Automated Analysis of Platelet Aggregation on Cultured Endothelium in a Microfluidic Chip Perfused with Human Whole Blood
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microfluidic Device Fabrication
2.2. Endothelial Cell Seeding and Introduction of Inflammatory Cytokines
2.3. Whole Blood Perfusion
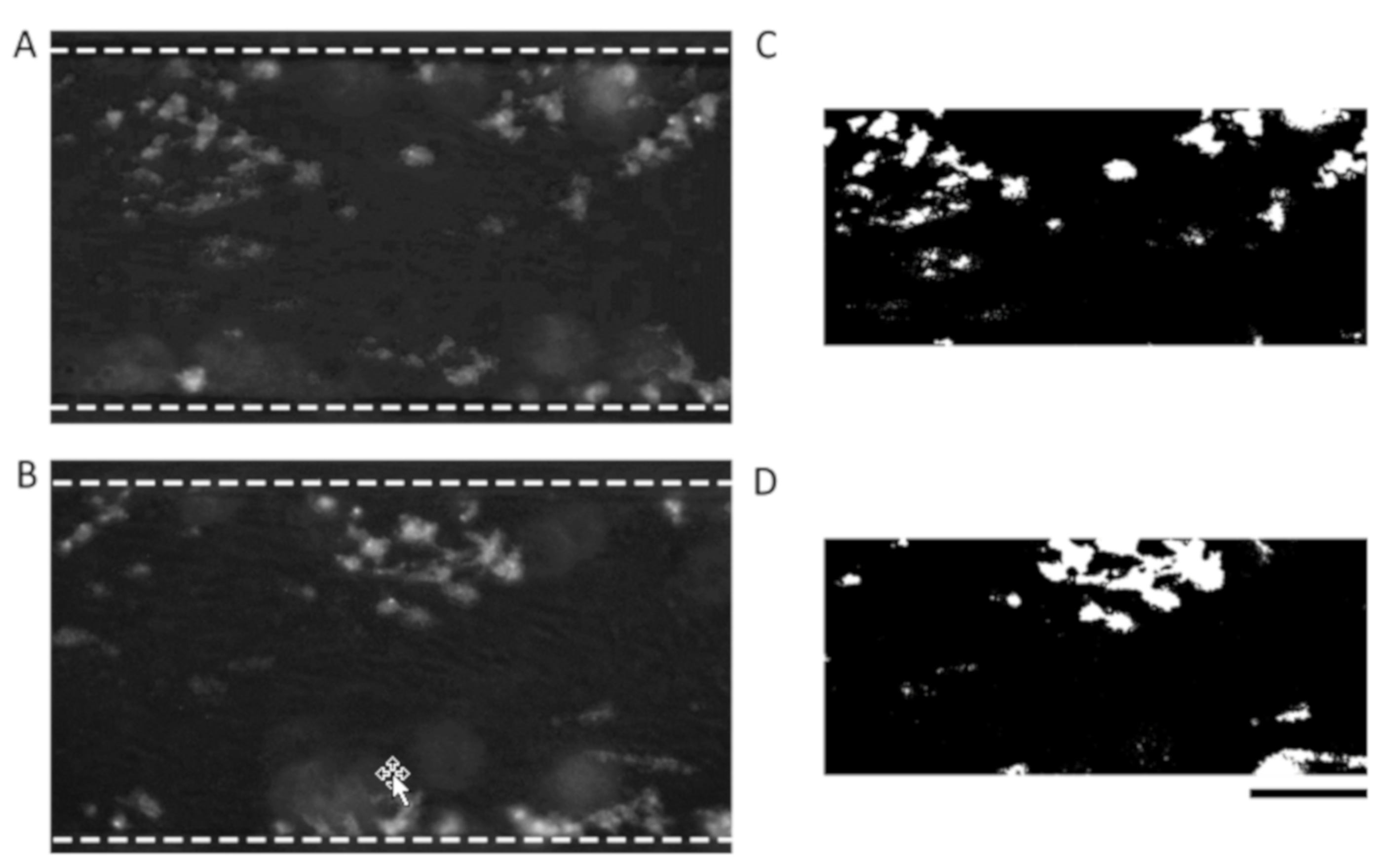
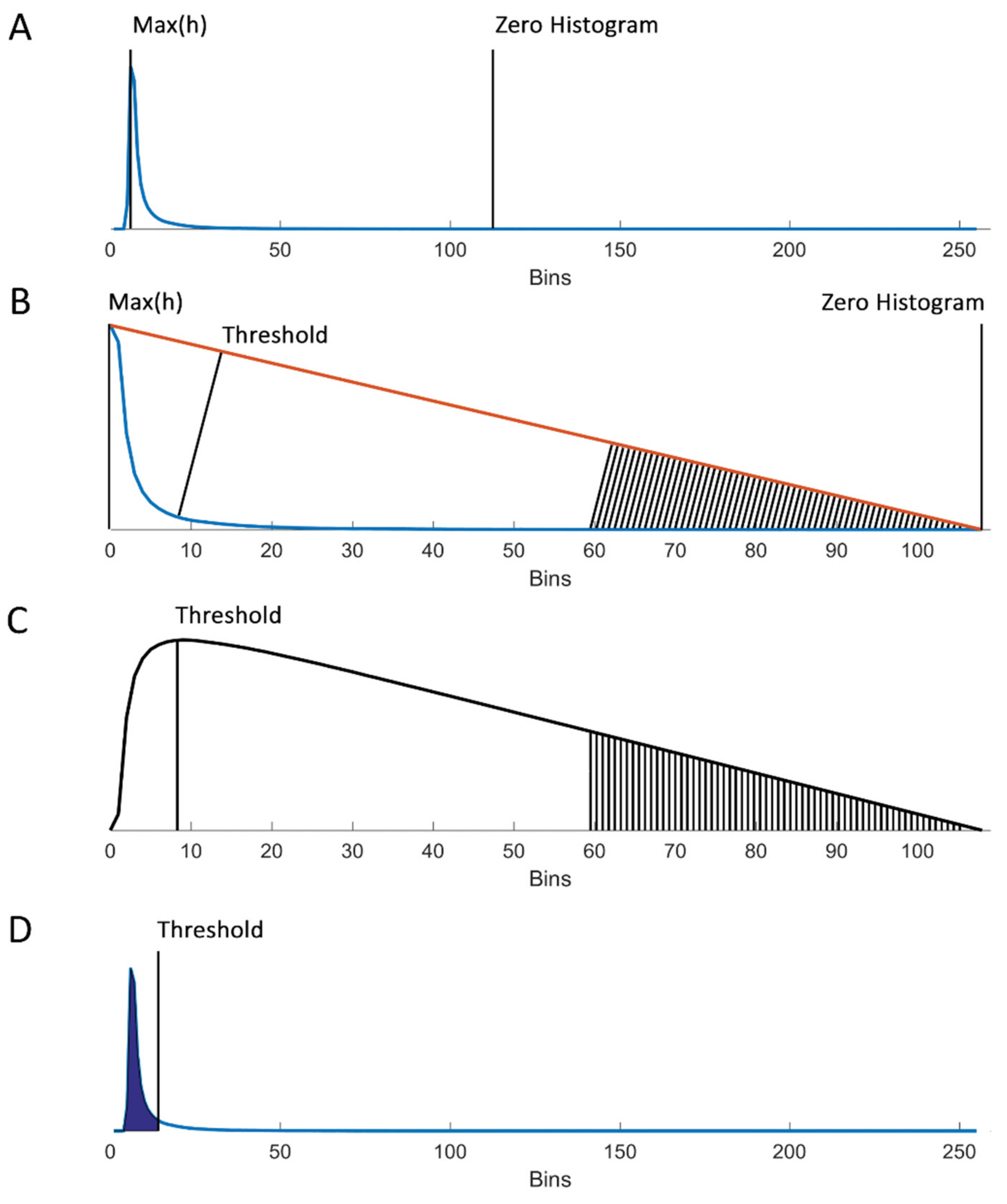
2.4. Determination of an ROI and Triangle Thresholding
2.5. Manual Identification of Platelet Aggregates and Statistics
