3D Printed Lab-on-a-Chip Platform for Chemical Stimulation and Parallel Analysis of Ion Channel Function
Abstract
:1. Introduction
2. Materials and Methods
2.1. 3D Printed Lab-on-a-Chip Platform
2.2. Pharmacological Reagents
2.3. Calcium Indicator
2.4. Cell Culture
2.5. Cell Line
2.6. Transient Transfection of HEK293 Cells for Calcium Imaging Experiments
2.7. PDL Coating of the 3D Printed Lab-on-a-Chip Platform for Functional Imaging Experiments
2.8. Cell Seeding for Functional Imaging Experiments
2.9. Staining of Cells with the Fluorescent Indicator Fluo-4 AM
2.10. Functional Imaging Experiments
2.11. Single Cell-Based Quantitative Image Analysis
2.12. Data Analysis and Visualization
2.13. Simulation of Fluid Dynamics
3. Results
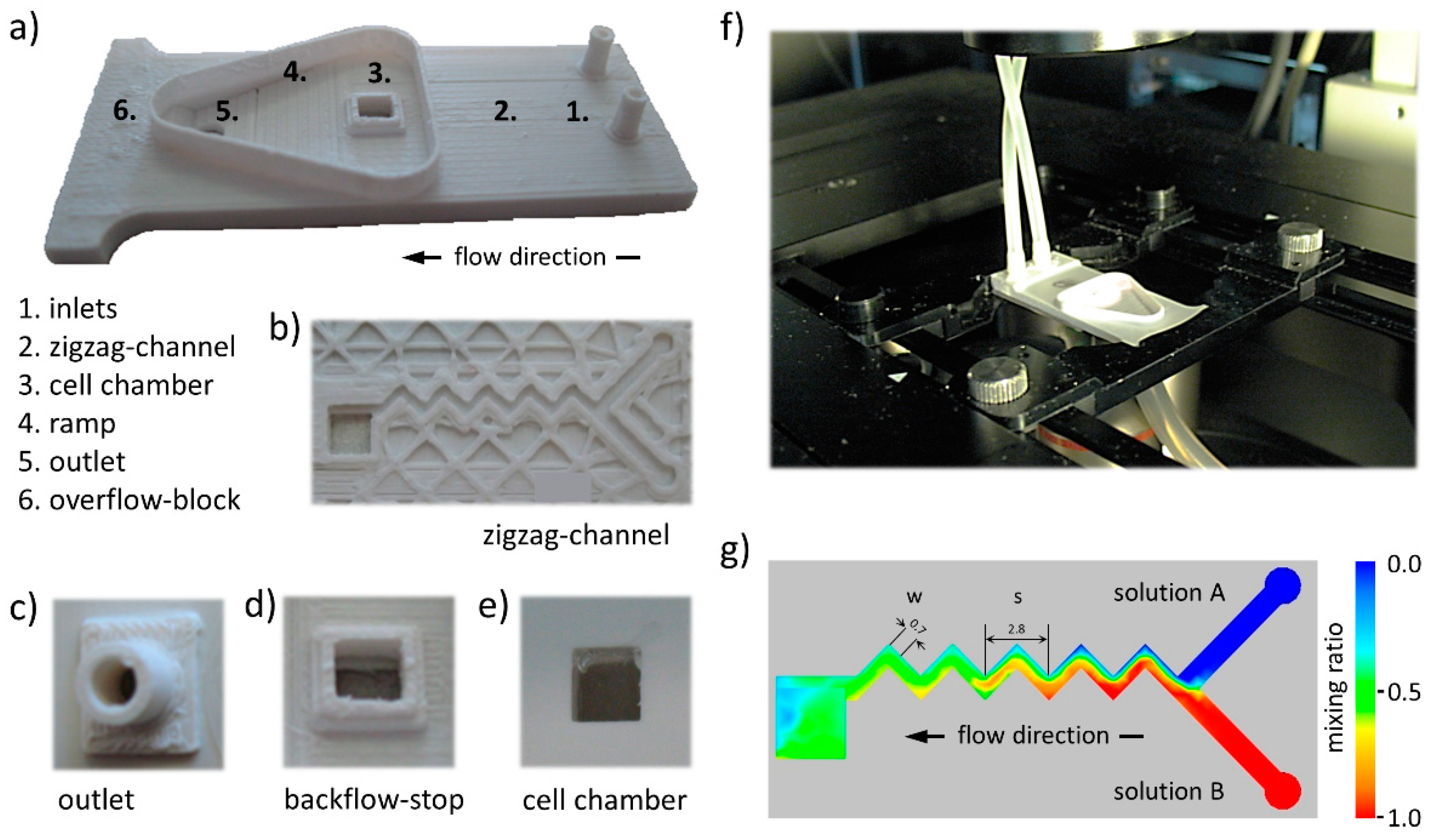
3.1. Ultra-Low-Cost 3D Printed Microfluidic Lab-on-a-Chip
3.2. Microfluidic Zigzag Channel for Generation of Homogeneous Mixtures
3.3. Chemical Stimulation and Parallel Microscopic Analysis of Ion Channel Function
3.4. Morphology-Based Viability Analysis of HEK293 Cells Cultured Inside the Microfluidic LOC
3.5. Example for Multiplication of the Platform in a Multichannel Microfluidic LOC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alexander, S.P.H.; Peters, J.A.; Kelly, E.; Marrion, N.; Benson, H.E.; Faccenda, E.; Pawson, A.J.; Sharman, J.L.; Southan, C.; Davies, J.A.; et al. The Concise Guide to PHARMACOLOGY 2015/16: Ligand-gated ion channels. Br. J. Pharmacol. 2015, 172, 5870–5903. [Google Scholar] [CrossRef] [PubMed]
- Wickenden, A.; Priest, B.; Erdemli, G. Ion channel drug discovery: Challenges and future directions. Future Med. Chem. 2012, 4, 661–679. [Google Scholar] [CrossRef] [PubMed]
- Becchetti, A.; Munaron, L.; Arcangeli, A. The role of ion channels and transporters in cell proliferation and cancer. Front. Physiol. 2013, 4, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kischel, P.; Girault, A.; Rodat-Despoix, L.; Chamlali, M.; Radoslavova, S.; Abou Daya, H.; Lefebvre, T.; Foulon, A.; Rybarczyk, P.; Hague, F.; et al. Ion channels: New actors playing in chemotherapeutic resistance. Cancers 2019, 11, 376. [Google Scholar] [CrossRef] [PubMed]
- Stoelzle, S.; Obergrussberger, A.; Bruggemann, A.; Haarmann, C.; George, M.; Kettenhofen, R.; Fertig, N. State-of-the-art automated patch clamp devices: Heat activation, action potentials, and high throughput in ion channel screening. Front. Pharmacol. 2011, 2, 76. [Google Scholar] [CrossRef] [PubMed]
- Hamill, O.P.; Marty, A.; Neher, E.; Sakmann, B.; Sigworth, F.J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Archiv: Eur. Physiol. 1981, 391, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Neher, E.; Sakmann, B. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature 1976, 260, 799–802. [Google Scholar] [CrossRef]
