Toxins of Prokaryotic Toxin-Antitoxin Systems with Sequence-Specific Endoribonuclease Activity
Abstract
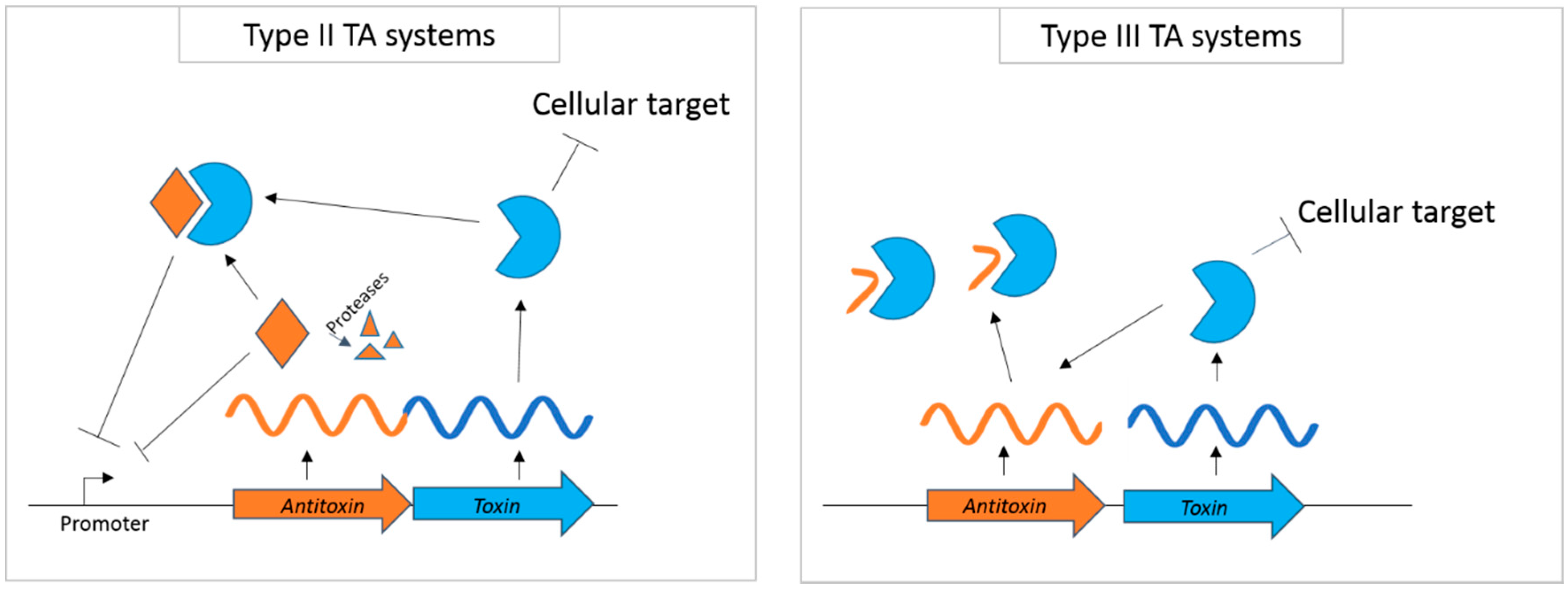
:1. Introduction
2. Families of Sequence-Specific Endoribonuclease TA Toxins
2.1. MazF-ec
2.2. MazF Homologues from M. tuberculosis
2.3. MazF Homologues from Other Gram Positive Bacteria
2.4. MazF-mx
2.5. MazF-hw
2.6. ChpBK
2.7. Kid (PemK)
2.8. VapC
2.9. MqsR
2.10. YhaV
2.11. HicA
2.12. RnlA
2.13. ToxNPa, ToxNBt, AbiQ
3. Physiological Roles
3.1. Plasmid Maintenance and Copy Number
3.2. Regulation of Translation
3.3. Auxiliary Transcriptional Regulation
3.4. Phage Abortive Infection
3.5. Cell Death Pathways (DNA Replication Related)
3.6. Stress Response
3.7. Persister
3.8. Biofilm
3.9. Virulence/Symbiosis and Metabolism
4. Network of TA Systems
4.1. Lon Dependent Degradation of Antitoxin
4.2. Transcriptional Cross-Activation
4.3. Other Regulation
5. Remarks
Acknowledgments
Conflicts of Interest
References
- Yamaguchi, Y.; Park, J.H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, M.G.; Pandey, D.P.; Jaskolska, M.; Gerdes, K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 2009, 191, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Hoeflich, K.P.; Ikura, M.; Qing, G.; Inouye, M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 2003, 12, 913–923. [Google Scholar] [CrossRef]
- Christensen, S.K.; Gerdes, K. RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 2003, 48, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Awano, N.; Inouye, M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol. Microbiol. 2011, 79, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Aakre, C.D.; Phung, T.N.; Huang, D.; Laub, M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell 2013, 52, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Pecota, D.C.; Osapay, G.; Selsted, M.E.; Wood, T.K. Antimicrobial properties of the Escherichia coli R1 plasmid host killing peptide. J. Biotechnol. 2003, 100, 1–12. [Google Scholar] [CrossRef]
- Pandey, D.P.; Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Cheng, H.Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Cook, G.M.; Robson, J.R.; Frampton, R.A.; McKenzie, J.; Przybilski, R.; Fineran, P.C.; Arcus, V.L. Ribonucleases in bacterial toxin-antitoxin systems. Biochim. Biophys. Acta 2013, 1829, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Inouye, M. RatA (YfjG), an Escherichia coli toxin, inhibits 70S ribosome association to block translation initiation. Mol. Microbiol. 2011, 79, 1418–1429. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.; Zavialov, A.V.; Pavlov, M.Y.; Elf, J.; Gerdes, K.; Ehrenberg, M. The bacterial toxin rele displays codon-specific cleavage of mRNAs in the ribosomal a site. Cell 2003, 112, 131–140. [Google Scholar] [CrossRef]
- Kawano, M.; Aravind, L.; Storz, G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.K.; Pedersen, K.; Hansen, F.G.; Gerdes, K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 2003, 332, 809–819. [Google Scholar] [CrossRef]
- Kamada, K.; Hanaoka, F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 2005, 19, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Inouye, M. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 2009, 284, 6627–6638. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yamaguchi, Y.; Inouye, M. Characterization of YafO, an Escherichia coli toxin. J. Biol. Chem. 2009, 284, 25522–25531. [Google Scholar] [CrossRef] [PubMed]
- Prysak, M.H.; Mozdzierz, C.J.; Cook, A.M.; Zhu, L.; Zhang, Y.; Inouye, M.; Woychik, N.A. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 2009, 71, 1071–1087. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Inouye, M. Characterization of the interactions within the MazEF addiction module of Escherichia coli. J. Biol. Chem. 2003, 278, 32300–32306. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhang, Y.; Teh, J.S.; Zhang, J.; Connell, N.; Rubin, H.; Inouye, M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 18638–18643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, L.; Zhang, J.; Inouye, M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 2005, 280, 26080–26088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Hara, H.; Kato, I.; Inouye, M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005, 280, 3143–3150. [Google Scholar] [CrossRef] [PubMed]