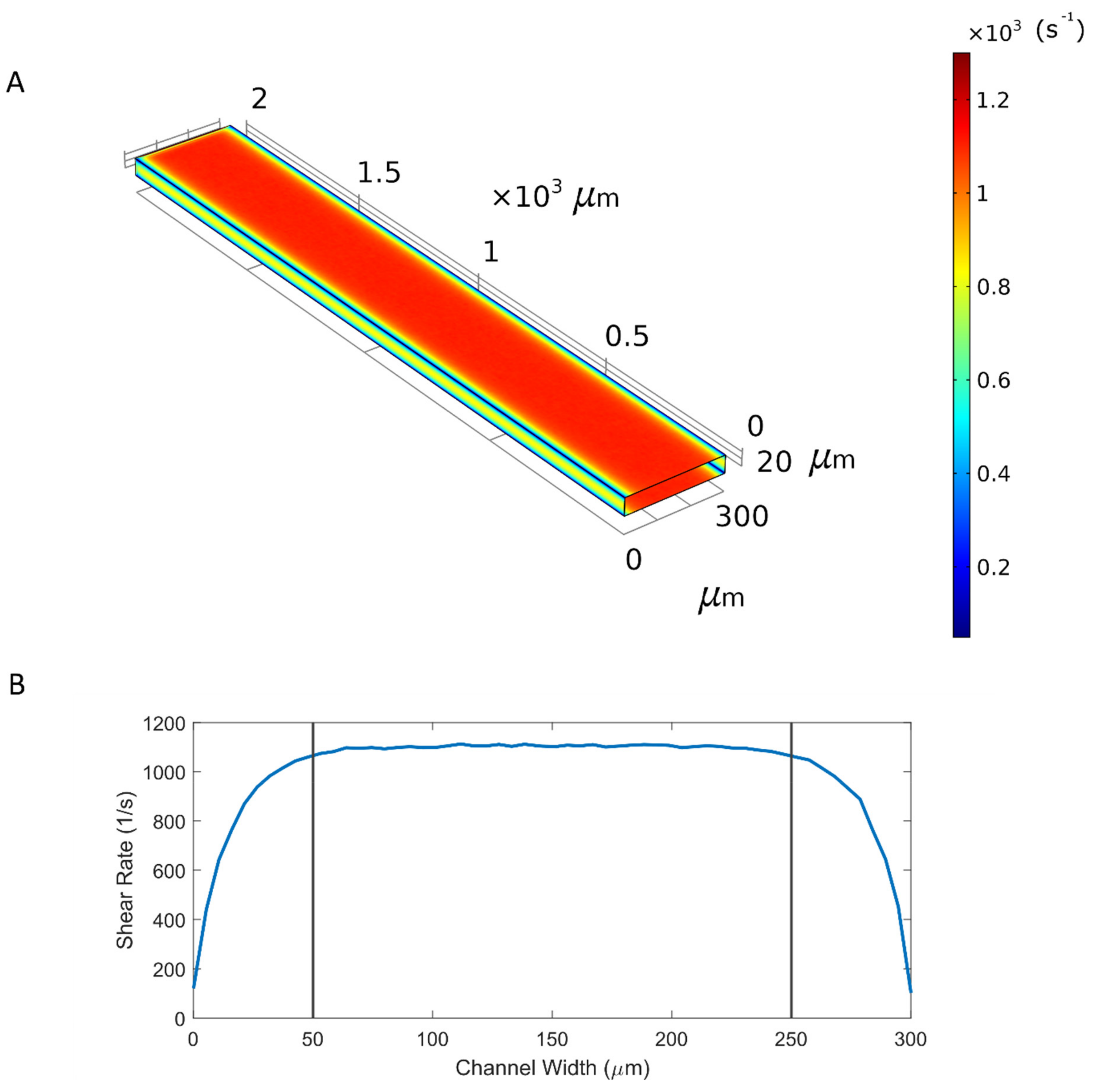
2.6. Computational Fluid Dynamics
3. Results
3.1. Microfluidic Device and Introduction of Data
3.2. Automated Threshold Using the Triangle Method
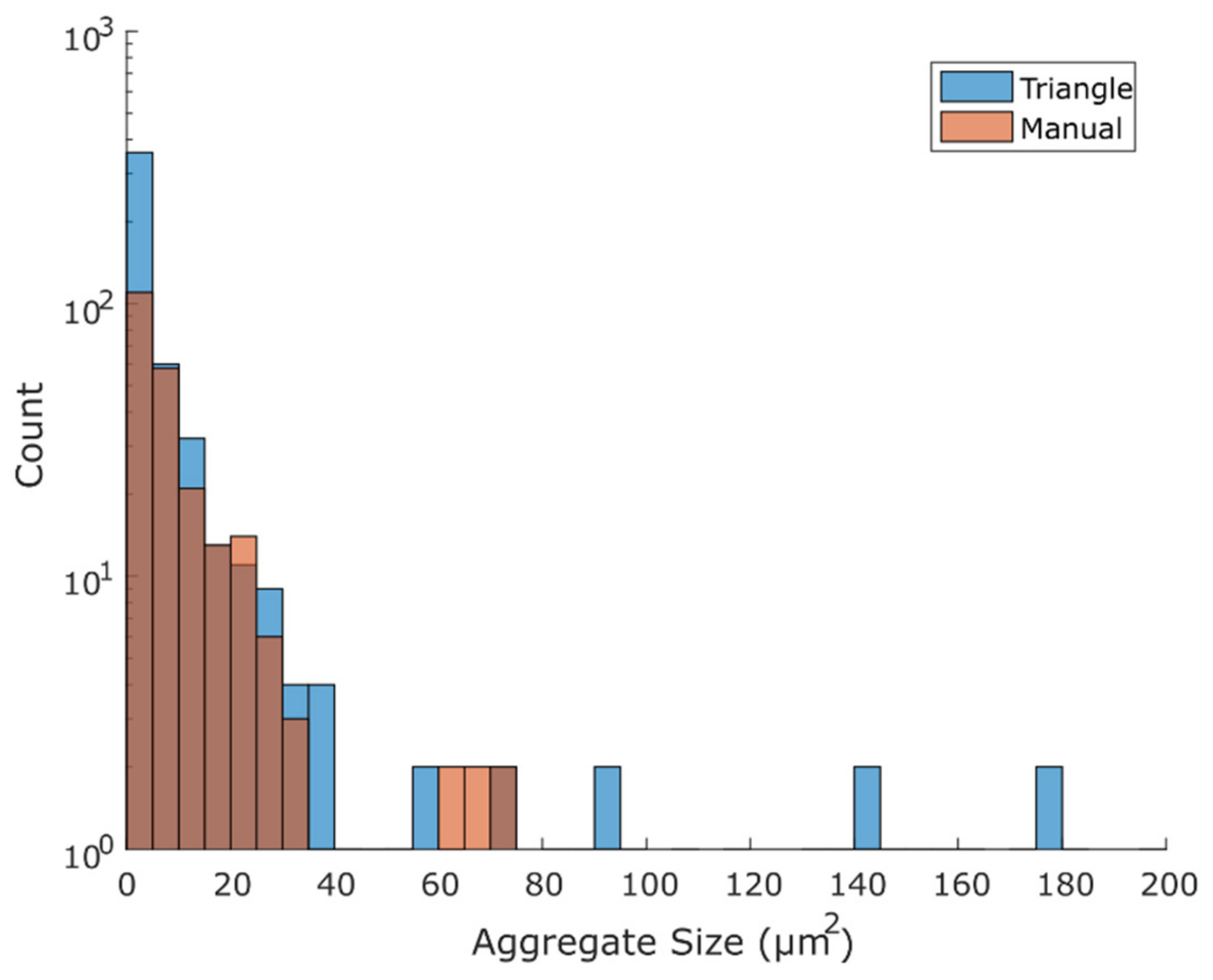
3.3. Comparison of Binary Output Using the Triangle Method with the Manual Method
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- WHO. Fact Sheet—Cardiovascular Diseases. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 15 July 2019).
