Portable Rice Disease Spores Capture and Detection Method Using Diffraction Fingerprints on Microfluidic Chip
Abstract
:1. Introduction
2. Materials and Methods
2.1. Design of Microfluidic Chip
2.2. Numerical Method
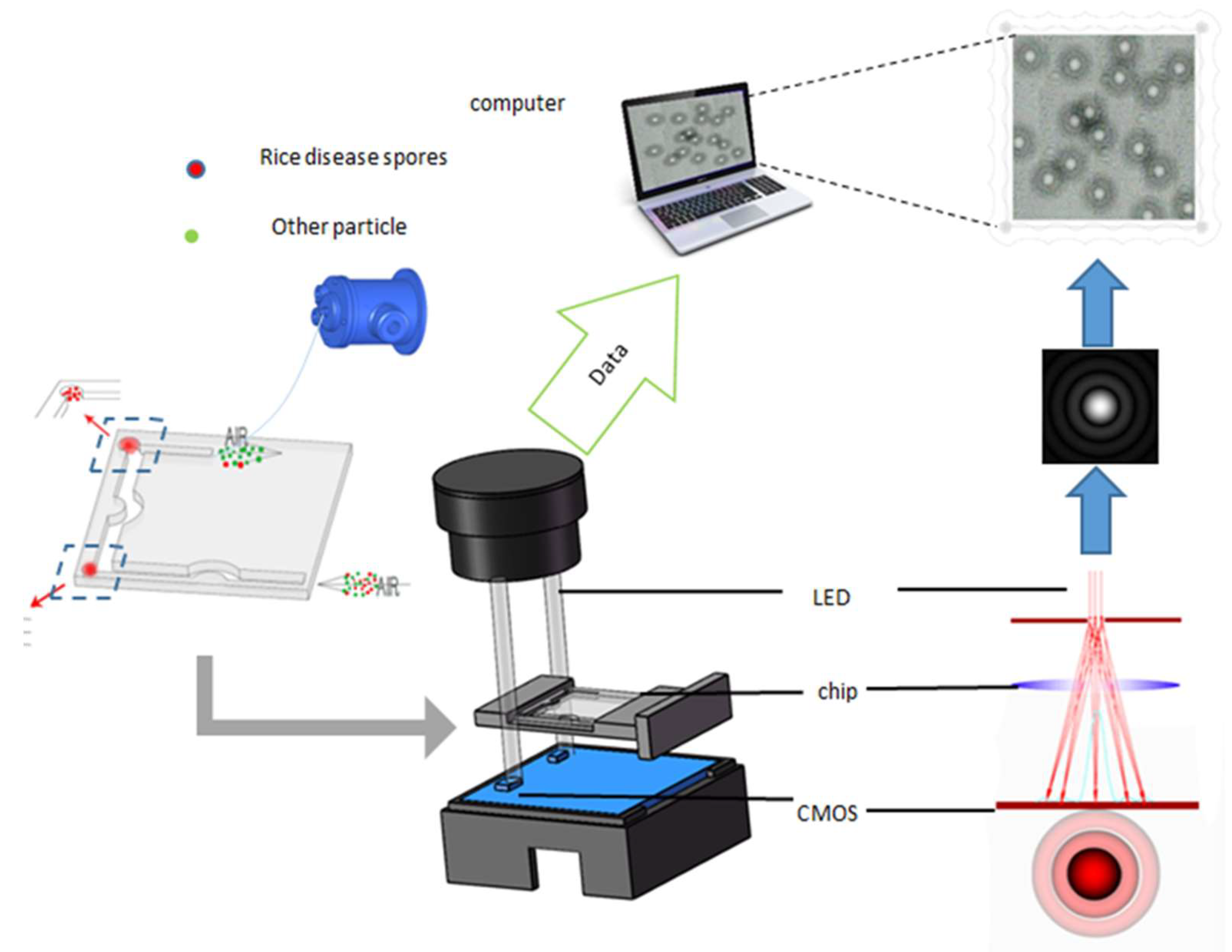
2.3. Diffraction Imaging Detection Platform Setup
2.4. Spore Detection Using Diffraction Parameters
3. Results and Discussion
3.1. Simulation and Experiment of Microfluidic Chip
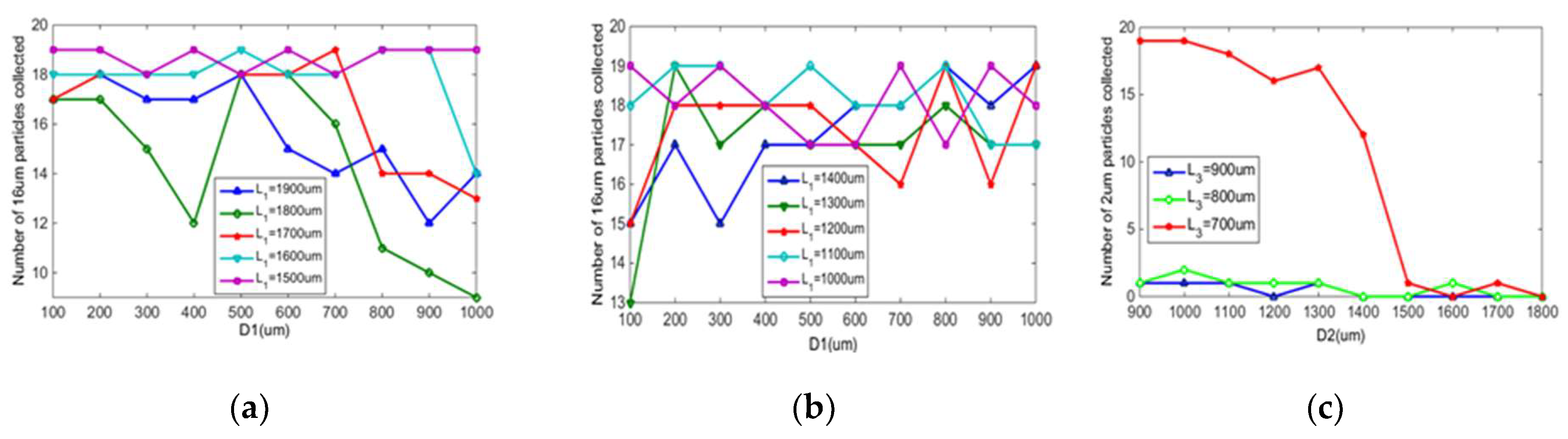
3.1.1. Particle Motion Simulation
3.1.2. Spore Collection Experiment
3.2. Diffraction Imaging Detection Platform
3.2.1. Diffraction Fingerprints Calculation and Investigation
3.2.2. Validation and Superiority of Proposed Approach
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wang, G.-L.; Valent, B. Durable resistance to rice blast. Science 2017, 355, 906–907. [Google Scholar] [CrossRef] [PubMed]
- Talbot, N.J. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. 2003, 57, 177–202. [Google Scholar] [CrossRef]
- Dai, X.; He, C.; Zhou, L.; Liang, M.; Fu, X.; Qin, P.; Yang, Y.; Chen, L. Identification of a specific molecular marker for the rice blast-resistant gene Pigm and molecular breeding of thermo-sensitive genic male sterile leaf-color marker lines. Mol. Breeding 2018, 386, 72. [Google Scholar] [CrossRef]
- Yangseon, K.; Jae-Hwan, R.; Ha, K. Early Forecasting of Rice Blast Disease Using Long Short-Term Memory Recurrent Neural Networks. Sustainability 2017, 10, 34. [Google Scholar]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Thomma, B.P.H.J.; et al. Effector-Mediated Suppression of Chitin-Triggered Immunity by Magnaporthe oryzae Is Necessary for Rice Blast Disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef]
- Li, Y.; Lu, Y.-G.; Shi, Y.; Wu, L.; Xu, Y.-J.; Huang, F.; Guo, X.-Y.; Zhang, Y.; Fan, J.; Zhao, J.-Q.; et al. Multiple Rice MicroRNAs Are Involved in Immunity against the Blast Fungus Magnaporthe oryzae. Plant Physiol. 2014, 164, 1077–1092. [Google Scholar] [CrossRef]
- Chen, J.J.; Li, K.T. Analysis of PCR Kinetics inside a Microfluidic DNA Amplification System. Micromachines 2018, 9, 48. [Google Scholar] [CrossRef]