- Gilbert, D.F.; Islam, R.; Lynagh, T.; Lynch, J.W.; Webb, T.I. High throughput techniques for discovering new glycine receptor modulators and their binding sites. Front. Mol. Neurosci. 2009, 2, 17. [Google Scholar] [CrossRef]
- Gilbert, D.F.; Wilson, J.C.; Nink, V.; Lynch, J.W.; Osborne, G.W. Multiplexed labeling of viable cells for high-throughput analysis of glycine receptor function using flow cytometry. Cytometry A. J. Int. Soc. Anal. Cytol. 2009, 75, 440–449. [Google Scholar] [CrossRef]
- Kruger, W.; Gilbert, D.; Hawthorne, R.; Hryciw, D.H.; Frings, S.; Poronnik, P.; Lynch, J.W. A yellow fluorescent protein-based assay for high-throughput screening of glycine and GABAA receptor chloride channels. Neurosci. Lett. 2005, 380, 340–345. [Google Scholar] [CrossRef]
- Kuenzel, K.; Friedrich, O.; Gilbert, D.F. A recombinant human pluripotent stem cell line stably expressing halide-sensitive YFP-I152L for GABAAR and GlyR-targeted high-throughput drug screening and toxicity testing. Front. Mol. Neurosci. 2016, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Kuenzel, K.; Mofrad, S.A.; Gilbert, D.F. Phenotyping cellular viability by functional analysis of ion channels: GlyR-targeted screening in NT2-N cells. In Cell Viability Assays: Methods and Protocols; Gilbert, D.F., Friedrich, O., Eds.; Springer: New York, NY, USA, 2017; pp. 205–214. [Google Scholar] [CrossRef]
- Milanos, S.; Kuenzel, K.; Gilbert Daniel, F.; Janzen, D.; Sasi, M.; Buettner, A.; Frimurer Thomas, M.; Villmann, C. Structural changes at the myrtenol backbone reverse its positive allosteric potential into inhibitory GABAA receptor modulation. Biolog. Chem. 2018, 399, 549. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.J.; Ko, E.A.; Namkung, W.; Verkman, A.S. Microfluidics platform for single-shot dose–response analysis of chloride channel-modulating compounds. Lab Chip 2013, 13, 3862–3867. [Google Scholar] [CrossRef] [PubMed]
- Dertinger, S.K.W.; Chiu, D.T.; Jeon, N.L.; Whitesides, G.M. Generation of gradients having complex shapes using microfluidic networks. Anal. Chem. 2001, 73, 1240–1246. [Google Scholar] [CrossRef]
- Jeon, N.L.; Dertinger, S.K.W.; Chiu, D.T.; Choi, I.S.; Stroock, A.D.; Whitesides, G.M. Generation of solution and surface gradients using microfluidic systems. Langmuir 2000, 16, 8311–8316. [Google Scholar] [CrossRef]
- Jin, B.J.; Lee, S.; Verkman, A.S. Hollow micropillar array method for high-capacity drug screening on filter-grown epithelial cells. Anal. Chem. 2018, 90, 7675–7681. [Google Scholar] [CrossRef] [PubMed]
- Grünberger, A.; Wiechert, W.; Kohlheyer, D. Single-cell microfluidics: Opportunity for bioprocess development. Curr. Opin. Biotechnol. 2014, 29, 15–23. [Google Scholar] [CrossRef]
