Microfluidic-Based Nucleic Acid Amplification Systems in Microbiology
Abstract
:1. Introduction
2. Spatial Polymerase Chain Reaction
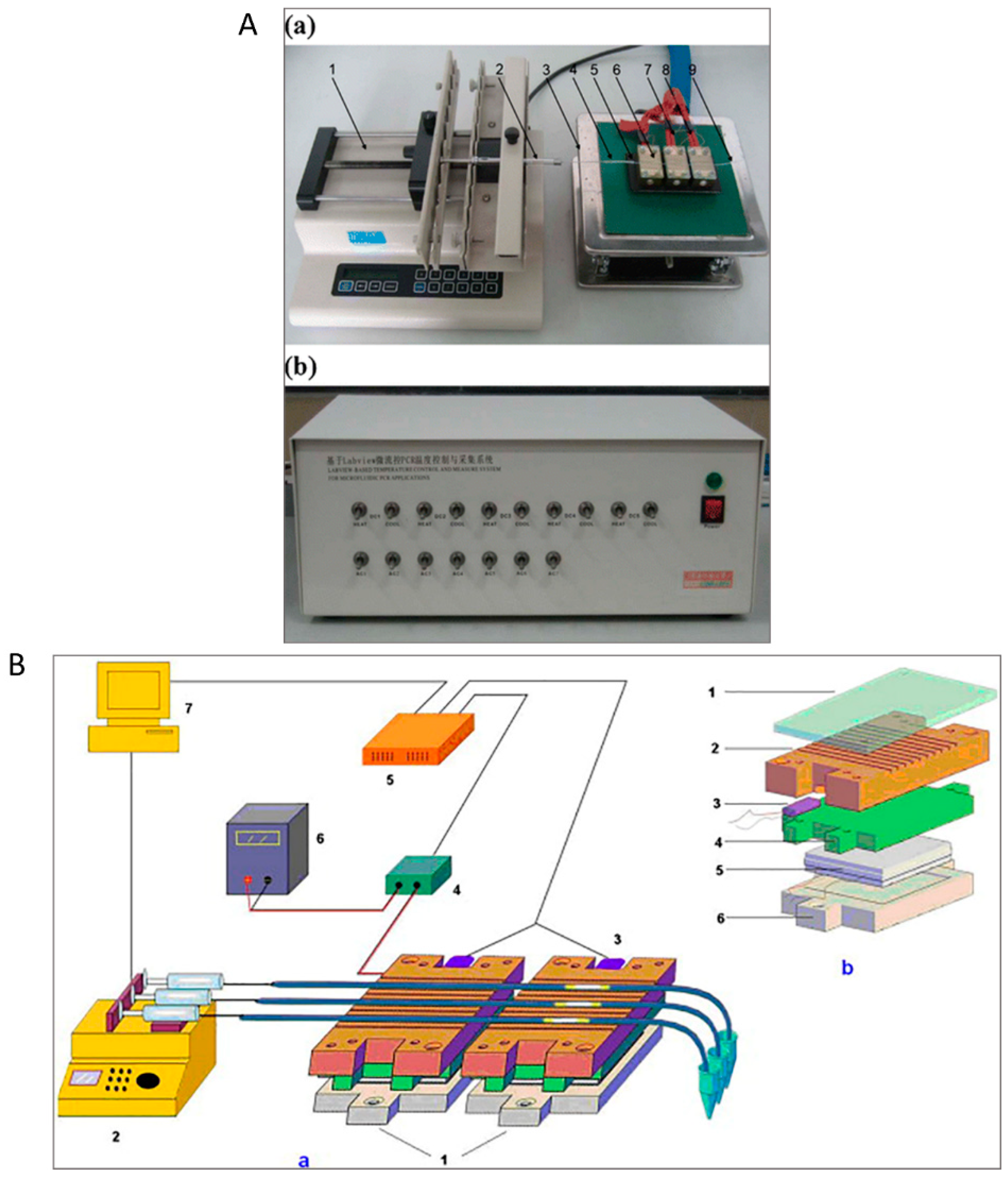
2.1. Serpentine Design
2.2. Oscillating-Flow Design
3. Transient PCR
3.1. Centrifugal Microfluidic Devices
3.2. Lab Disk
4. Array
5. Isothermal Amplification-Based Microfluidic Devices
5.1. RPA-Based Microfluidic Devices
5.2. HDA-Based Microfluidic Devices
5.3. LAMP-Based Microfluidic Devices
5.3.1. End-Point Colorimetric Detection
5.3.2. Real-Time Optical Detection
6. Design Considerations for PCR Devices
6.1. Sealing
6.2. Valving
6.3. Detection
7. Droplet-Based Microfluidics
7.1. Continuous-Flow Microfluidic
7.2. Digital Microfluidics
8. Microfluidic Sample Preparation for Amplification
9. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Pahlow, S.; Meisel, S.; Cialla-May, D.; Weber, K.; Rösch, P.; Popp, J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv. Drug Deliv. Rev. 2015, 89, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, Y.; Li, M.; Tang, L.; He, J.-J. DNAzyme-integrated plasmonic nanosensor for bacterial sample-to-answer detection. Biosens. Bioelectron. 2017, 89, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Murugan, V.; Sankaran, K. Bacterial Lipid Modification of ICP11 and a New ELISA System Applicable for WSSV Infection Detection. Mar. Biotechnol. 2018, 20, 375–384. [Google Scholar] [CrossRef]
- Webster, R.P.; Cohen, C.F.; Saeed, F.O.; Wetzel, H.N.; Ball, W.J., Jr.; Kirley, T.L.; Norman, A.B. Evaluation of methods to reduce background using the Python-based ELISA_QC program. J. Immunol. Methods 2018, 456, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Nacef, M.; Chevalier, M.; Chollet, S.; Drider, D.; Flahaut, C. MALDI-TOF mass spectrometry for the identification of lactic acid bacteria isolated from a French cheese: The Maroilles. Int. J. Food Microbiol. 2017, 247, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Carballo, B.L.; McGuiness, I.; McBeth, C.; Kalashnikov, M.; Borrós, S.; Sharon, A.; Sauer-Budge, A.F. Low-cost, real-time, continuous flow PCR system for pathogen detection. Biomed. Microdevices 2016, 18, 34. [Google Scholar] [CrossRef]
- Stedtfeld, R.D.; Stedtfeld, T.M.; Samhan, F.; Kanitkar, Y.H.; Hatzinger, P.B.; Cupples, A.M.; Hashsham, S.A. Direct loop mediated isothermal amplification on filters for quantification of Dehalobacter in groundwater. J. Microbiol. Methods 2016, 131, 61–67. [Google Scholar] [CrossRef] [Green Version]
- Eyre, D.W.; Golubchik, T.; Gordon, N.C.; Bowden, R.; Piazza, P.; Batty, E.M.; Ip, C.L.; Wilson, D.J.; Didelot, X.; O’Connor, L. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2012, 2, e001124. [Google Scholar] [CrossRef]
- Claydon, M.A.; Davey, S.N.; Edwards-Jones, V.; Gordon, D.B. The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol. 1996, 14, 1584. [Google Scholar] [CrossRef]
- Ziegler, D.; Pothier, J.F.; Ardley, J.; Fossou, R.K.; Pflüger, V.; De Meyer, S.; Vogel, G.; Tonolla, M.; Howieson, J.; Reeve, W. Ribosomal protein biomarkers provide root nodule bacterial identification by MALDI-TOF MS. Appl. Microbiol. Biotechnol. 2015, 99, 5547–5562. [Google Scholar] [CrossRef]
- Zhu, Y.; Qiao, L.; Prudent, M.; Bondarenko, A.; Gasilova, N.; Möller, S.B.; Lion, N.; Pick, H.; Gong, T.; Chen, Z. Sensitive and fast identification of bacteria in blood samples by immunoaffinity mass spectrometry for quick BSI diagnosis. Chem. Sci. 2016, 7, 2987–2995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, L.E.; Hunfeld, K.-P.; Steinbrucker, M.; Brade, V.; Book, M.; Seifert, H.; Bingold, T.; Hoeft, A.; Wissing, H.; Stüber, F. Improved detection of blood stream pathogens by real-time PCR in severe sepsis. Intensive Care Med. 2010, 36, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Notomi, T. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Lefever, S.; Pattyn, F.; De Wilde, B.; Coppieters, F.; De Keulenaer, S.; Hellemans, J.; Vandesompele, J. High-throughput PCR assay design for targeted resequencing using primerXL. BMC Bioinform. 2017, 18, 400. [Google Scholar] [CrossRef]
- Schadt, E.E.; Turner, S.; Kasarskis, A. A window into third-generation sequencing. Hum. Mol. Gen 2010, 19, R227–R240. [Google Scholar] [CrossRef]
- Higgins, D.; Pal, C.; Sulaiman, I.M.; Jia, C.; Zerwekh, T.; Dowd, S.E.; Banerjee, P. Application of high-throughput pyrosequencing in the analysis of microbiota of food commodities procured from small and large retail outlets in a US metropolitan area—A pilot study. Food Res. Int. 2018, 105, 29–40. [Google Scholar] [CrossRef]
- Vu, T.; Davidson, S.-L.; Borgesi, J.; Maksudul, M.; Jeon, T.-J.; Shim, J. Piecing together the puzzle: Nanopore technology in detection and quantification of cancer biomarkers. RSC Adv. 2017, 7, 42653–42666. [Google Scholar] [CrossRef]
- Kou, S.; Cheng, D.; Sun, F.; Hsing, I.-M. Microfluidics and microbial engineering. Lab Chip 2016, 16, 432–446. [Google Scholar] [CrossRef]
- Ahrberg, C.D.; Manz, A.; Chung, B.G. Polymerase chain reaction in microfluidic devices. Lab Chip 2016, 16, 3866–3884. [Google Scholar] [CrossRef] [Green Version]
- Manz, A.; Graber, N.; Widmer, H.á. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Gravesen, P.; Branebjerg, J.; Jensen, O.S. Microfluidics—A review. J. Micromech. Microeng. 1993, 3, 168. [Google Scholar] [CrossRef]
- Beebe, D.J.; Mensing, G.A.; Walker, G.M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 2002, 4, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, I.; Babenko, V.; Martínez-Rodríguez, S.; Gavira, J. Protein separation under a microfluidic regime. Analyst 2018, 143, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Wilding, P.; Shoffner, M.A.; Kricka, L.J. PCR in a silicon microstructure. Clin. Chem. 1994, 40, 1815–1818. [Google Scholar]
- Nguyen, N.-T.; Hejazian, M.; Ooi, C.H.; Kashaninejad, N. Recent advances and future perspectives on microfluidic liquid handling. Micromachines 2017, 8, 186. [Google Scholar] [CrossRef]
- Kopp, M.U.; De Mello, A.J.; Manz, A. Chemical amplification: Continuous-flow PCR on a chip. Science 1998, 280, 1046–1048. [Google Scholar] [CrossRef]
- Strohmeier, O.; Keller, M.; Schwemmer, F.; Zehnle, S.; Mark, D.; von Stetten, F.; Zengerle, R.; Paust, N. Centrifugal microfluidic platforms: Advanced unit operations and applications. Chem. Soc. Rev. 2015, 44, 6187–6229. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Abalde-Cela, S.; Carbó-Argibay, E.; Diéguez, L.; Piotrowski, M.; Kolen’ko, Y.V.; Prado, M. Combination of microfluidic loop-mediated isothermal amplification with gold nanoparticles for rapid detection of Salmonella spp. in food samples. Front. Microbiol. 2017, 8, 2159. [Google Scholar] [CrossRef]
- Ahmad, F.; Stedtfeld, R.D.; Waseem, H.; Williams, M.R.; Cupples, A.M.; Tiedje, J.M.; Hashsham, S.A. Most probable number-loop mediated isothermal amplification (MPN-LAMP) for quantifying waterborne pathogens in <25 min. J. Microbiol. Methods 2017, 132, 27–33. [Google Scholar]
- Jiang, X.; Jing, W.; Sun, X.; Liu, Q.; Yang, C.; Liu, S.; Qin, K.; Sui, G. High-throughput microfluidic device for LAMP analysis of airborne bacteria. ACS Sens. 2016, 1, 958–962. [Google Scholar] [CrossRef]