- Schifano, J.M.; Edifor, R.; Sharp, J.D.; Ouyang, M.; Konkimalla, A.; Husson, R.N.; Woychik, N.A. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal a site. Proc. Natl. Acad. Sci. USA 2013, 110, 8501–8506. [Google Scholar] [CrossRef] [PubMed]
- Schifano, J.M.; Cruz, J.W.; Vvedenskaya, I.O.; Edifor, R.; Ouyang, M.; Husson, R.N. tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res. 2016, 44, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Schifano, J.M.; Vvedenskaya, I.O.; Knoblauch, J.G.; Ouyang, M.; Nickels, B.E.; Woychik, N.A. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Yamaguchi, Y.; Inouye, M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 2011, 585, 2526–2532. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Nariya, H.; Park, J.H.; Inouye, M. Inhibition of specific gene expressions by protein-mediated mRNA interference. Nat. Commun. 2012, 3, 607. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Inoue, K.; Yoshizumi, S.; Kobayashi, H.; Zhang, Y.; Ouyang, M.; Kato, F.; Sugai, M.; Inouye, M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 2009, 191, 3248–3255. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.F.; Park, J.H.; Prax, M.; Herbig, A.; Nieselt, K.; Rosenstein, R.; Inouye, M.; Bertram, R. Characterization of a MazEF toxin-antitoxin homologue from Staphylococcus equorum. J. Bacteriol. 2013, 195, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Bhatnagar, R. Moxt toxin of Bacillus anthracis exhibits sequence specific ribonuclease activity. Biochem. Biophys. Res. Commun. 2014, 450, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Vesper, O.; Amitai, S.; Belitsky, M.; Byrgazov, K.; Kaberdina, A.C.; Engelberg-Kulka, H.; Moll, I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 2011, 147, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Oron-Gottesman, A.; Sauert, M.; Moll, I.; Engelberg-Kulka, H. A stress-induced bias in the reading of the genetic code in Escherichia coli. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Yoshizumi, S.; Zhang, Y.; Yamaguchi, Y.; Chen, L.; Kreiswirth, B.N.; Inouye, M. Staphylococcus aureus YoeB homologues inhibit translation initiation. J. Bacteriol. 2009, 191, 5868–5872. [Google Scholar] [CrossRef] [PubMed]
- Winther, K.S.; Gerdes, K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. USA 2011, 108, 7403–7407. [Google Scholar] [CrossRef] [PubMed]
- Winther, K.S.; Brodersen, D.E.; Brown, A.K.; Gerdes, K. VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat. Commun. 2013, 4, 2796. [Google Scholar] [CrossRef] [PubMed]
- Temmel, H.; Muller, C.; Sauert, M.; Vesper, O.; Reiss, A.; Popow, J.; Martinez, J.; Moll, I. The RNA ligase RtcB reverses MazF-induced ribosome heterogeneity in Escherichia coli. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kamada, K.; Hanaoka, F.; Burley, S.K. Crystal structure of the MazE/MazF complex: Molecular bases of antidote-toxin recognition. Mol. Cell 2003, 11, 875–884. [Google Scholar] [CrossRef]
- Brown, B.L.; Grigoriu, S.; Kim, Y.; Arruda, J.M.; Davenport, A.; Wood, T.K.; Peti, W.; Page, R. Three dimensional structure of the MqsR:MqsA complex: A novel ta pair comprised of a toxin homologous to rele and an antitoxin with unique properties. PLoS Pathog. 2009, 5, e1000706. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, M.; Borch, J.; Jorgensen, M.G.; Gerdes, K. Messenger RNA interferase RelE controls RelBE transcription by conditional cooperativity. Mol. Microbiol. 2008, 69, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wang, X.; Zhang, X.S.; Grigoriu, S.; Page, R.; Peti, W.; Wood, T.K. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 2010, 12, 1105–1121. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Benedik, M.J.; Wood, T.K. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environ. Microbiol. 2012, 14, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Zhu, L.; Suzuki, M.; Inouye, M. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 2004, 279, 20678–20684. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, B.; Madine, M.A.; de la Cueva-Mendez, G. Kid cleaves specific mRNAs at UUACU sites to rescue the copy number of plasmid R1. EMBO J. 2005, 24, 3459–3469. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Phadtare, S.; Nariya, H.; Ouyang, M.; Husson, R.N.; Inouye, M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 2008, 69, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Nariya, H.; Inouye, M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 2008, 132, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Short, F.L.; Pei, X.Y.; Blower, T.R.; Ong, S.L.; Fineran, P.C.; Luisi, B.F.; Salmond, G.P. Selectivity and self-assembly in the control of a bacterial toxin by an antitoxic noncoding RNA pseudoknot. Proc. Natl. Acad. Sci. USA 2013, 110, E241–249. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, J.L.; Robson, J.; Berney, M.; Smith, T.C.; Ruthe, A.; Gardner, P.P.; Arcus, V.L.; Cook, G.M. A VapBC toxin-antitoxin module is a posttranscriptional regulator of metabolic flux in Mycobacteria. J. Bacteriol. 2012, 194, 2189–2204. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Park, J.H.; Inouye, M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 2009, 284, 28746–28753. [Google Scholar] [CrossRef] [PubMed]
- Schifano, J.M.; Woychik, N.A. 23S rRNA as an a-Maz-ing new bacterial toxin target. RNA Biol. 2014, 11, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Donegan, N.P.; Memmi, G.; Cheung, A.L. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 2007, 189, 8871–8879. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Mao, L.; Inouye, M. Single protein production (SPP) system in Escherichia coli. Nat. Protoc. 2007, 2, 1802–1810. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Zhang, J.; Liu, M.; Woychik, N.A.; Inouye, M. Single protein production in living cells facilitated by an mRNA interferase. Mol. Cell 2005, 18, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Shell, S.S.; Wang, J.; Lapierre, P.; Mir, M.; Chase, M.R.; Pyle, M.M.; Gawande, R.; Ahmad, R.; Sarracino, D.A.; Ioerger, T.R.; et al. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015, 11, e1005641. [Google Scholar] [CrossRef] [PubMed]
- Mets, T.; Lippus, M.; Schryer, D.; Liiv, A.; Kasari, V.; Paier, A.; Maivali, U.; Remme, J.; Tenson, T.; Kaldalu, N. Toxins MazF and MqsR cleave Escherichia coli rRNA precursors at multiple sites. RNA Biol. 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Y.; Zhang, Y.; Chan, M.C.; Mal, T.K.; Hoeflich, K.P.; Inouye, M.; Ikura, M. Characterization of dual substrate binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. J. Mol. Biol. 2006, 357, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Marianovsky, I.; Aizenman, E.; Engelberg-Kulka, H.; Glaser, G. The regulation of the Escherichia coli MazEF promoter involves an unusual alternating palindrome. J. Biol. Chem. 2001, 276, 5975–5984. [Google Scholar] [CrossRef] [PubMed]
- Aizenman, E.; Engelberg-Kulka, H.; Glaser, G. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: A model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 1996, 93, 6059–6063. [Google Scholar] [CrossRef] [PubMed]
- Leviev, I.; Levieva, S.; Garrett, R.A. Role for the highly conserved region of domain IV of 23S-like rRNA in subunit-subunit interactions at the peptidyl transferase centre. Nucleic Acids Res. 1995, 23, 1512–1517. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.K.; Sharma, M.R.; Kiel, M.C.; Hirokawa, G.; Booth, T.M.; Spahn, C.M.; Grassucci, R.A.; Kaji, A.; Frank, J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc. Natl. Acad. Sci. USA 2004, 101, 8900–8905. [Google Scholar] [CrossRef] [PubMed]
- Boynton, T.O.; McMurry, J.L.; Shimkets, L.J. Characterization of Myxococcus xanthus MazF and implications for a new point of regulation. Mol. Microbiol. 2013, 87, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Holkenbrink, C.; Treuner-Lange, A.; Higgs, P.I. Myxococcus xanthus developmental cell fate production: Heterogeneous accumulation of developmental regulatory proteins and reexamination of the role of MazF in developmental lysis. J. Bacteriol. 2012, 194, 3058–3068. [Google Scholar] [CrossRef] [PubMed]