- Gilbert, D.F.; Mofrad, S.A.; Friedrich, O.; Wiest, J. Proliferation characteristics of cells cultured under periodic versus static conditions. Cytotechnology 2019, 71, 443–452. [Google Scholar] [CrossRef]
- Lembong, J.; Lerman, M.J.; Kingsbury, T.J.; Civin, C.I.; Fisher, J.P. A fluidic culture platform for spatially patterned cell growth, differentiation, and cocultures. Tissue Eng. Part A 2018, 24, 1715–1732. [Google Scholar] [CrossRef]
- Schneidereit, D.; Tschernich, J.; Friedrich, O.; Scharin-Mehlmann, M.; Gilbert, D.F. 3D-printed reusable cell culture chamber with integrated electrodes for electrical stimulation and parallel microscopic evaluation. 3D Print. Add. Manuf. 2018, 5, 115–125. [Google Scholar] [CrossRef]
- Landrain, T.; Meyer, M.; Perez, A.M.; Sussan, R. Do-it-yourself biology: challenges and promises for an open science and technology movement. Syst. Synth. Biol. 2013, 7, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Seyfried, G.; Pei, L.; Schmidt, M. European do-it-yourself (DIY) biology: Beyond the hope, hype and horror. Bioessays 2014, 36, 548–551. [Google Scholar] [CrossRef]
- Ferretti, F. Mapping do-it-yourself science. Life Sci. Soc. Policy 2019, 15, 1. [Google Scholar] [CrossRef]
- Kahl, M.; Gertig, M.; Hoyer, P.; Friedrich, O.; Gilbert, D.F. Ultra-low-cost 3D bioprinting: modification & application of an off-the-shelf desktop 3D-printer for biofabrication. Front. Bioeng. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Schmitt, M.A.; Friedrich, O.; Gilbert, D.F. Portoporator (c): A portable low-cost electroporation device for gene transfer to cultured cells in biotechnology, biomedical research and education. Biosens. Bioelectron. 2019, 131, 95–103. [Google Scholar] [CrossRef]
- Caterina, M.J.; Julius, D. The vanilloid receptor: A molecular gateway to the pain pathway. Annu. Rev. Neurosci. 2001, 24, 487–517. [Google Scholar] [CrossRef]
- Caterina, M.J.; Schumacher, M.A.; Tominaga, M.; Rosen, T.A.; Levine, J.D.; Julius, D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 1997, 389, 816–824. [Google Scholar] [CrossRef]
- Gee, K.R.; Brown, K.A.; Chen, W.N.; Bishop-Stewart, J.; Gray, D.; Johnson, I. Chemical and physiological characterization of fluo-4 Ca(2+)-indicator dyes. Cell calcium 2000, 27, 97–106. [Google Scholar] [CrossRef]
- Gilbert, D.; Esmaeili, A.; Lynch, J.W. Optimizing the expression of recombinant alphabetagamma GABAA receptors in HEK293 cells for high-throughput screening. J. Biomol. Screening 2009, 14, 86–91. [Google Scholar] [CrossRef]
- Gilbert, D.F.; Meinhof, T.; Pepperkok, R.; Runz, H. DetecTiff: A novel image analysis routine for high-content screening microscopy. J. Biomol. Screening 2009, 14, 944–955. [Google Scholar] [CrossRef]