- Safavieh, M.; Kanakasabapathy, M.K.; Tarlan, F.; Ahmed, M.U.; Zourob, M.; Asghar, W.; Shafiee, H. Emerging loop-mediated isothermal amplification-based microchip and microdevice technologies for nucleic acid detection. ACS Biomater.-Sci. Eng. 2016, 2, 278–294. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, H.-R. A review on continuous-flow microfluidic PCR in droplets: Advances, challenges and future. Anal. Chim. Acta 2016, 914, 7–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.-W.; Zhu, Y.; Feng, Y.-M.; Fang, J.; Fang, Q. Droplet-Based Multivolume Digital Polymerase Chain Reaction by a Surface-Assisted Multifactor Fluid Segmentation Approach. Anal. Chem. 2016, 89, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Park, B.H.; Choi, G.; Seo, J.H.; Jung, J.H.; Choi, J.S.; Seo, T.S. Fully automated and colorimetric foodborne pathogen detection on an integrated centrifugal microfluidic device. Lab Chip 2016, 16, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xu, Y.; Liu, H.; Peng, J.; Irudayaraj, J.; Cui, F. An integrated microsystem with dielectrophoresis enrichment and impedance detection for detection of Escherichia coli. Biomed. Microdevices 2017, 19, 34. [Google Scholar] [CrossRef] [PubMed]
- Weibel, D.B.; DiLuzio, W.R.; Whitesides, G.M. Microfabrication meets microbiology. Nat. Rev. Microbiol. 2007, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Asiello, P.J.; Baeumner, A.J. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip 2011, 11, 1420–1430. [Google Scholar] [CrossRef]
- Bridle, H.; Miller, B.; Desmulliez, M.P. Application of microfluidics in waterborne pathogen monitoring: A review. Water Res. 2014, 55, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Castillo, F.Y.; Loera-Muro, A.; Jacques, M.; Garneau, P.; Avelar-González, F.J.; Harel, J.; Guerrero-Barrera, A.L. Waterborne pathogens: Detection methods and challenges. Pathogens 2015, 4, 307–334. [Google Scholar] [CrossRef]
- Kaminski, T.S.; Scheler, O.; Garstecki, P. Droplet microfluidics for microbiology: Techniques, applications and challenges. Lab Chip 2016, 16, 2168–2187. [Google Scholar] [CrossRef]
- Zhang, D.; Bi, H.; Liu, B.; Qiao, L. Detection of Pathogenic Microorganisms by Microfluidics Based Analytical Methods; ACS Publications: Washington, DC, USA, 2018. [Google Scholar]
- Zulkifli, S.N.; Rahim, H.A.; Lau, W.J. Detection of Contaminants in Water Supply: A Review on State-of-the-Art Monitoring Technologies and Their Applications. Sens. Actuators B Chem. 2018, 255, 2657–2689. [Google Scholar] [CrossRef]
- Polini, A.; Mele, E.; Sciancalepore, A.G.; Girardo, S.; Biasco, A.; Camposeo, A.; Cingolani, R.; Weitz, D.A.; Pisignano, D. Reduction of water evaporation in polymerase chain reaction microfluidic devices based on oscillating-flow. Biomicrofluidics 2010, 4, 036502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.J.; Liao, M.H.; Li, K.T.; Shen, C.M. One-heater flow-through polymerase chain reaction device by heat pipes cooling. Biomicrofluidics 2015, 9, 014107. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Shen, C.M.; Ko, Y.W. Analytical study of a microfludic DNA amplification chip using water cooling effect. Biomed. Microdevices 2013, 15, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fozdar, D.Y.; Ali, M.F.; Li, H.; Shao, D.; Vykoukal, D.M.; Vykoukal, J.; Floriano, P.N.; Olsen, M.; McDevitt, J.T. A continuous-flow polymerase chain reaction microchip with regional velocity control. J. Microelectromech. Syst. 2006, 15, 223–236. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Garling, D.J. Rapid cycle DNA amplification: Time and temperature optimization. BioTechniques 1991, 10, 76–83. [Google Scholar]
- Wittwer, C.T.; Herrmann, M.G.; Gundry, C.N.; Elenitoba-Johnson, K.S. Real-time multiplex PCR assays. Methods 2001, 25, 430–442. [Google Scholar] [CrossRef]
- Sun, K.; Yamaguchi, A.; Ishida, Y.; Matsuo, S.; Misawa, H. A heater-integrated transparent microchannel chip for continuous-flow PCR. Sens. Actuators B Chem. 2002, 84, 283–289. [Google Scholar] [CrossRef]
- Hashimoto, M.; Chen, P.-C.; Mitchell, M.W.; Nikitopoulos, D.E.; Soper, S.A.; Murphy, M.C. Rapid PCR in a continuous flow device. Lab Chip 2004, 4, 638–645. [Google Scholar] [CrossRef] [Green Version]
- Trinh, K.T.L.; Lee, N.Y. Glass-polytetrafluoroethylene-glass based sandwich microdevice for continuous-flow polymerase chain reaction and its application for fast identification of foodborne pathogens. Talanta 2018, 176, 544–550. [Google Scholar] [CrossRef]
- Niu, Z.Q.; Chen, W.Y.; Shao, S.Y.; Jia, X.Y.; Zhang, W.P. DNA amplification on a PDMS–glass hybrid microchip. J. Micromech. Microeng. 2006, 16, 425. [Google Scholar] [CrossRef]
- Andersson, H.; Van Der Wijngaart, W.; Nilsson, P.; Enoksson, P.; Stemme, G. A valve-less diffuser micropump for microfluidic analytical systems. Sens. Actuators B Chem. 2001, 72, 259–265. [Google Scholar] [CrossRef]
- Xie, J.; Shih, J.; Lin, Q.; Yang, B.; Tai, Y.-C. Surface micromachined electrostatically actuated micro peristaltic pump. Lab Chip 2004, 4, 495–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gervais, L.; Hitzbleck, M.; Delamarche, E. Capillary-driven multiparametric microfluidic chips for one-step immunoassays. Biosens. Bioelectron. 2011, 27, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Delamarche, E.; Juncker, D.; Schmid, H. Microfluidics for processing surfaces and miniaturizing biological assays. Adv. Mater. 2005, 17, 2911–2933. [Google Scholar] [CrossRef]
- Tachibana, H.; Saito, M.; Tsuji, K.; Yamanaka, K.; Tamiya, E. Self-propelled continuous-flow PCR in capillary-driven microfluidic device: Microfluidic behavior and DNA amplification. Sens. Actuators B Chem. 2015, 206, 303–310. [Google Scholar] [CrossRef]
- Tachibana, H.; Saito, M.; Shibuya, S.; Tsuji, K.; Miyagawa, N.; Yamanaka, K.; Tamiya, E. On-chip quantitative detection of pathogen genes by autonomous microfluidic PCR platform. Biosens. Bioelectron. 2015, 74, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, H.; Xing, D. Multichannel oscillatory-flow multiplex PCR microfluidics for high-throughput and fast detection of foodborne bacterial pathogens. Biomed. Microdevices 2011, 13, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; West, J.; Auroux, P.-A.; Manz, A.; Day, P.J. Ultrasensitive PCR and real-time detection from human genomic samples using a bidirectional flow microreactor. Anal. Chem. 2007, 79, 9185–9190. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liang, G.; Lei, X.; Chen, B.; Wang, W.; Zhou, X. Highly efficient capillary polymerase chain reaction using an oscillation droplet microreactor. Anal. Chim. Acta 2012, 718, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Kim, H.-Y.; Kang, J.Y.; Kim, T.S. How the capillary burst microvalve works. J. Colloid Interface Sci. 2007, 306, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M.; Huang, P.-C.; Lin, M.-G. Analysis and experiment of capillary valves for microfluidics on a rotating disk. Microfluid Nanofluidics 2008, 4, 427–437. [Google Scholar] [CrossRef]
- Sayad, A.; Ibrahim, F.; Uddin, S.M.; Cho, J.; Madou, M.; Thong, K.L. A microdevice for rapid, monoplex and colorimetric detection of foodborne pathogens using a centrifugal microfluidic platform. Biosens. Bioelectron. 2018, 100, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Brennan, D.; Coughlan, H.; Clancy, E.; Dimov, N.; Barry, T.; Kinahan, D.; Ducrée, J.; Smith, T.J.; Galvin, P. Development of an on-disc isothermal in vitro amplification and detection of bacterial RNA. Sens. Actuators B Chem. 2017, 239, 235–242. [Google Scholar] [CrossRef]