- Tsuchimoto, S.; Nishimura, Y.; Ohtsubo, E. The stable maintenance system Pem of plasmid R100: Degradation of PemI protein may allow PemK protein to inhibit cell growth. J. Bacteriol. 1992, 174, 4205–4211. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Echevarria, M.J.; Berzal-Herranz, A.; Gerdes, K.; Diaz-Orejas, R. The kis and kid genes of the ParD maintenance system of plasmid R1 form an operon that is autoregulated at the level of transcription by the co-ordinated action of the Kis and Kid proteins. Mol. Microbiol. 1991, 5, 2685–2693. [Google Scholar] [CrossRef] [PubMed]
- Tsuchimoto, S.; Ohtsubo, H.; Ohtsubo, E. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J. Bacteriol. 1988, 170, 1461–1466. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; de Torrontegui, G.; Diaz, R. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol. Genet. Genom. MGG 1987, 210, 101–110. [Google Scholar] [CrossRef]
- Arcus, V.L.; McKenzie, J.L.; Robson, J.; Cook, G.M. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng. Des. Sel. PEDS 2011, 24, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Wilbur, J.S.; Chivers, P.T.; Mattison, K.; Potter, L.; Brennan, R.G.; So, M. Neisseria gonorrhoeae FitA interacts with FitB to bind DNA through its ribbon-helix-helix motif. Biochemistry 2005, 44, 12515–12524. [Google Scholar] [CrossRef] [PubMed]
- Katz, M.E.; Strugnell, R.A.; Rood, J.I. Molecular characterization of a genomic region associated with virulence in Dichelobacter nodosus. Infect. Immun. 1992, 60, 4586–4592. [Google Scholar] [PubMed]
- Arcus, V.L.; Rainey, P.B.; Turner, S.J. The PIN-domain toxin-antitoxin array in Mycobacteria. Trends Microbiol. 2005, 13, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Ramage, H.R.; Connolly, L.E.; Cox, J.S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: Implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009, 5, e1000767. [Google Scholar] [CrossRef] [PubMed]
- Robson, J.; McKenzie, J.L.; Cursons, R.; Cook, G.M.; Arcus, V.L. The VapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 2009, 390, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Barrios, A.F.; Zuo, R.; Hashimoto, Y.; Yang, L.; Bentley, W.E.; Wood, T.K. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, b3022). J. Bacteriol. 2006, 188, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Bedzyk, L.A.; Thomas, S.M.; Ye, R.W.; Wood, T.K. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 2004, 64, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 2009, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.L.; Wood, T.K.; Peti, W.; Page, R. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J. Biol. Chem. 2011, 286, 2285–2296. [Google Scholar] [CrossRef] [PubMed]
- Kasari, V.; Kurg, K.; Margus, T.; Tenson, T.; Kaldalu, N. The Escherichia coli mqsR and ygiT genes encode a new toxin-antitoxin pair. J. Bacteriol. 2010, 192, 2908–2919. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, O.; Schuenemann, V.J.; Hand, N.J.; Silhavy, T.J.; Martin, J.; Lupas, A.N.; Djuranovic, S. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J. Mol. Biol. 2007, 372, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Arbing, M.A.; Handelman, S.K.; Kuzin, A.P.; Verdon, G.; Wang, C.; Su, M.; Rothenbacher, F.P.; Abashidze, M.; Liu, M.; Hurley, J.M.; et al. Crystal structures of Phd-Doc, HigA, and YeeU establish multiple evolutionary links between microbial growth-regulating toxin-antitoxin systems. Structure 2010, 18, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Leplae, R.; Geeraerts, D.; Hallez, R.; Guglielmini, J.; Dreze, P.; Van Melderen, L. Diversity of bacterial type II toxin-antitoxin systems: A comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011, 39, 5513–5525. [Google Scholar] [CrossRef] [PubMed]
- Kai, T.; Selick, H.E.; Yonesaki, T. Destabilization of bacteriophage T4 mRNAs by a mutation of gene 61.5. Genetics 1996, 144, 7–14. [Google Scholar] [PubMed]
