov, I.; Muñoz-Gómez, S.A.; Antunes, A.; Fry, B.G. Evolution stings: The origin and diversification of scorpion toxin peptide scaffolds. Toxins 2013, 5, 2456–2487. [Google Scholar] [CrossRef] [PubMed]
- Peigneur, S.; Béress, L.; Möller, C.; Marí, F.; Forssmann, W.G.; Tytgat, J. A natural point mutation changes both target selectivity and mechanism of action of sea anemone toxins. FASEB J. 2012, 26, 5141–5151. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Vetter, I.; Lewis, R.J.; Peigneur, S.; Tytgat, J.; Lam, A.; Gallant, E.M.; Beard, N.A.; Alewood, P.F.; Dulhunty, A.F. Multiple actions of phi-LITX-Lw1a on ryanodine receptors reveal a functional link between scorpion DDH and ICK toxins. Proc. Natl. Acad. Sci. USA 2013, 110, 8906–8911. [Google Scholar] [CrossRef] [PubMed]
- Vandendriessche, T.; Kopljar, I.; Jenkins, D.P.; Diego-Garcia, E.; Abdel-Mottaleb, Y.; Vermassen, E.; Clynen, E.; Schoofs, L.; Wulff, H.; Snyders, D.; et al. Purification, molecular cloning and functional characterization of HelaTx1 (Heterometrus laoticus): The first member of a new kappa-KTX subfamily. Biochem. Pharmacol. 2012, 83, 1307–1317. [Google Scholar] [CrossRef] [PubMed]




© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Gao, B.; Zhu, S. Exon Shuffling and Origin of Scorpion Venom Biodiversity. Toxins 2017, 9, 10. https://doi.org/10.3390/toxins9010010
Wang X, Gao B, Zhu S. Exon Shuffling and Origin of Scorpion Venom Biodiversity. Toxins. 2017; 9(1):10. https://doi.org/10.3390/toxins9010010
Chicago/Turabian StyleWang, Xueli, Bin Gao, and Shunyi Zhu. 2017. "Exon Shuffling and Origin of Scorpion Venom Biodiversity" Toxins 9, no. 1: 10. https://doi.org/10.3390/toxins9010010
APA StyleWang, X., Gao, B., & Zhu, S. (2017). Exon Shuffling and Origin of Scorpion Venom Biodiversity. Toxins, 9(1), 10. https://doi.org/10.3390/toxins9010010