- Law, I.; Loo, J.; Kwok, H.; Yeung, H.; Leung, C.; Hui, M.; Wu, S.; Chan, H.; Kwan, Y.; Ho, H. Automated real-time detection of drug-resistant Mycobacterium tuberculosis on a lab-on-a-disc by Recombinase Polymerase Amplification. Anal. Biochem. 2018, 544, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Ma, K.-S.; Kim, J.; Zoval, J.V.; Peytavi, R.; Bergeron, M.G.; Madou, M.J. Dynamic automated DNA hybridization on a CD (compact disc) fluidic platform. Sens. Actuators B Chem. 2006, 114, 173–181. [Google Scholar] [CrossRef]
- Furutani, S.; Nagai, H.; Takamura, Y.; Kubo, I. Compact disk (CD)-shaped device for single cell isolation and PCR of a specific gene in the isolated cell. Anal. Bioanal. Chem. 2010, 398, 2997–3004. [Google Scholar] [CrossRef] [PubMed]
- Amasia, M.; Cozzens, M.; Madou, M.J. Centrifugal microfluidic platform for rapid PCR amplification using integrated thermoelectric heating and ice-valving. Sens. Actuators B Chem. 2012, 161, 1191–1197. [Google Scholar] [CrossRef]
- Strohmeier, O.; Marquart, N.; Mark, D.; Roth, G.; Zengerle, R.; von Stetten, F. Real-time PCR based detection of a panel of food-borne pathogens on a centrifugal microfluidic “LabDisk” with on-disk quality controls and standards for quantification. Anal. Methods 2014, 6, 2038–2046. [Google Scholar] [CrossRef]
- Czilwik, G.; Schwarz, I.; Keller, M.; Wadle, S.; Zehnle, S.; von Stetten, F.; Mark, D.; Zengerle, R.; Paust, N. Microfluidic vapor-diffusion barrier for pressure reduction in fully closed PCR modules. Lab Chip 2015, 15, 1084–1091. [Google Scholar] [CrossRef]
- Huang, G.; Huang, Q.; Xie, L.; Xiang, G.; Wang, L.; Xu, H.; Ma, L.; Luo, X.; Xin, J.; Zhou, X. A rapid, low-cost, and microfluidic chip-based system for parallel identification of multiple pathogens related to clinical pneumonia. Sci. Rep. 2017, 7, 6441. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.J.; Docker, P.T.; Yelland, J.V.; Dyer, C.E.; Greenman, J.; Greenway, G.M.; Haswell, S.J. Rapid PCR amplification using a microfluidic device with integrated microwave heating and air impingement cooling. Lab Chip 2010, 10, 1725–1728. [Google Scholar] [CrossRef] [PubMed]
- Pal, D.; Venkataraman, V. A portable battery-operated chip thermocycler based on induction heating. Sens. Actuators A Phys. 2002, 102, 151–156. [Google Scholar] [CrossRef]
- Burger, J.; Gross, A.; Mark, D.; von Stetten, F.; Zengerle, R.; Roth, G. IR thermocycler for centrifugal microfluidic platform with direct on-disk wireless temperature measurement system. In Proceedings of the 16th International Solid-State Sensors, Actuators and Microsystems Conference (TRANSDUCERS), Beijing, China, 5–9 June 2011; pp. 2867–2870. [Google Scholar]
- Hettiarachchi, K.; Kim, H.; Faris, G.W. Optical manipulation and control of real-time PCR in cell encapsulating microdroplets by IR laser. Microfluid Nanofluidics 2012, 13, 967–975. [Google Scholar] [CrossRef]
- Focke, M.; Stumpf, F.; Faltin, B.; Reith, P.; Bamarni, D.; Wadle, S.; Müller, C.; Reinecke, H.; Schrenzel, J.; Francois, P. Microstructuring of polymer films for sensitive genotyping by real-time PCR on a centrifugal microfluidic platform. Lab Chip 2010, 10, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Focke, M.; Stumpf, F.; Roth, G.; Zengerle, R.; von Stetten, F. Centrifugal microfluidic system for primary amplification and secondary real-time PCR. Lab Chip 2010, 10, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Park, B.H.; Jung, J.H.; Choi, G.; Lee, D.C.; Seo, T.S. Centrifugal loop-mediated isothermal amplification microdevice for rapid, multiplex and colorimetric foodborne pathogen detection. Biosens. Bioelectron. 2016, 75, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.H.; Park, B.H.; Oh, S.J.; Choi, G.; Lee, E.Y.; Seo, T.S. Development of a high-throughput centrifugal loop-mediated isothermal amplification microdevice for multiplex foodborne pathogenic bacteria detection. Sens. Actuators B Chem. 2017, 246, 146–153. [Google Scholar] [CrossRef]
- Keller, M.; Wadle, S.; Paust, N.; Dreesen, L.; Nuese, C.; Strohmeier, O.; Zengerle, R.; von Stetten, F. Centrifugo-thermopneumatic fluid control for valving and aliquoting applied to multiplex real-time PCR on off-the-shelf centrifugal thermocycler. RSC Adv. 2015, 5, 89603–89611. [Google Scholar] [CrossRef] [Green Version]
- Czilwik, G.; Messinger, T.; Strohmeier, O.; Wadle, S.; Von Stetten, F.; Paust, N.; Roth, G.; Zengerle, R.; Saarinen, P.; Niittymäki, J. Rapid and fully automated bacterial pathogen detection on a centrifugal-microfluidic LabDisk using highly sensitive nested PCR with integrated sample preparation. Lab Chip 2015, 15, 3749–3759. [Google Scholar] [CrossRef]
- Roy, E.; Stewart, G.; Mounier, M.; Malic, L.; Peytavi, R.; Clime, L.; Madou, M.; Bossinot, M.; Bergeron, M.G.; Veres, T. From cellular lysis to microarray detection, an integrated thermoplastic elastomer (TPE) point of care Lab on a Disc. Lab Chip 2015, 15, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhu, Y.; Zhang, Y.; Wang, L.; Chen, J.; Lu, Y.; Xu, Y.; Xing, W. Multiplex detection of bacteria on an integrated centrifugal disk using bead-beating lysis and loop-mediated amplification. Sci. Rep. 2017, 7, 1460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, F.; Liu, C.; Feng, Q.; Ma, T.; Zhao, Z.; Li, T.; Jiang, X.; Sun, J. Hand-powered centrifugal microfluidic platform inspired by the spinning top for sample-to-answer diagnostics of nucleic acids. Lab Chip 2018, 18, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-Y.; Shannon, K.; Beaudette, L.A. Detection of bacterial pathogens in municipal wastewater using an oligonucleotide microarray and real-time quantitative PCR. J. Microbiol. Methods 2006, 65, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Yauk, C.L.; Berndt, M.L.; Williams, A.; Douglas, G.R. Comprehensive comparison of six microarray technologies. Nucleic Acids Res. 2004, 32, e124. [Google Scholar] [CrossRef] [PubMed]
- Mitterer, G.; Huber, M.; Leidinger, E.; Kirisits, C.; Lubitz, W.; Mueller, M.W.; Schmidt, W.M. Microarray-based identification of bacteria in clinical samples by solid-phase PCR amplification of 23S ribosomal DNA sequences. J. Clin. Microbiol. 2004, 42, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Strizhkov, B.N.; Drobyshev, A.L.; Mikhailovich, V.M.; Mirzabekov, A.D. PCR amplification on a microarray of gel-immobilized oligonucleotides: Detection of bacterial toxin-and drug-resistant genes and their mutations. BioTechniques 2000, 29, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Bing, D.H.; Boles, C.; Rehman, F.N.; Audeh, M.; Belmarsh, M.; Kelley, B.; Adams, C.P. Bridge amplification: A solid phase PCR system for the amplification and detection of allelic differences in single copy genes. In Proceedings of the Seventh International Symposium on Human Identification, Scottsdale, AZ, USA, 18 Septmber 1996; pp. 18–20. [Google Scholar]
- Nagai, H.; Murakami, Y.; Morita, Y.; Yokoyama, K.; Tamiya, E. Development of a microchamber array for picoliter PCR. Anal. Chem. 2001, 73, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, Y.; Kerman, K.; Kobayashi, M.; Yamamura, S.; Morita, Y.; Tamiya, E. Microchamber array based DNA quantification and specific sequence detection from a single copy via PCR in nanoliter volumes. Biosens. Bioelectron. 2005, 20, 1482–1490. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.; Hurley, J.; Garcia, J.; Yoder, K.; Katz, A.; Roberts, D.; Cho, J.; Kanigan, T.; Ilyin, S.E.; Horowitz, D. Nanoliter high throughput quantitative PCR. Nucleic Acids Res. 2006, 34, e123. [Google Scholar] [CrossRef] [PubMed]