- Wei, Y.; Gao, Z.Q.; Otsuka, Y.; Naka, K.; Yonesaki, T.; Zhang, H.; Dong, Y.H. Structure-function studies of Escherichia coli RnlA reveal a novel toxin structure involved in bacteriophage resistance. Mol. Microbiol. 2013, 90, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Koga, M.; Iwamoto, A.; Yonesaki, T. A role of RnlA in the RNase LS activity from Escherichia coli. Genes Genet. Syst. 2007, 82, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Naka, K.; Koga, M.; Yonesaki, T.; Otsuka, Y. RNase HI stimulates the activity of RnlA toxin in Escherichia coli. Mol. Microbiol. 2014, 91, 596–605. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Yonesaki, T. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 2012, 83, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Goeders, N.; Chai, R.; Chen, B.; Day, A.; Salmond, G.P. Structure, evolution, and functions of bacterial type III toxin-antitoxin systems. Toxins 2016, 8, 282. [Google Scholar] [CrossRef] [PubMed]
- Blower, T.R.; Pei, X.Y.; Short, F.L.; Fineran, P.C.; Humphreys, D.P.; Luisi, B.F.; Salmond, G.P. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat. Struct. Mol. Biol. 2011, 18, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P. The phage abortive infection system, toxin, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Rao, F.; Short, F.L.; Voss, J.E.; Blower, T.R.; Orme, A.L.; Whittaker, T.E.; Luisi, B.F.; Salmond, G.P. Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein-RNA toxin-antitoxin system. Nucleic Acids Res. 2015, 43, 9529–9540. [Google Scholar] [CrossRef] [PubMed]
- Samson, J.E.; Spinelli, S.; Cambillau, C.; Moineau, S. Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type III toxin-antitoxin system. Mol. Microbiol. 2013, 87, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef] [PubMed]
- Hayes, F. Toxins-antitoxins: Plasmid maintenance, programmed cell death, and cell cycle arrest. Science 2003, 301, 1496–1499. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.; Sat, B.; Reches, M.; Engelberg-Kulka, H. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 2001, 183, 2046–2050. [Google Scholar] [CrossRef] [PubMed]
- Tsilibaris, V.; Maenhaut-Michel, G.; Mine, N.; Van Melderen, L. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 2007, 189, 6101–6108. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wood, T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011, 77, 5577–5583. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kim, Y.; Hong, S.H.; Ma, Q.; Brown, B.L.; Pu, M.; Tarone, A.M.; Benedik, M.J.; Peti, W.; Page, R.; et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 2011, 7, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef] [PubMed]
- Lehnherr, H.; Maguin, E.; Jafri, S.; Yarmolinsky, M.B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 1993, 233, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Hanawalt, P. Sedimentation analysis of deoxyribonucleic acid from thymine-starved Escherichia coli. J. Bacteriol. 1975, 121, 537–547. [Google Scholar] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Pecota, D.C.; Wood, T.K. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 1996, 178, 2044–2050. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Yonesaki, T. A novel endoribonuclease, RNase LS, in Escherichia coli. Genetics 2005, 169, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Yonesaki, T. Recognition and specific degradation of bacteriophage T4 mRNAs. Genetics 2001, 158, 7–17. [Google Scholar] [PubMed]
- Koga, M.; Otsuka, Y.; Lemire, S.; Yonesaki, T. Escherichia coli RnlA and RnlB compose a novel toxin-antitoxin system. Genetics 2011, 187, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Gao, Z.; Zhang, H.; Dong, Y. Structural characterizations of phage antitoxin Dmd and its interactions with bacterial toxin RnlA. Biochem. Biophys. Res. Commun. 2016, 472, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.; Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genom. MGG 2004, 272, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Alawneh, A.M.; Qi, D.; Yonesaki, T.; Otsuka, Y. An ADP-ribosyltransferase Alt of bacteriophage T4 negatively regulates the Escherichia coli MazF toxin of a toxin-antitoxin module. Mol. Microbiol. 2016, 99, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Otsuka, Y.; Gao, Z.Q.; Wei, Y.; Chen, Z.; Masuda, M.; Yonesaki, T.; Zhang, H.; Dong, Y.H. Structural insights into the inhibition mechanism of bacterial toxin LsoA by bacteriophage antitoxin Dmd. Mol. Microbiol. 2016, 101, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Blower, T.R.; Evans, T.J.; Przybilski, R.; Fineran, P.C.; Salmond, G.P. Viral evasion of a bacterial suicide system by RNA-based molecular mimicry enables infectious altruism. PLoS Genet. 2012, 8, e1003023. [Google Scholar] [CrossRef] [PubMed]