- Chesmore, D.; Bernard, T.; Inman, A.J.; Bowyer, R.J. Image analysis for the identification of the quarantine pest Tilletia indica*. EPPO Bull. 2003, 33, 495–499. [Google Scholar] [CrossRef]
- Osimani, A.; Milanovi, V.; Cardinali, F.; Garofalo, C.; Clementi, F.; Pasquini, M.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Loreto, N.; et al. The bacterial biota of laboratory-reared edible mealworms (Tenebrio molitor, L.): From feed to frass. Int. J Food Microbiol. 2018, 272, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Nugaeva, N.; Gfeller, K.Y.; Backmann, N.; Lang, H.P.; Düggelin, M.; Hegner, M.; et al. Micromechanical cantilever array sensors for selective fungal immobilization and fast growth detection. Biosens. Bioelectron. 2006, 21, 849–856. [Google Scholar] [CrossRef]
- Peterson, T.S.; Spitsbergen, J.M.; Feist, S.W.; Kent, M.L. Luna stain, an improved selective stain for detection of microsporidian spores in histologic sections. Dis. Aquat. Org. 2011, 95, 175–180. [Google Scholar] [CrossRef]
- Morel, N.; Volland, H.; Dano, J.; Lamourette, P.; Sylvestre, P.; Mock, M.; Créminon, C. Fast and Sensitive Detection of Bacillus anthracis Spores by Immunoassay. Appl. Environ. Microbiol. 2012, 78, 6491–6498. [Google Scholar] [CrossRef]
- Roy, M.; Jin, G.; Seo, D.; Nam, M.-H.; Seo, S. A simple and low-cost device performing blood cell counting based on lens-free shadow imaging technique. Sens. Actuators B 2014, 201, 321–328. [Google Scholar] [CrossRef]
- Li, G.; Zhang, R.; Yang, N.; Yin, C.; Wei, M.; Zhang, Y.; Sun, J. An approach for cell viability online detection based on the characteristics of lensfree cell diffraction fingerprint. Biosens. Bioelectron. 2018, 107, 163–169. [Google Scholar] [CrossRef]
- Roy, M.; Seo, D.; Oh, C.-H.; Nam, M.-H.; Kim, Y.J.; Seo, S. Low-cost telemedicine device performing cell and particle size measurement based on lens-free shadow imaging technology. Biosens. Bioelectron. 2015, 67, 715–723. [Google Scholar] [CrossRef]