- Zaragoza, C.; Gomez-Guerrero, C.; Martin-Ventura, J.L.; Blanco-Colio, L.; Lavin, B.; Mallavia, B.; Tarin, C.; Mas, S.; Ortiz, A.; Egido, J. Animal models of cardiovascular diseases. J. Biomed. Biotechnol. 2011, 2011, 497841. [Google Scholar] [CrossRef]
- Jirouskova, M.; Shet, A.S.; Johnson, G.J. A guide to murine platelet structure, function, assays, and genetic alterations. J. Thromb. Haemost. 2007, 5, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Suo, J.; Ferrara, D.E.; Sorescu, D.; Guldberg, R.E.; Taylor, W.R.; Giddens, D.P. Hemodynamic shear stresses in mouse aortas: Implications for atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 346–351. [Google Scholar] [CrossRef]
- Costa, P.F.; Albers, H.J.; Linssen, J.E.A.; Middelkamp, H.H.T.; van der Hout, L.; Passier, R.; van den Berg, A.; Malda, J.; van der Meer, A.D. Mimicking arterial thrombosis in a 3D-printed microfluidic in vitro vascular model based on computed tomography angiography data. Lab Chip 2017, 17, 2785–2792. [Google Scholar] [CrossRef]
- Westein, E.; van der Meer, A.D.; Kuijpers, M.J.E.; Frimat, J.-P.; van den Berg, A.; Heemskerk, J.W.M. Atherosclerotic geometries exacerbate pathological thrombus formation poststenosis in a von Willebrand factor-dependent manner. Proc. Natl. Acad. Sci. USA 2013, 110, 1357–1362. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Davoudi, F.; Walch, P.; Manbachi, A.; Luo, X.; Dell’Erba, V.; Miri, A.K.; Albadawi, H.; Arneri, A.; Li, X.; et al. Bioprinted thrombosis-on-a-chip. Lab Chip 2016, 16, 4097–4105. [Google Scholar] [CrossRef]
- Jain, A.; van der Meer, A.D.; Papa, A.-L.; Barrile, R.; Lai, A.; Schlechter, B.L.; Otieno, M.A.; Louden, C.S.; Hamilton, G.A.; Michelson, A.D.; et al. Assessment of whole blood thrombosis in a microfluidic device lined by fixed human endothelium. Biomed. Microdevices 2016, 18, 73. [Google Scholar] [CrossRef]
- Jain, A.; Barrile, R.; van der Meer, A.D.; Mammoto, A.; Mammoto, T.; De Ceunynck, K.; Aisiku, O.; Otieno, M.A.; Louden, C.S.; Hamilton, G.A.; et al. Primary Human Lung Alveolus-on-a-chip Model of Intravascular Thrombosis for Assessment of Therapeutics. Clin. Pharmacol. Ther. 2018, 103, 332–340. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, J.; Craven, M.; Choi, N.W.; Totorica, S.; Diaz-Santana, A.; Kermani, P.; Hempstead, B.; Fischbach-Teschl, C.; López, J.A.; et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc. Natl. Acad. Sci. USA 2012, 109, 9342–9347. [Google Scholar] [CrossRef]
- Barrile, R.; van der Meer, A.D.; Park, H.; Fraser, J.P.; Simic, D.; Teng, F.; Conegliano, D.; Nguyen, J.; Jain, A.; Zhou, M.; et al. Organ-on-Chip Recapitulates Thrombosis Induced by an anti-CD154 Monoclonal Antibody: Translational Potential of Advanced Microengineered Systems. Clin. Pharmacol. Ther. 2018, 104, 1240–1248. [Google Scholar] [CrossRef]
- Mathur, T.; Singh, K.A.; R. Pandian, N.K.; Tsai, S.-H.; Hein, T.W.; Gaharwar, A.K.; Flanagan, J.M.; Jain, A. Organ-on-chips made of blood: Endothelial progenitor cells from blood reconstitute vascular thromboinflammation in vessel-chips. Lab Chip 2019, 19, 2500–2511. [Google Scholar] [CrossRef]
- Menon, N.V.; Tay, H.M.; Wee, S.N.; Li, K.H.H.; Hou, H.W. Micro-engineered perfusable 3D vasculatures for cardiovascular diseases. Lab Chip 2017, 17, 2960–2968. [Google Scholar] [CrossRef]
- Mannino, R.G.; Myers, D.R.; Ahn, B.; Wang, Y.; Rollins, M.; Gole, H.; Lin, A.S.; Guldberg, R.E.; Giddens, D.P.; Timmins, L.H.; et al. “Do-it-yourself in vitro vasculature that recapitulates in vivo geometries for investigating endothelial-blood cell interactions”. Sci. Rep. 2015, 5, 12401. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.; Friman, O.; Guertin, D.A.; Chang, J.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef]