- Sberro, H.; Leavitt, A.; Kiro, R.; Koh, E.; Peleg, Y.; Qimron, U.; Sorek, R. Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol. Cell 2013, 50, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Barner, H.D.; Cohen, S.S. The induction of thymine synthesis by T2 infection of a thymine requiring mutant of Escherichia coli. J. Bacteriol. 1954, 68, 80–88. [Google Scholar] [PubMed]
- Sangurdekar, D.P.; Hamann, B.L.; Smirnov, D.; Srienc, F.; Hanawalt, P.C.; Khodursky, A.B. Thymineless death is associated with loss of essential genetic information from the replication origin. Mol. Microbiol. 2010, 75, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Freifelder, D. Single-strand breaks in bacterial DNA associated with thymine starvation. J. Mol. Biol. 1969, 45, 1–7. [Google Scholar] [CrossRef]
- Guarino, E.; Salguero, I.; Jimenez-Sanchez, A.; Guzman, E.C. Double-strand break generation under deoxyribonucleotide starvation in Escherichia coli. J. Bacteriol. 2007, 189, 5782–5786. [Google Scholar] [CrossRef] [PubMed]
- Fonville, N.C.; Bates, D.; Hastings, P.J.; Hanawalt, P.C.; Rosenberg, S.M. Role of reca and the sos response in thymineless death in Escherichia coli. PLoS Genet. 2010, 6, e1000865. [Google Scholar] [CrossRef] [PubMed]
- Erental, A.; Sharon, I.; Engelberg-Kulka, H. Two programmed cell death systems in Escherichia coli: An apoptotic-like death is inhibited by the mazef-mediated death pathway. PLoS Biol. 2012, 10, e1001281. [Google Scholar] [CrossRef] [PubMed]
- Sat, B.; Reches, M.; Engelberg-Kulka, H. The Escherichia coli mazef suicide module mediates thymineless death. J. Bacteriol. 2003, 185, 1803–1807. [Google Scholar] [CrossRef] [PubMed]
- Timson, J. Hydroxyurea. Mutat. Res. 1975, 32, 115–132. [Google Scholar] [CrossRef]
- Foti, J.J.; Schienda, J.; Sutera, V.A., Jr.; Lovett, S.T. A bacterial g protein-mediated response to replication arrest. Mol. Cell 2005, 17, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.W.; Kohanski, M.A.; Simmons, L.A.; Winkler, J.A.; Collins, J.J.; Walker, G.C. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol. Cell 2009, 36, 845–860. [Google Scholar] [CrossRef] [PubMed]
- Godoy, V.G.; Jarosz, D.F.; Walker, F.L.; Simmons, L.A.; Walker, G.C. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J. 2006, 25, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Christensen-Dalsgaard, M.; Jorgensen, M.G.; Gerdes, K. Three new rele-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 2010, 75, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Zhang, Z.; Khodursky, A.; Kaldalu, N.; Kurg, K.; Lewis, K. Persisters: A distinct physiological state of E. coli. BMC Microbiol. 2006, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Dorr, T.; Vulic, M.; Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Hong, S.H.; Peti, W.; Benedik, M.J.; Page, R.; Wood, T.K. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 2013, 15, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Kwan, B.W.; Lord, D.M.; Peti, W.; Page, R.; Benedik, M.J.; Wood, T.K. The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ. Microbiol. 2015, 17, 3168–3181. [Google Scholar] [CrossRef] [PubMed]
- Hengge, R. The two-component network and the general stress sigma factor Rpos (σs) in Escherichia coli. In Bacterial Signal Transduction: Networks and Drug Target; Utsumi, R., Ed.; Springer: New York, NY, USA, 2008; pp. 40–53. [Google Scholar]
- Maezato, Y.; Daugherty, A.; Dana, K.; Soo, E.; Cooper, C.; Tachdjian, S.; Kelly, R.M.; Blum, P. VapC6, a ribonucleolytic toxin regulates thermophilicity in the crenarchaeote Sulfolobus solfataricus. RNA 2011, 17, 1381–1392. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Georgopoulos, C. Identification, cloning, and characterization of the Escherichia coli sohA gene, a suppressor of the htrA (degP) null phenotype. J. Bacteriol. 1990, 172, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Hand, N.J.; Silhavy, T.J. Null mutations in a nudix gene, ygdP, implicate an alarmone response in a novel suppression of hybrid jamming. J. Bacteriol. 2003, 185, 6530–6539. [Google Scholar] [CrossRef] [PubMed]