- Kwak, Y.H.; Lee, J.; Lee, J.; Kwak, S.H.; Oh, S.; Paek, S.-H.; Ha, U.-H.; Seo, S. A simple and low-cost biofilm quantification method using LED and CMOS image sensor. J. Microbiol. Methods 2014, 107, 150–156. [Google Scholar] [CrossRef]
- Ishida, T.; Shimamoto, T.; Kaminaga, M.; Kuchimaru, T.; Kizaka-Kondoh, S.; Omata, T. Microfluidic High-Migratory Cell Collector Suppressing Artifacts Caused by Microstructures. Micromachines 2019, 10, 116. [Google Scholar] [CrossRef]
- Yang, N.; Zhang, R.; Xu, P.; Xiang, Z. Optimised Microfluidic Device for Assessing A Rheumatoid Factor in Patients. Electronics World 2013, 119, 40–43. [Google Scholar]
- Xu, J.; Zhang, J.; Wang, H.; Mi, J. Fine Particle Behavior in the Air Flow Past a Triangular Cylinder. Aerosol Sci Tech. 2013, 47, 875–884. [Google Scholar] [CrossRef]
- Zhang, T.; Takahashi, H.; Hata, M.; Toriba, A.; Ikeda, T.; Otani, Y.; Furuuchi, M. Development of a Sharp-Cut Inertial Filter Combined with an Impactor. Aerosol. Air Qual. Res. 2017, 17, 644–652. [Google Scholar] [CrossRef]
- Chung, A.J.; Gossett, D.R.; Carlo, D.D. Three Dimensional, Sheathless, and High-Throughput Microparticle Inertial Focusing Through Geometry-Induced Secondary Flows. Small 2013, 9, 685–690. [Google Scholar] [CrossRef]
- Seo, J.; Lean, M.H.; Kole, A. Membrane-free microfiltration by asymmetric inertial migration. Appl. Phys. Lett. 2007, 91, 033901. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, N.; Chen, C.; Li, T.; Li, Z.; Zou, L.; Zhang, R.; Mao, H. Portable Rice Disease Spores Capture and Detection Method Using Diffraction Fingerprints on Microfluidic Chip. Micromachines 2019, 10, 289. https://doi.org/10.3390/mi10050289
Yang N, Chen C, Li T, Li Z, Zou L, Zhang R, Mao H. Portable Rice Disease Spores Capture and Detection Method Using Diffraction Fingerprints on Microfluidic Chip. Micromachines. 2019; 10(5):289. https://doi.org/10.3390/mi10050289
Chicago/Turabian StyleYang, Ning, Chiyuan Chen, Tao Li, Zhuo Li, Lirong Zou, Rongbiao Zhang, and Hanping Mao. 2019. "Portable Rice Disease Spores Capture and Detection Method Using Diffraction Fingerprints on Microfluidic Chip" Micromachines 10, no. 5: 289. https://doi.org/10.3390/mi10050289