- Sala, L.; van Meer, B.J.; Tertoolen, L.G.J.; Bakkers, J.; Bellin, M.; Davis, R.P.; Denning, C.; Dieben, M.A.E.; Eschenhagen, T.; Giacomelli, E.; et al. MUSCLEMOTION. Circ. Res. 2018, 122, e5–e16. [Google Scholar] [CrossRef]
- Probst, C.; Schneider, S.; Loskill, P. High-throughput organ-on-a-chip systems: Current status and remaining challenges. Curr. Opin. Biomed. Eng. 2018, 6, 33–41. [Google Scholar] [CrossRef]
- Six, K.R.; Devloo, R.; Van Aelst, B.; Vandekerckhove, P.; Feys, H.B.; Compernolle, V. A Microfluidic Flow Chamber Model for Platelet Transfusion and Hemostasis Measures Platelet Deposition and Fibrin Formation in Real-time. J. Vis. Exp. 2017, e55351. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, A.; Ju, L.; Passam, F. Straight Channel Microfluidic Chips for the Study of Platelet Adhesion under Flow. Bio. Protoc. 2019, 9. [Google Scholar] [CrossRef]
- de Witt, S.M.; Swieringa, F.; Cavill, R.; Lamers, M.M.E.; van Kruchten, R.; Mastenbroek, T.; Baaten, C.; Coort, S.; Pugh, N.; Schulz, A.; et al. Identification of platelet function defects by multi-parameter assessment of thrombus formation. Nat. Commun. 2014, 5, 4257. [Google Scholar] [CrossRef]
- Hansen, R.R.; Tipnis, A.A.; White-Adams, T.C.; Di Paola, J.A.; Neeves, K.B. Characterization of collagen thin films for von Willebrand factor binding and platelet adhesion. Langmuir 2011, 27, 13648–13658. [Google Scholar] [CrossRef] [PubMed]
- Neeves, K.B.; Onasoga, A.A.; Hansen, R.R.; Lilly, J.J.; Venckunaite, D.; Sumner, M.B.; Irish, A.T.; Brodsky, G.; Manco-Johnson, M.J.; Di Paola, J.A. Sources of Variability in Platelet Accumulation on Type 1 Fibrillar Collagen in Microfluidic Flow Assays. PLoS ONE 2013, 8, e54680. [Google Scholar] [CrossRef]
- Matsui, H.; Sugimoto, M.; Mizuno, T.; Tsuji, S.; Miyata, S.; Matsuda, M.; Yoshioka, A. Distinct and concerted functions of von Willebrand factor and fibrinogen in mural thrombus growth under high shear flow. Blood 2002, 100, 3604–3610. [Google Scholar] [CrossRef] [PubMed]
- Colace, T.; Falls, E.; Zheng, X.L.; Diamond, S.L. Analysis of morphology of platelet aggregates formed on collagen under laminar blood flow. Ann. Biomed. Eng. 2011, 39, 922–929. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man. Cybern. 1979, C, 62–66. [Google Scholar] [CrossRef]
- Zack, G.W.; Rogers, E.; Latt, S.A. Automatic Measurement of Sister Chromatid Exchange Frequency. J. Histochem. Cytochem. 1977, 25, 741–753. [Google Scholar] [CrossRef]
- van der Helm, M.W.; van der Meer, A.D.; Eijkel, J.C.T.; van den Berg, A.; Segerink, L.I. Microfluidic organ-on-chip technology for blood-brain barrier research. Tissue Barriers 2016, 4, e1142493. [Google Scholar] [CrossRef]
- Hromada, C.; Mühleder, S.; Grillari, J.; Redl, H.; Holnthoner, W. Endothelial Extracellular Vesicles—Promises and Challenges. Front. Physiol. 2017, 8, 275. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albers, H.J.; Passier, R.; van den Berg, A.; van der Meer, A.D. Automated Analysis of Platelet Aggregation on Cultured Endothelium in a Microfluidic Chip Perfused with Human Whole Blood. Micromachines 2019, 10, 781. https://doi.org/10.3390/mi10110781
Albers HJ, Passier R, van den Berg A, van der Meer AD. Automated Analysis of Platelet Aggregation on Cultured Endothelium in a Microfluidic Chip Perfused with Human Whole Blood. Micromachines. 2019; 10(11):781. https://doi.org/10.3390/mi10110781
Chicago/Turabian StyleAlbers, Hugo J., Robert Passier, Albert van den Berg, and Andries D. van der Meer. 2019. "Automated Analysis of Platelet Aggregation on Cultured Endothelium in a Microfluidic Chip Perfused with Human Whole Blood" Micromachines 10, no. 11: 781. https://doi.org/10.3390/mi10110781
APA StyleAlbers, H. J., Passier, R., van den Berg, A., & van der Meer, A. D. (2019). Automated Analysis of Platelet Aggregation on Cultured Endothelium in a Microfluidic Chip Perfused with Human Whole Blood. Micromachines, 10(11), 781. https://doi.org/10.3390/mi10110781