- Snyder, W.B.; Silhavy, T.J. Enhanced export of beta-galactosidase fusion proteins in prlF mutants is Lon dependent. J. Bacteriol. 1992, 174, 5661–5668. [Google Scholar] [CrossRef] [PubMed]
- Daimon, Y.; Narita, S.; Akiyama, Y. Activation of toxin-antitoxin system toxins suppresses lethality caused by the loss of sigmaE in Escherichia coli. J. Bacteriol. 2015, 197, 2316–2324. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Castro-Camargo, M.; Gerdes, K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 2013, 154, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef] [PubMed]
- Germain, E.; Roghanian, M.; Gerdes, K.; Maisonneuve, E. Stochastic induction of persister cells by HipA through (p)ppgpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. USA 2015, 112, 5171–5176. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Moyed, H.S.; Bertrand, K.P. HipA, a newly recognized gene of Escherichia coli k-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983, 155, 768–775. [Google Scholar] [PubMed]
- Falla, T.J.; Chopra, I. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of HipA. Antimicrob. Agents Chemother. 1998, 42, 3282–3284. [Google Scholar] [PubMed]
- Rotem, E.; Loinger, A.; Ronin, I.; Levin-Reisman, I.; Gabay, C.; Shoresh, N.; Biham, O.; Balaban, N.Q. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. USA 2010, 107, 12541–12546. [Google Scholar] [CrossRef] [PubMed]
- Kaspy, I.; Rotem, E.; Weiss, N.; Ronin, I.; Balaban, N.Q.; Glaser, G. Hipa-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 2013, 4, 3001. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Awano, N.; Masuda, H.; Park, J.H.; Inouye, M. Transcriptional repressor HipB regulates the multiple promoters in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2013, 23, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Shakespeare, L.J.; Jorgensen, M.G.; Gerdes, K. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. USA 2011, 108, 13206–13211. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, N.; Kwan, B.W.; Wood, T.K. Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci. Rep. 2016, 6, 20519. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Wang, X.; O’Connor, H.F.; Benedik, M.J.; Wood, T.K. Bacterial persistence increases as environmental fitness decreases. Microb. Biotechnol. 2012, 5, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Hammar, M.; Arnqvist, A.; Bian, Z.; Olsen, A.; Normark, S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli k-12. Mol. Microbiol. 1995, 18, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Brombacher, E.; Baratto, A.; Dorel, C.; Landini, P. Gene expression regulation by the curli activator csgd protein: Modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 2006, 188, 2027–2037. [Google Scholar] [CrossRef] [PubMed]
- Da Re, S.; Ghigo, J.M. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 2006, 188, 3073–3087. [Google Scholar] [CrossRef] [PubMed]
- Bloom-Ackermann, Z.; Steinberg, N.; Rosenberg, G.; Oppenheimer-Shaanan, Y.; Pollack, D.; Ely, S.; Storzi, N.; Levy, A.; Kolodkin-Gal, I. Toxin-antitoxin systems eliminate defective cells and preserve symmetry in Bacillus subtilis biofilms. Environ. Microbiol. 2016, 18, 5032–5047. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.J.; Wade, W.D.; Akierman, S.; Vacchi-Suzzi, C.; Stremick, C.A.; Turner, R.J.; Ceri, H. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 2009, 53, 2253–2258. [Google Scholar] [CrossRef] [PubMed]
- Hopper, S.; Wilbur, J.S.; Vasquez, B.L.; Larson, J.; Clary, S.; Mehr, I.J.; Seifert, H.S.; So, M. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect. Immun. 2000, 68, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.J.; Delogu, G. The PE multigene family: A ‘molecular mantra’ for Mycobacteria. Trends Microbiol. 2002, 10, 246–249. [Google Scholar] [CrossRef]
- Van Pittius, N.C.G.; Sampson, S.L.; Lee, H.; Kim, Y.; van Helden, P.D.; Warren, R.M. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the RSAT-6 (esx) gene cluster regions. BMC Evol. Biol. 2006, 6, 95. [Google Scholar]