- Stedtfeld, R.D.; Baushke, S.W.; Tourlousse, D.M.; Miller, S.M.; Stedtfeld, T.M.; Gulari, E.; Tiedje, J.M.; Hashsham, S.A. Development and experimental validation of a predictive threshold cycle equation for quantification of virulence and marker genes by high-throughput nanoliter-volume PCR on the OpenArray platform. J. Appl. Environ. Microbiol. 2008, 74, 3831–3838. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Jing, F.; Li, G.; Fan, X.; Jia, C.; Zhou, H.; Jin, Q.; Zhao, J. A microfluidic droplet digital PCR for simultaneous detection of pathogenic Escherichia coli O157 and Listeria monocytogenes. Biosens. Bioelectron. 2015, 74, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.G.; Tan, W.; Zhao, M.-Q.; Ricco, A.J.; Fan, Z.H. Integrating polymerase chain reaction, valving, and electrophoresis in a plastic device for bacterial detection. Anal. Chem. 2003, 75, 4591–4598. [Google Scholar] [CrossRef] [PubMed]
- Lagally, E.T.; Simpson, P.C.; Mathies, R.A. Monolithic integrated microfluidic DNA amplification and capillary electrophoresis analysis system. Sens. Actuators B Chem. 2000, 63, 138–146. [Google Scholar] [CrossRef]
- Ramalingam, N.; Rui, Z.; Liu, H.-B.; Dai, C.-C.; Kaushik, R.; Ratnaharika, B.; Gong, H.-Q. Real-time PCR-based microfluidic array chip for simultaneous detection of multiple waterborne pathogens. Sens. Actuators B Chem. 2010, 145, 543–552. [Google Scholar] [CrossRef]
- Ramalingam, N.; San, T.C.; Kai, T.J.; Mak, M.Y.M.; Gong, H.-Q. Microfluidic devices harboring unsealed reactors for real-time isothermal helicase-dependent amplification. Microfluid Nanofluidics 2009, 7, 325. [Google Scholar] [CrossRef]
- Ishii, S.; Segawa, T.; Okabe, S. Simultaneous quantification of multiple food and waterborne pathogens by use of microfluidic quantitative PCR. J. Appl. Environ. Microbiol. 2013, 79. [Google Scholar] [CrossRef]
- Kleyer, H.; Tecon, R.; Or, D. Resolving species level changes in a representative soil bacterial community using microfluidic quantitative PCR. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Vincent, M.; Xu, Y.; Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Shi, C.; Liu, Q.; Ma, C.; Zhong, W. Exponential strand-displacement amplification for detection of microRNAs. Anal. Chem. 2013, 86, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.; Wharam, S.; Weston, A.; Cardy, D.; Wilson, W. Use of signal-mediated amplification of RNA technology (SMART) to detect marine cyanophage DNA. BioTechniques 2002, 32, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Han, D.; Deng, M.; Wang, J.; Shi, C. Single primer-triggered isothermal amplification for double-stranded DNA detection. ChemComm 2015, 51, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Hu, L.; Zhong, H.; Wang, H.; Yusa, S.-I.; Weiss, T.C.; Romaniuk, P.J.; Pickerill, S.; You, Q. Cross priming amplification: Mechanism and optimization for isothermal DNA amplification. Sci. Rep. 2012, 2, 246. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Tsaloglou, M.-N.; Watson, R.; Rushworth, C.; Zhao, Y.; Niu, X.; Sutton, J.; Morgan, H. Real-time microfluidic recombinase polymerase amplification for the toxin B gene of Clostridium difficile on a SlipChip platform. Analyst 2015, 140, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Park, J.; Kim, C.-J.; Cho, Y.-K. Fully integrated lab-on-a-disc for nucleic acid analysis of food-borne pathogens. Anal. Chem. 2014, 86, 3841–3848. [Google Scholar] [CrossRef]
- Lutz, S.; Weber, P.; Focke, M.; Faltin, B.; Hoffmann, J.; Müller, C.; Mark, D.; Roth, G.; Munday, P.; Armes, N. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA). Lab Chip 2010, 10, 887–893. [Google Scholar] [CrossRef]
- Hakenberg, S.; Hügle, M.; Weidmann, M.; Hufert, F.; Dame, G.; Urban, G.A. A phaseguided passive batch microfluidic mixing chamber for isothermal amplification. Lab Chip 2012, 12, 4576–4580. [Google Scholar] [CrossRef] [Green Version]
- Shen, F.; Davydova, E.K.; Du, W.; Kreutz, J.E.; Piepenburg, O.; Ismagilov, R.F. Digital isothermal quantification of nucleic acids via simultaneous chemical initiation of recombinase polymerase amplification reactions on SlipChip. Anal. Chem. 2011, 83, 3533–3540. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, S.; Valiadi, M.; Turner, C.; Sutton, M.; Morgan, H. Sample pre-concentration on a digital microfluidic platform for rapid AMR detection in urine. Lab Chip 2019, 19, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Mahalanabis, M.; Do, J.; ALMuayad, H.; Zhang, J.Y.; Klapperich, C.M. An integrated disposable device for DNA extraction and helicase dependent amplification. Biomed. Microdevices 2010, 12, 353–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barreda-García, S.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Helicase-dependent isothermal amplification: A novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal. Bioanal. Chem. 2018, 410, 679–693. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Seyrig, G.; Ahmad, F.; Stedtfeld, R.D.; Tourlousse, D.M.; Hashsham, S.A. Simple, powerful, and smart: Using LAMP for low-cost screening of multiple waterborne pathogens. Environ. Microbiol. Curr. Technol. Water Appl. 2011, 4, 103–125. [Google Scholar]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Abdul-Ghani, R.; Al-Mekhlafi, A.M.; Karanis, P. Loop-mediated isothermal amplification (LAMP) for malarial parasites of humans: Would it come to clinical reality as a point-of-care test? Acta Trop. 2012, 122, 233–240. [Google Scholar] [CrossRef]
- Kiddle, G.; Hardinge, P.; Buttigieg, N.; Gandelman, O.; Pereira, C.; McElgunn, C.J.; Rizzoli, M.; Jackson, R.; Appleton, N.; Moore, C. GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnol. 2012, 12, 15. [Google Scholar] [CrossRef]
- Craw, P.; Balachandran, W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: A critical review. Lab Chip 2012, 12, 2469–2486. [Google Scholar] [CrossRef]
- Goto, M.; Honda, E.; Ogura, A.; Nomoto, A.; Hanaki, K.-I. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. BioTechniques 2009, 46, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Fu, K.; Liu, Y.; Ding, X.; Hu, J.; Wu, W.; Xu, K.; Song, X.; Wang, J.; Mu, Y. Development of a self-priming PDMS/paper hybrid microfluidic chip using mixed-dye-loaded loop-mediated isothermal amplification assay for multiplex foodborne pathogens detection. Anal. Chim. Acta 2018, 1040, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Kong, J.; Li, X.; Fang, X.; Chen, Q. Colorimetric LAMP microfluidic chip for detecting three allergens: Peanut, sesame and soybean. Sci. Rep. 2018, 8, 8682. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Liu, Z.; Yan, S.; Yin, F.; Feng, X.; Liu, B.-F. Identifying multiple bacterial pathogens by loop-mediated isothermal amplification on a rotate & react slipchip. Sens. Actuators B Chem. 2016, 228, 491–499. [Google Scholar] [CrossRef]
- Dou, M.; Dominguez, D.C.; Li, X.; Sanchez, J.; Scott, G. A versatile PDMS/paper hybrid microfluidic platform for sensitive infectious disease diagnosis. Anal. Chem. 2014, 86, 7978–7986. [Google Scholar] [CrossRef] [PubMed]
- Connelly, J.T.; Rolland, J.P.; Whitesides, G.M. “Paper machine” for molecular diagnostics. Anal. Chem. 2015, 87, 7595–7601. [Google Scholar] [CrossRef]