- Lambert, J.W. Molecular Study of Capsaicin in Aqueous and Hydrophobic Environments; Virginia Polytechnic Institute and State University: Blacksburg, VA, USA, 2006. [Google Scholar]
- Su, Y.; Chen, G.; Kenig, E.Y. An experimental study on the numbering-up of microchannels for liquid mixing. Lab Chip 2015, 15, 179–187. [Google Scholar] [CrossRef]
- Li, F.; Zheng, Y.; Wu, J.; Zhao, L.; Shui, L.; Pu, Q.; Liu, S. Smartphone assisted immunodetection of HIV p24 antigen using reusable, centrifugal microchannel array chip. Talanta 2019, 203, 83–89. [Google Scholar] [CrossRef]
- Mengeaud, V.; Josserand, J.; Girault, H.H. Mixing processes in a zigzag microchannel: Finite element simulations and optical study. Anal. Chem. 2002, 74, 4279–4286. [Google Scholar] [CrossRef]
- Peterson, S.L.; McDonald, A.; Gourley, P.L.; Sasaki, D.Y. Poly(dimethylsiloxane) thin films as biocompatible coatings for microfluidic devices: Cell culture and flow studies with glial cells. J. Biomed. Mater. Res. Part A 2005, 72, 10–18. [Google Scholar] [CrossRef]
- Varga, M.; Wolff, P.; Wolter, K.J. Biocompatibility study of three distinct carbon pastes for application as electrode material in neural stimulations and recordings. J. Mater. Sci.-Mater. Med. 2017, 28, 30. [Google Scholar] [CrossRef]
- Chen, Q.; Mangadlao, J.D.; Wallat, J.; De Leon, A.; Pokorski, J.K.; Advincula, R.C. 3D printing biocompatible Polyurethane/Poly(lactic acid)/Graphene Oxide nanocomposites: Anisotropic properties. ACS Appl. Mater. Interfaces 2017, 9, 4015–4023. [Google Scholar] [CrossRef]
- Tajbakhsh, S.; Hajiali, F. A comprehensive study on the fabrication and properties of biocomposites of poly(lactic acid)/ceramics for bone tissue engineering. Mater. Sci. Eng. C Mater. Boil. App. 2017, 70, 897–912. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aschenbrenner, D.; Friedrich, O.; Gilbert, D.F. 3D Printed Lab-on-a-Chip Platform for Chemical Stimulation and Parallel Analysis of Ion Channel Function. Micromachines 2019, 10, 548. https://doi.org/10.3390/mi10080548
Aschenbrenner D, Friedrich O, Gilbert DF. 3D Printed Lab-on-a-Chip Platform for Chemical Stimulation and Parallel Analysis of Ion Channel Function. Micromachines. 2019; 10(8):548. https://doi.org/10.3390/mi10080548
Chicago/Turabian StyleAschenbrenner, Daniel, Oliver Friedrich, and Daniel F. Gilbert. 2019. "3D Printed Lab-on-a-Chip Platform for Chemical Stimulation and Parallel Analysis of Ion Channel Function" Micromachines 10, no. 8: 548. https://doi.org/10.3390/mi10080548
APA StyleAschenbrenner, D., Friedrich, O., & Gilbert, D. F. (2019). 3D Printed Lab-on-a-Chip Platform for Chemical Stimulation and Parallel Analysis of Ion Channel Function. Micromachines, 10(8), 548. https://doi.org/10.3390/mi10080548