- Bodogai, M.; Ferenczi, S.; Bashtovyy, D.; Miclea, P.; Papp, P.; Dusha, I. The ntrPR operon of Sinorhizobium meliloti is organized and functions as a toxin-antitoxin module. Mol. Plant-Microbe Interact. MPMI 2006, 19, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Olah, B.; Kiss, E.; Gyorgypal, Z.; Borzi, J.; Cinege, G.; Csanadi, G.; Batut, J.; Kondorosi, A.; Dusha, I. Mutation in the ntrR gene, a member of the vap gene family, increases the symbiotic efficiency of Sinorhizobium meliloti. Mol. Plant-Microbe Interact. MPMI 2001, 14, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Kasari, V.; Mets, T.; Tenson, T.; Kaldalu, N. Transcriptional cross-activation between toxin-antitoxin systems of Escherichia coli. BMC Microbiol. 2013, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Winther, K.S.; Gerdes, K. Ectopic production of VapCs from enterobacteria inhibits translation and trans-activates YoeB mRNA interferase. Mol. Microbiol. 2009, 72, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.K.; Mikkelsen, M.; Pedersen, K.; Gerdes, K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 2001, 98, 14328–14333. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.K.; Maenhaut-Michel, G.; Mine, N.; Gottesman, S.; Gerdes, K.; Van Melderen, L. Overproduction of the lon protease triggers inhibition of translation in Escherichia coli: Involvement of the YefM- YoeB toxin-antitoxin system. Mol. Microbiol. 2004, 51, 1705–1717. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pino, A.; Balasubramanian, S.; Wyns, L.; Gazit, E.; De Greve, H.; Magnuson, R.D.; Charlier, D.; van Nuland, N.A.; Loris, R. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 2010, 142, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Sharp, J.D.; Kobayashi, H.; Woychik, N.A.; Inouye, M. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J. Biol. Chem. 2010, 285, 39732–39738. [Google Scholar] [CrossRef] [PubMed]
| Toxins | Types | Sources | Consensus Sequences (Reference) | Substrates | |||
|---|---|---|---|---|---|---|---|
| mRNA | rRNA | tRNA | tmRNA | ||||
| MazF/ChpBK | |||||||
| MazF-ec | II | E. coli | A^CA [3] | + | 16S | ||
| ChpBK | E. coli | ^ACY and A^CY [22] | + | ||||
| Kid (PemK) | R100 and R1 plasmid | U^AH and UA^H [43] | + | ||||
| UU^ACU [44] | |||||||
| MazF-mt1 | M. tuberculosis | U^AC [21] *1 | + | ||||
| MazF-mt3 | M. tuberculosis | U^CCUU [26] | + | 16S, 23S | |||
| MazF-mt6 | M. tuberculosis | UU^CCU [24] | + | 23S | |||
| MazF-mt7 | M. tuberculosis | U^CGCU [45] *2 | + | ||||
| MazF-mt9 | M. tuberculosis | UU^U [25] | + | + | |||
| MazF-mx | M. xanthus | GU^UGC [46] *3 | + | ||||
| MazF-hw | H. walsbyi | UU^ACUCA [28] | + | ||||
| MazF-bs | B. subtilis | U^ACAU [27] | + | ||||
| MoxT | B. anthracis | U^ACAU [31] | + | ||||
| MazF | S. aureus | UACAU [29] | + | ||||
| MazF | S. equorum | U^ACAU [30] | + | ||||
| ToxNPa | III | P. atrosepticum | AA^AK [47] | + | |||
| ToxNBt | B. thuringiensis | A^AAAA [47] | + | ||||
| AbiQ | L. lactis | n.d. | + | ||||
| Homologue of phage t4 RNase H with PIN domain | |||||||
| VapC | II | M. tuberculosis | AU^AW-hairpin-G- [48] | + | |||
| VapC | S. flexneri 2a and S. enterica | Anticodon stem-loop of tRNAfMet [35] | - | - | + | - | |
| RelE super family | |||||||
| MqsR | II | E. coli | GCU and GCA [49] *4 | + | |||
| YhaV | E. coli | n.d. | + | 16S, 23S | |||
| No known homologues | |||||||
| HicA | II | E. coli | n.d. for mRNA; | + | + | ||
| A^AAC in tmRNA [2] | |||||||
| RnlA | E. coli | n.d. | + | ||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masuda, H.; Inouye, M. Toxins of Prokaryotic Toxin-Antitoxin Systems with Sequence-Specific Endoribonuclease Activity. Toxins 2017, 9, 140. https://doi.org/10.3390/toxins9040140
Masuda H, Inouye M. Toxins of Prokaryotic Toxin-Antitoxin Systems with Sequence-Specific Endoribonuclease Activity. Toxins. 2017; 9(4):140. https://doi.org/10.3390/toxins9040140
Chicago/Turabian StyleMasuda, Hisako, and Masayori Inouye. 2017. "Toxins of Prokaryotic Toxin-Antitoxin Systems with Sequence-Specific Endoribonuclease Activity" Toxins 9, no. 4: 140. https://doi.org/10.3390/toxins9040140
APA StyleMasuda, H., & Inouye, M. (2017). Toxins of Prokaryotic Toxin-Antitoxin Systems with Sequence-Specific Endoribonuclease Activity. Toxins, 9(4), 140. https://doi.org/10.3390/toxins9040140