- Safavieh, M.; Ahmed, M.U.; Sokullu, E.; Ng, A.; Braescu, L.; Zourob, M. A simple cassette as point-of-care diagnostic device for naked-eye colorimetric bacteria detection. Analyst 2014, 139, 482–487. [Google Scholar] [CrossRef]
- Tang, R.; Yang, H.; Choi, J.R.; Gong, Y.; Hu, J.; Wen, T.; Li, X.; Xu, B.; Mei, Q.; Xu, F. based device with on-chip reagent storage for rapid extraction of DNA from biological samples. Microchim. Acta 2017, 184, 2141–2150. [Google Scholar] [CrossRef]
- Choi, J.R.; Hu, J.; Tang, R.; Gong, Y.; Feng, S.; Ren, H.; Wen, T.; Li, X.; Abas, W.A.B.W.; Pingguan-Murphy, B. An integrated paper-based sample-to-answer biosensor for nucleic acid testing at the point of care. Lab Chip 2016, 16, 611–621. [Google Scholar] [CrossRef]
- Trinh, K.T.L.; Trinh, T.N.D.; Lee, N.Y. Fully integrated and slidable paper-embedded plastic microdevice for point-of-care testing of multiple foodborne pathogens. Biosens. Bioelectron. 2019, 135, 120–128. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, Y.; Jiang, X.; Xu, S.; Sui, G. Microfluidic chip for rapid analysis of cerebrospinal fluid infected with Staphylococcus aureus. Anal. Methods 2014, 6, 2015–2019. [Google Scholar] [CrossRef]
- Fang, X.; Chen, H.; Yu, S.; Jiang, X.; Kong, J. Predicting viruses accurately by a multiplex microfluidic loop-mediated isothermal amplification chip. Anal. Chem. 2010, 83, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Chen, H.; Xu, L.; Jiang, X.; Wu, W.; Kong, J. A portable and integrated nucleic acid amplification microfluidic chip for identifying bacteria. Lab Chip 2012, 12, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.K.; Yip, S.P.; Lee, T.M. Ultrasensitive and Closed-Tube Colorimetric Loop-Mediated Isothermal Amplification Assay Using Carboxyl-Modified Gold Nanoparticles. Small 2014, 10, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Liu, Y.; Kong, J.; Jiang, X. Loop-mediated isothermal amplification integrated on microfluidic chips for point-of-care quantitative detection of pathogens. Anal. Chem. 2010, 82, 3002–3006. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-H.; Lien, K.-Y.; Wu, J.-J.; Lee, G.-B. A magnetic bead-based assay for the rapid detection of methicillin-resistant Staphylococcus aureus by using a microfluidic system with integrated loop-mediated isothermal amplification. Lab Chip 2011, 11, 1521–1531. [Google Scholar] [CrossRef]
- Chuang, T.-L.; Wei, S.-C.; Lee, S.-Y.; Lin, C.-W. A polycarbonate based surface plasmon resonance sensing cartridge for high sensitivity HBV loop-mediated isothermal amplification. Biosens. Bioelectron. 2012, 32, 89–95. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Huang, J.-G.; Chuang, T.-L.; Sheu, J.-C.; Chuang, Y.-K.; Holl, M.; Meldrum, D.R.; Lee, C.-N.; Lin, C.-W. Compact optical diagnostic device for isothermal nucleic acids amplification. Sens. Actuators B Chem. 2008, 133, 493–501. [Google Scholar] [CrossRef]
- Ahmad, F.; Seyrig, G.; Tourlousse, D.M.; Stedtfeld, R.D.; Tiedje, J.M.; Hashsham, S.A. A CCD-based fluorescence imaging system for real-time loop-mediated isothermal amplification-based rapid and sensitive detection of waterborne pathogens on microchips. Biomed. Microdevices 2011, 13, 929. [Google Scholar] [CrossRef]
- Stedtfeld, R.D.; Tourlousse, D.M.; Seyrig, G.; Stedtfeld, T.M.; Kronlein, M.; Price, S.; Ahmad, F.; Gulari, E.; Tiedje, J.M.; Hashsham, S.A. Gene-Z: A device for point of care genetic testing using a smartphone. Lab Chip 2012, 12, 1454–1462. [Google Scholar] [CrossRef]
- Chang, W.-H.; Yang, S.-Y.; Wang, C.-H.; Tsai, M.-A.; Wang, P.-C.; Chen, T.-Y.; Chen, S.-C.; Lee, G.-B. Rapid isolation and detection of aquaculture pathogens in an integrated microfluidic system using loop-mediated isothermal amplification. Sens. Actuators B Chem. 2013, 180, 96–106. [Google Scholar] [CrossRef]
- Chiu, N.-F.; Huang, T.-Y.; Kuo, C.-C.; Lee, W.-C.; Hsieh, M.-H.; Lai, H.-C. Single-layer graphene based SPR biochips for tuberculosis bacillus detection. In Proceedings of the Biophotonics: Photonic Solutions for Better Health Care III, Brussels, Belgium, 16–19 April 2012. [Google Scholar]
- Zhou, Q.-J.; Wang, L.; Chen, J.; Wang, R.-N.; Shi, Y.-H.; Li, C.-H.; Zhang, D.-M.; Yan, X.-J.; Zhang, Y.-J. Development and evaluation of a real-time fluorogenic loop-mediated isothermal amplification assay integrated on a microfluidic disc chip (on-chip LAMP) for rapid and simultaneous detection of ten pathogenic bacteria in aquatic animals. J. Microbiol. Methods 2014, 104, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, P.; Zhao, X.; Du, W.; Feng, X.; Liu, B.-F. A self-contained microfluidic in-gel loop-mediated isothermal amplification for multiplexed pathogen detection. Sens. Actuators B Chem. 2017, 239, 1–8. [Google Scholar] [CrossRef]
- Verpoorte, E.; De Rooij, N.F. Microfluidics meets MEMS. Proc. IEEE 2003, 91, 930–953. [Google Scholar] [CrossRef] [Green Version]
- Abgrall, P.; Gue, A. Lab-on-chip technologies: Making a microfluidic network and coupling it into a complete microsystem—A review. J. Micromech. Microeng. 2007, 17, R15. [Google Scholar] [CrossRef]
- Nge, P.N.; Rogers, C.I.; Woolley, A.T. Advances in microfluidic materials, functions, integration, and applications. Chem. Rev. 2013, 113, 2550–2583. [Google Scholar] [CrossRef]
- Fujii, T. PDMS-based microfluidic devices for biomedical applications. Microelectron. Eng. 2002, 61, 907–914. [Google Scholar] [CrossRef]
- Oblath, E.; Henley, W.; Alarie, J.; Ramsey, J. A microfluidic chip combining DNA extraction and real-time PCR for identifying bacteria in saliva. In Proceedings of the 15th International Conference on Miniaturized Systems for Chemistry and Life Sciences 2011, Charlottesville, VA, USA, 2–6 October 2011. [Google Scholar]
- Rezai, P.; Selvaganapathy, P.R.; Wohl, G.R. Plasma enhanced bonding of polydimethylsiloxane with parylene and its optimization. J. Micromech. Microeng. 2011, 21. [Google Scholar] [CrossRef]
- Fu, Y.; Zhou, H.; Jia, C.; Jing, F.; Jin, Q.; Zhao, J.; Li, G. A microfluidic chip based on surfactant-doped polydimethylsiloxane (PDMS) in a sandwich configuration for low-cost and robust digital PCR. Sens. Actuators B Chem. 2017, 245, 414–422. [Google Scholar] [CrossRef]
- Xu, T.; Wu, L.; Wang, X.; Zhu, X.; Bao, Y.; Cai, S.; Li, G.; Li, X. A PDMS-based digital PCR chip with vacuum aspiration and water-filling cavity integrated for sample loading and evaporation reduction. In Proceedings of the 2018 IEEE Micro Electro Mechanical Systems (MEMS), Belfast, UK, 21–25 Jannuary 2018; pp. 1142–1145. [Google Scholar]
- Mukhopadhyay, R. When PDMS Isn’t the Best; ACS Publications: Washington, DC, USA, 2007. [Google Scholar]
- Sollier, E.; Murray, C.; Maoddi, P.; Di Carlo, D. Rapid prototyping polymers for microfluidic devices and high pressure injections. Lab Chip 2011, 11, 3752–3765. [Google Scholar] [CrossRef]
- Kinahan, D.J.; Kearney, S.M.; Faneuil, O.P.; Glynn, M.T.; Dimov, N.; Ducrée, J. Paper imbibition for timing of multi-step liquid handling protocols on event-triggered centrifugal microfluidic lab-on-a-disc platforms. RSC Adv. 2015, 5, 1818–1826. [Google Scholar] [CrossRef]
- Gorkin, R.; Park, J.; Siegrist, J.; Amasia, M.; Lee, B.S.; Park, J.-M.; Kim, J.; Kim, H.; Madou, M.; Cho, Y.-K. Centrifugal microfluidics for biomedical applications. Lab Chip 2010, 10, 1758–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, S.; Wang, S.; Luo, J.; Lee, L.J.; Yang, S.-T.; Madou, M.J. Design of a compact disk-like microfluidic platform for enzyme-linked immunosorbent assay. Anal. Chem. 2004, 76, 1832–1837. [Google Scholar] [CrossRef] [PubMed]
- Zehnle, S.; Schwemmer, F.; Bergmann, R.; von Stetten, F.; Zengerle, R.; Paust, N. Pneumatic siphon valving and switching in centrifugal microfluidics controlled by rotational frequency or rotational acceleration. Microfluid Nanofluidics 2015, 19, 1259–1269. [Google Scholar] [CrossRef]
- Gorkin, R., III; Nwankire, C.E.; Gaughran, J.; Zhang, X.; Donohoe, G.G.; Rook, M.; O’Kennedy, R.; Ducrée, J. Centrifugo-pneumatic valving utilizing dissolvable films. Lab Chip 2012, 12, 2894–2902. [Google Scholar] [CrossRef] [PubMed]
- Carpentras, D.; Kulinsky, L.; Madou, M. A novel magnetic active valve for lab-on-CD technology. J. Microelectromech. Syst. 2015, 24, 1322–1330. [Google Scholar] [CrossRef]
- Aeinehvand, M.M.; Ibrahim, F.; Harun, S.W.; Kazemzadeh, A.; Rothan, H.A.; Yusof, R.; Madou, M. Reversible thermo-pneumatic valves on centrifugal microfluidic platforms. Lab Chip 2015, 15, 3358–3369. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, S.; Szilágyi, A.; Sumaru, K.; Hattori, K.; Takagi, T.; Filipcsei, G.; Zrínyi, M.; Kanamori, T. On-demand microfluidic control by micropatterned light irradiation of a photoresponsive hydrogel sheet. Lab Chip 2009, 9, 196–198. [Google Scholar] [CrossRef]
- Abi-Samra, K.; Hanson, R.; Madou, M.; Gorkin III, R.A. Infrared controlled waxes for liquid handling and storage on a CD-microfluidic platform. Lab Chip 2011, 11, 723–726. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.H.; Bonanno, J.; Yang, J.; Lenigk, R.; Grodzinski, P. Single-use, thermally actuated paraffin valves for microfluidic applications. Sens. Actuators B Chem. 2004, 98, 328–336. [Google Scholar] [CrossRef]
- Liu, C.; Mauk, M.G.; Bau, H.H. A disposable, integrated loop-mediated isothermal amplification cassette with thermally actuated valves. Microfluid Nanofluidics 2011, 11, 209–220. [Google Scholar] [CrossRef]
- Brennan, D.; Glynn, B.; Keegan, G.; McDonagh, C.; Barry, T.; Galvin, P. Incorporating asymmetric PCR and microarray hybridization protocols onto an integrated microfluidic device, screening for the Escherichia coli ssrA gene. Sens. Actuators B Chem. 2018, 261, 325–334. [Google Scholar] [CrossRef]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Boitard, L.; Cottinet, D.; Bremond, N.; Baudry, J.; Bibette, J. Growing microbes in millifluidic droplets. Eng. Life Sci. 2015, 15, 318–326. [Google Scholar] [CrossRef]
- Damodaran, S.P.; Eberhard, S.; Boitard, L.; Rodriguez, J.G.; Wang, Y.; Bremond, N.; Baudry, J.; Bibette, J.; Wollman, F.-A. A millifluidic study of cell-to-cell heterogeneity in growth-rate and cell-division capability in populations of isogenic cells of Chlamydomonas reinhardtii. PLoS ONE 2015, 10, e0118987. [Google Scholar] [CrossRef]
- Jiang, L.; Boitard, L.; Broyer, P.; Chareire, A.-C.; Bourne-Branchu, P.; Mahé, P.; Tournoud, M.; Franceschi, C.; Zambardi, G.; Baudry, J. Digital antimicrobial susceptibility testing using the MilliDrop technology. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 415–422. [Google Scholar] [CrossRef]
- Chong, Z.Z.; Tan, S.H.; Gañán-Calvo, A.M.; Tor, S.B.; Loh, N.H.; Nguyen, N.-T. Active droplet generation in microfluidics. Lab Chip 2016, 16, 35–58. [Google Scholar] [CrossRef]
- Churski, K.; Korczyk, P.; Garstecki, P. High-throughput automated droplet microfluidic system for screening of reaction conditions. Lab Chip 2010, 10, 816–818. [Google Scholar] [CrossRef]
- Thorsen, T.; Roberts, R.W.; Arnold, F.H.; Quake, S.R. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 2001, 86, 4163. [Google Scholar] [CrossRef]
- Anna, S.L.; Bontoux, N.; Stone, H.A. Formation of dispersions using “flow focusing” in microchannels. Appl. Phys. Lett. 2003, 82, 364–366. [Google Scholar] [CrossRef]
- Garstecki, P.; Fuerstman, M.J.; Stone, H.A.; Whitesides, G.M. Formation of droplets and bubbles in a microfluidic T-junction—Scaling and mechanism of break-up. Lab Chip 2006, 6, 437–446. [Google Scholar] [CrossRef] [PubMed]
- De Menech, M.; Garstecki, P.; Jousse, F.; Stone, H. Transition from squeezing to dripping in a microfluidic T-shaped junction. J. Fluid Mech. 2008, 595, 141–161. [Google Scholar] [CrossRef]
- Garstecki, P.; Stone, H.A.; Whitesides, G.M. Mechanism for flow-rate controlled breakup in confined geometries: A route to monodisperse emulsions. Phys. Rev. Lett. 2005, 94, 164501. [Google Scholar] [CrossRef] [PubMed]
- Huebner, A.; Bratton, D.; Whyte, G.; Yang, M.; Abell, C.; Hollfelder, F. Static microdroplet arrays: A microfluidic device for droplet trapping, incubation and release for enzymatic and cell-based assays. Lab Chip 2009, 9, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Link, D.; Anna, S.L.; Weitz, D.; Stone, H. Geometrically mediated breakup of drops in microfluidic devices. Phys. Rev. Lett. 2004, 92. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Gulati, S.; Edel, J.B. Pillar-induced droplet merging in microfluidic circuits. Lab Chip 2008, 8, 1837–1841. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Caen, O.; Vrignon, J.; Konrad, M.; Taly, V.; Baret, J.-C. Parallelized ultra-high throughput microfluidic emulsifier for multiplex kinetic assays. Biomicrofluidics 2015, 9. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Pan, M.; Gai, Y.; Pang, S.; Han, C.; Yang, C.; Tang, S.K. Optofluidic ultrahigh-throughput detection of fluorescent drops. Lab Chip 2015, 15, 1417–1423. [Google Scholar] [CrossRef] [Green Version]
- Novak, R.; Zeng, Y.; Shuga, J.; Venugopalan, G.; Fletcher, D.A.; Smith, M.T.; Mathies, R.A. Single-Cell Multiplex Gene Detection and Sequencing with Microfluidically Generated Agarose Emulsions. Angew. Chem. 2011, 50, 390–395. [Google Scholar] [CrossRef]
- Leng, X.; Zhang, W.; Wang, C.; Cui, L.; Yang, C.J. Agarose droplet microfluidics for highly parallel and efficient single molecule emulsion PCR. Lab Chip 2010, 10, 2841–2843. [Google Scholar] [CrossRef] [Green Version]
- Dorfman, K.D.; Chabert, M.; Codarbox, J.-H.; Rousseau, G.; De Cremoux, P.; Viovy, J.-L. Contamination-free continuous flow microfluidic polymerase chain reaction for quantitative and clinical applications. Anal. Chem. 2005, 77, 3700–3704. [Google Scholar] [CrossRef] [PubMed]
- Hartung, R.; Brösing, A.; Sczcepankiewicz, G.; Liebert, U.; Häfner, N.; Dürst, M.; Felbel, J.; Lassner, D.; Köhler, J. Application of an asymmetric helical tube reactor for fast identification of gene transcripts of pathogenic viruses by micro flow-through PCR. Biomed. Microdevices 2009, 11, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Markey, A.L.; Mohr, S.; Day, P.J. High-throughput droplet PCR. Methods 2010, 50, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, T.; Kuyama, H.; Hanafusa, N.; Togawa, Y. A simple device using magnetic transportation for droplet-based PCR. Biomed. Microdevices 2007, 9, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Kinz, E.; Leiherer, A.; Lang, A.; Drexel, H.; Muendlein, A. Accurate quantitation of JAK2 V617F allele burden by array-based digital PCR. Int. J. Lab. Hematol. 2015, 37, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Hatch, A.C.; Fisher, J.S.; Tovar, A.R.; Hsieh, A.T.; Lin, R.; Pentoney, S.L.; Yang, D.L.; Lee, A.P. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip 2011, 11, 3838–3845. [Google Scholar] [CrossRef] [PubMed]
- Beneyton, T.; Coldren, F.; Baret, J.-C.; Griffiths, A.D.; Taly, V. CotA laccase: High-throughput manipulation and analysis of recombinant enzyme libraries expressed in E. coli using droplet-based microfluidics. Analyst 2014, 139, 3314–3323. [Google Scholar] [CrossRef]
- Lim, S.W.; Tran, T.M.; Abate, A.R. PCR-activated cell sorting for cultivation-free enrichment and sequencing of rare microbes. PLoS ONE 2015, 10, e0113549. [Google Scholar] [CrossRef]
- Sidore, A.M.; Lan, F.; Lim, S.W.; Abate, A.R. Enhanced sequencing coverage with digital droplet multiple displacement amplification. Nucleic Acids Res. 2015, 44, e66. [Google Scholar] [CrossRef]
- Easley, C.J.; Karlinsey, J.M.; Bienvenue, J.M.; Legendre, L.A.; Roper, M.G.; Feldman, S.H.; Hughes, M.A.; Hewlett, E.L.; Merkel, T.J.; Ferrance, J.P. A fully integrated microfluidic genetic analysis system with sample-in–answer-out capability. Proc. Natl. Acad. Sci. USA 2006, 103, 19272–19277. [Google Scholar] [CrossRef]
- Hua, Z.; Rouse, J.L.; Eckhardt, A.E.; Srinivasan, V.; Pamula, V.K.; Schell, W.A.; Benton, J.L.; Mitchell, T.G.; Pollack, M.G. Multiplexed real-time polymerase chain reaction on a digital microfluidic platform. Anal. Chem. 2010, 82, 2310–2316. [Google Scholar] [CrossRef] [PubMed]
- Mellors, J.W.; Rinaldo, C.R.; Gupta, P.; White, R.M.; Todd, J.A.; Kingsley, L.A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 1996, 272, 1167–1170. [Google Scholar] [CrossRef] [PubMed]
- Heredia, N.J.; Belgrader, P.; Wang, S.; Koehler, R.; Regan, J.; Cosman, A.M.; Saxonov, S.; Hindson, B.; Tanner, S.C.; Brown, A.S. Droplet Digital™ PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods 2013, 59, S20–S23. [Google Scholar] [CrossRef] [PubMed]
- Heyries, K.A.; Tropini, C.; VanInsberghe, M.; Doolin, C.; Petriv, I.; Singhal, A.; Leung, K.; Hughesman, C.B.; Hansen, C.L. Megapixel digital PCR. Nat. Methods 2011, 8, 649. [Google Scholar] [CrossRef] [PubMed]
- Kreutz, J.E.; Munson, T.; Huynh, T.; Shen, F.; Du, W.; Ismagilov, R.F. Theoretical design and analysis of multivolume digital assays with wide dynamic range validated experimentally with microfluidic digital PCR. Anal. Chem. 2011, 83, 8158–8168. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Sun, B.; Kreutz, J.E.; Davydova, E.K.; Du, W.; Reddy, P.L.; Joseph, L.J.; Ismagilov, R.F. Multiplexed quantification of nucleic acids with large dynamic range using multivolume digital RT-PCR on a rotational SlipChip tested with HIV and hepatitis C viral load. J. Am. Chem. Soc. 2011, 133, 17705–17712. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, S.; Valiadi, M.; Tsaloglou, M.-N.; Parry-Jones, L.; Jacobs, A.; Watson, R.; Turner, C.; Amos, R.; Hadwen, B.; Buse, J. Rapid and sensitive detection of antibiotic resistance on a programmable digital microfluidic platform. Lab Chip 2015, 15, 3065–3075. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, S.; Sellars, S.L.; Turner, C.; Sutton, J.M.; Morgan, H. A programmable digital microfluidic assay for the simultaneous detection of multiple anti-microbial resistance genes. Micromachines 2017, 8, 111. [Google Scholar] [CrossRef]
- Pompano, R.R.; Liu, W.; Du, W.; Ismagilov, R.F. Microfluidics using spatially defined arrays of droplets in one, two, and three dimensions. Annu. Rev. Anal. Chem. 2011, 4, 59–81. [Google Scholar] [CrossRef]
- Xu, B.; Nguyen, N.-T.; Wong, T.N. Temperature-induced droplet coalescence in microchannels. Biomicrofluidics 2012, 6. [Google Scholar] [CrossRef]
- Ma, Y.-D.; Luo, K.; Chang, W.-H.; Lee, G.-B. A microfluidic chip capable of generating and trapping emulsion droplets for digital loop-mediated isothermal amplification analysis. Lab Chip 2018, 18, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Byrnes, S.A.; Chang, T.C.; Huynh, T.; Astashkina, A.; Weigl, B.H.; Nichols, K.P. Simple Polydisperse Droplet Emulsion Polymerase Chain Reaction with Statistical Volumetric Correction Compared with Microfluidic Droplet Digital Polymerase Chain Reaction. Anal. Chem. 2018, 90, 9374–9380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Mathies, R.A. Integrated microfluidic systems for high-performance genetic analysis. Trends Biotechnol. 2009, 27, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Mariella, R. Sample preparation: The weak link in microfluidics-based biodetection. Biomed. Microdevices 2008, 10, 777. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Mauk, M.; Qiu, X.; Liu, C.; Kim, J.; Ramprasad, S.; Ongagna, S.; Abrams, W.R.; Malamud, D.; Corstjens, P.L. An integrated, self-contained microfluidic cassette for isolation, amplification, and detection of nucleic acids. Biomed. Microdevices 2010, 12, 705–719. [Google Scholar] [CrossRef] [Green Version]
- Lien, K.-Y.; Liu, C.-J.; Kuo, P.-L.; Lee, G.-B. Microfluidic system for detection of α-thalassemia-1 deletion using saliva samples. Anal. Chem. 2009, 81, 4502–4509. [Google Scholar] [CrossRef] [PubMed]
- Oblath, E.A.; Henley, W.H.; Alarie, J.P.; Ramsey, J.M. A microfluidic chip integrating DNA extraction and real-time PCR for the detection of bacteria in saliva. Lab Chip 2013, 13, 1325–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-H.; Wang, C.-H.; Wu, J.-J.; Lee, G.-B. Rapid detection of live methicillin-resistant Staphylococcus aureus by using an integrated microfluidic system capable of ethidium monoazide pre-treatment and molecular diagnosis. Biomicrofluidics 2012, 6. [Google Scholar] [CrossRef]
- Chang, W.-H.; Wang, C.-H.; Lin, C.-L.; Wu, J.-J.; Lee, M.S.; Lee, G.-B. Rapid detection and typing of live bacteria from human joint fluid samples by utilizing an integrated microfluidic system. Biosens. Bioelectron. 2015, 66, 148–154. [Google Scholar] [CrossRef]
- Chen, S.-L.; Chang, W.-H.; Wang, C.-H.; You, H.-L.; Wu, J.-J.; Liu, T.-H.; Lee, M.S.; Lee, G.-B. An integrated microfluidic system for live bacteria detection from human joint fluid samples by using ethidium monoazide and loop-mediated isothermal amplification. Microfluid Nanofluidics 2017, 21, 87. [Google Scholar] [CrossRef]
- Karle, M.; Vashist, S.K.; Zengerle, R.; von Stetten, F. Microfluidic solutions enabling continuous processing and monitoring of biological samples: A review. Anal. Chim. Acta 2016, 929, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.-Y.; Wang, C.-H.; Che, Y.-J.; Kao, C.-Y.; Wu, J.-J.; Lee, G.-B. An integrated microfluidic system for diagnosis of the resistance of Helicobacter pylori to quinolone-based antibiotics. Biosens. Bioelectron. 2016, 78, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.L.; Zhang, Y.; Lee, N.Y. A functionally integrated thermoplastic microdevice for one-step solid-phase-based nucleic acid purification and isothermal amplification for facile detection of foodborne pathogen. Biotechnol. Bioeng. 2016, 113, 2614–2623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tran, H.H.; Chung, B.H.; Lee, N.Y. Solid-phase based on-chip DNA purification through a valve-free stepwise injection of multiple reagents employing centrifugal force combined with a hydrophobic capillary barrier pressure. Analyst 2013, 138, 1750–1757. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; De Jonghe, J.; Kulesa, A.B.; Feldman, D.; Vatanen, T.; Bhattacharyya, R.P.; Berdy, B.; Gomez, J.; Nolan, J.; Epstein, S. High-throughput automated microfluidic sample preparation for accurate microbial genomics. Nat. Commun. 2017, 8, 13919. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, P.; Evander, M.; Petersson, K.; Mellhammar, L.; Lehmusvuori, A.; Karhunen, U.; Soikkeli, M.; Seppä, T.; Tuunainen, E.; Spangar, A. Integrated acoustic separation, enrichment, and microchip polymerase chain reaction detection of bacteria from blood for rapid sepsis diagnostics. Anal. Chem. 2016, 88, 9403–9411. [Google Scholar] [CrossRef] [PubMed]
- Ip, K.-U.; Chang, J.-R.; Liu, T.-H.; Dou, H.-Y.; Lee, G.-B. An integrated microfluidic system for identification of live mycobacterium tuberculosis by real-time polymerase chain reaction. In Proceedings of the 2018 IEEE Micro Electro Mechanical Systems (MEMS), Belfast, UK, 21–25 Jannuary 2018; pp. 1124–1127. [Google Scholar]
- Yu, J.-C.; Hu, C.-C.; Chang, W.-H.; Chen, P.-C.; Lee, M.S.; Peng, K.-T.; Lee, G.-B. An integrated microfluidic system using mannose-binding lectin for bacteria isolation and biofilm-related gene detection. Microfluid Nanofluidics 2018, 22, 13. [Google Scholar] [CrossRef]






| Main Focus of the Review | Year |
|---|---|
| Review of few studies reporting microfabrication of PCR in microbiology [37] | 2007 |
| A comprehensive review paper of miniaturized isothermal nucleic acid amplification [38] | 2011 |
| A review on the general use of microfluidics in bacterial pathogens monitoring with less focus on amplification methods [39] | 2014 |
| A review of works based on centrifugal microfluidic platforms for NA detection and amplification in microbial samples [28] | 2015 |
| A review of a few studies on microfluidic PCR for bacterial pathogens identification [40] | 2015 |
| A comprehensive and systematic review of loop-mediated isothermal amplification-based microfluidics for pathogenic nucleic acid detection [32] | 2016 |
| A review of microfluidics based microbial engineering with a brief explanation of bacterial genotyping [19] | 2016 |
| A review of general PCR microfluidic devices [20] | 2016 |
| A review describing applications of droplet microfluidics in microbiology [41] | 2016 |
| A comprehensive review focusing on detection of microorganisms using microfluidic-based analytical approaches [42] | 2018 |
| A review of a few studies on micro-scale bacterial NA amplification [43] | 2018 |
| Target Bacteria | Sensitivity | Time (cycles) | Detection Technology | Heating Technology | Ref. |
|---|---|---|---|---|---|
| Staphylococcus aureus | Below 10 copies of DNA per well | 110 min (50 cycles) | FAM-labeled hydrolysis probes; real-time fluorescence detection | Air mediated in commercially available PCR thermocycler | [78] |
| Staphylococcus aureus | Less than 7 copies per sample | 17 min (10 cycles) primary PCR; 52 min (50 cycles) main PCR | FAM-labeled hydrolysis probes; real-time fluorescence detection | Air mediated in commercially available PCR thermocycler | [79] |
| Salmonella enterica | Amplification of one gene from single cell | 8.33–20.83 min (20–50 cycles) | FAM-labeled hydrolysis probes; post-PCR fluorescence detection | Contact | [69] |
| Bacillus anthracis, Bacillus cereus | Not mentioned | 53 min (35 cycles) | Off-chip (analysis of PCR products by gel electrophoresis) | Contact; with thermoelectric modules | [70] |
| Listeria monocytogenes, Salmonella typhimurium, EHEC, S. aureus, Citrobacter freundii, and Campylobacter jejuni | 0.1 pg DNA per well for Salmonella and Listeria | Around 2 h (50 cycles) | FAM-labeled hydrolysis probes; real-time fluorescence detection | Air mediated in commercially available PCR thermocycler | [71] |
| Escherichia coli | Not mentioned | Around 54 min (30 cycles) | Agarose gel pre-stained with ethidium bromide | Convection of hot air and ambient air in commercially available PCR thermocycler | [72] |
| Haemophilus influenzae | Fluorescence sensitivity down to 100 CE with tmRNA | 70 min (Isothermal) | FAM-labeled beacon probes; fluorescence detection | Non-contact infrared (IR) heating | [66] |
| 24 Pneumonia-related pathogens | As few as 10 copies | 45 min (Isothermal) | Real-time fluorescence detection | Contact | [73] |
| Mycobacterium tuberculosis | 102 colony-forming unit per millilitre | 15 min (Isothermal) | Real-time fluorescence detection | Contact (printed circuit board heater) | [67] |
| E. coli O157:H7, S. typhimurium, and Vibrio parahaemolyticus | 380 copies | 60 min (Isothermal) | Colorimetric detection using eriochrome black T; naked eye | Contact | [80] |
| E. coli O157:H7, S. typhimurium, and V. parahaemolyticus | 500 copies | 60 min (Isothermal) | Colorimetric detection using eriochrome black T; naked eye | Contact | [81] |
| E. coli, Salmonella spp, and Vibrio cholerae | 3 × 10−5 ng·μL−1 | 60 min (Isothermal) | Calcein colorimetric method; smart phone | Contact | [65] |
| Dye | Colour Before Amplification | Colour After Amplification | Prevents LAMP | Ref |
|---|---|---|---|---|
| Hydroxynaphtol blue (HNB) | Violet | blue | no | [125] |
| Mixed-dye (HNB + SYBR Green I) | Orange-red | green | no | [126] |
| NeuRed | Light brown | pink | no | [127] |
| Gold nanoparticles | Purple | red | no | [29] |
| Calcin | Yellow | green | no | [31,128,129] |
| SYBR GREEN | Dark orange | green | no | [130] |
| HNB Calcein | Purple Yellow | Blue green | no | [131] |
| Platform | Complexity | Sample Volume | Assay Time | Throughput | Sensitivity | Utility |
|---|---|---|---|---|---|---|
| Serpentine [59] | -Several heaters and pumping are required-Complex channel design | 0.35 μL | 18 min | Low | 0.031 pg/μL | On-site gene testing |
| Oscillating-flow [62] | -Pumping is required-Complex design-Low detection speed | 2 μL | 12 min | Low | 10 DNA copies | On-the-spot analysis |
| Centrifugal [66] | -Elaborate designs and rotating platforms-Complex electronic components | 40 μL | 70 min | Low | 100 CE with tmRNA | Clinical application |
| Lab disk [86] | -Very complex design-Long analysis time-Mostly unsuitable for multiplexing | 4.8 μL of the sample plus preloaded primers and LAMP reagents | 60 min | Low | 2 × 102 cells per μL | Nucleic acid diagnostics in resource-limited settings particularly in clinical stage |
| Array [102] | -Robotic liquid handling is required-Complex and expensive | 20 uL | ~60 min | High | High | Water distribution systems, clinical field |
| LAMP-based [139] | -Difficult naked eye detection in a few microliters of sample-Instruments for visualisation are required | 600 nL | ~70 min | Low | 3 copies/μL | Applications in point-of-care settings |
| Droplet-based [201] | -Difficult droplet manipulation-Evaporation during thermal cycling-Droplet-to-droplet coalescence-Limited to laboratories with trained personnel-Expensive | Droplets ranging in diameter from ~1.5 to 13,117 μm with a median diameter of ∼56 μm (90 pL) | ~70 min | High | 0.682 copies/μL | Variety of settings for the quantification of nucleic acids in complex samples |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorgannezhad, L.; Stratton, H.; Nguyen, N.-T. Microfluidic-Based Nucleic Acid Amplification Systems in Microbiology. Micromachines 2019, 10, 408. https://doi.org/10.3390/mi10060408
Gorgannezhad L, Stratton H, Nguyen N-T. Microfluidic-Based Nucleic Acid Amplification Systems in Microbiology. Micromachines. 2019; 10(6):408. https://doi.org/10.3390/mi10060408
Chicago/Turabian StyleGorgannezhad, Lena, Helen Stratton, and Nam-Trung Nguyen. 2019. "Microfluidic-Based Nucleic Acid Amplification Systems in Microbiology" Micromachines 10, no. 6: 408. https://doi.org/10.3390/mi10060408
APA StyleGorgannezhad, L., Stratton, H., & Nguyen, N.-T. (2019). Microfluidic-Based Nucleic Acid Amplification Systems in Microbiology. Micromachines, 10(6), 408. https://doi.org/10.3390/mi10060408