A Prominent Cell Manipulation Technique in BioMEMS: Dielectrophoresis
Abstract
:1. Introduction
2. DEP Background
3. Applications of DEP
3.1. Dielectric Characterization of Cells
3.2. Multidrug Resistance (MDR) Detection in Cancer Cells
3.3. Separation of Cells
3.4. DEP for other Purposes
4. Challenges, Conclusions, and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Menon, K.; Joy, R.A.; Sood, N.; Mittal, R.K. The Applications of BioMEMS in Diagnosis, Cell Biology, and Therapy: A Review. Bionanoscience 2013, 3, 356–366. [Google Scholar] [CrossRef]
- James, T.; Mannoor, M.S.; Ivanov, D.V. BioMEMS—Advancing the frontiers of medicine. Sensors 2008, 8, 6077–6107. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.K.; Dubey, V.; Mishra, P.M.; Khan, I. MEMS Technology: A Review. J. Eng. Res. Rep. 2019, 4, 1–24. [Google Scholar] [CrossRef]
- Ma, J. Advanced MEMS-based technologies and displays. Displays 2015, 37, 2–10. [Google Scholar] [CrossRef]
- Nisar, A.; Afzulpurkar, N.; Mahaisavariya, B.; Tuantranont, A. MEMS-based micropumps in drug delivery and biomedical applications. Sens. Actuators B Chem. 2008, 130, 917–942. [Google Scholar] [CrossRef]
- Villanueva, L.G.; Bausells, J.; Brugger, J. Grand Challenge in N/MEMS. Front. Mech. Eng. 2016, 1. [Google Scholar] [CrossRef] [Green Version]
- Madou, M.J. Fundamentals of Microfabrication The Science of Miniaturization; CRC Press: Boca Raton, FL, USA, 2002; ISBN 9780072483116. [Google Scholar]
- Gardner, J.W.; Varadan, V.K.; Awadelkarim, O.O. Microsensors, MEMS, and Smart Devices; John Wiley & Sons: West Sussex, UK, 2002; ISBN 047186109X. [Google Scholar]
- Villarruel Mendoza, L.A.; Scilletta, N.A.; Bellino, M.G.; Desimone, M.F.; Catalano, P.N. Recent Advances in Micro-Electro-Mechanical Devices for Controlled Drug Release Applications. Front. Bioeng. Biotechnol. 2020, 8, 1–28. [Google Scholar] [CrossRef]
- Mohammadi Aria, M.; Erten, A.; Yalcin, O. Technology Advancements in Blood Coagulation Measurements for Point-of-Care Diagnostic Testing. Front. Bioeng. Biotechnol. 2019, 7, 1–18. [Google Scholar] [CrossRef]
- Rebeiz, G.M. RF MEMS: Theory, Design and Technology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2003; ISBN 9781439833230. [Google Scholar]
- Ho, C.; Tai, Y.-C. Micro-Electro-Mechanical-Systems (MEMS) and Fluid Flows. Annu. Rev. Fluid Mech. 1998, 30, 579–612. [Google Scholar] [CrossRef] [Green Version]
- Liu, A.-Q. Photonic MEMS Devices Design, Fabrication and Control; Thompson, B.J., Ed.; CRC Press: Boca Raton, FL, USA, 2011; ISBN 978-1-4398-0279-3. [Google Scholar]
- Bashir, R. BioMEMS: State-of-the-art in detection, opportunities and prospects. Adv. Drug Deliv. Rev. 2004, 56, 1565–1586. [Google Scholar] [CrossRef]
- Azizipour, N.; Avazpour, R.; Rosenzweig, D.H.; Sawan, M.; Ajji, A. Evolution of biochip technology: A review from lab-on-a-chip to organ-on-a-chip. Micromachines 2020, 11, 599. [Google Scholar] [CrossRef]
- Polla, D.L.; Erdman, A.G.; Robbins, W.P.; Markus, D.T.; Diaz-diaz, J.; Rizq, R.; Wang, A.; Krulevitch, P. Microdevices in Medicine. Annu. Rev. Biomed. Eng. 2000, 2, 551–576. [Google Scholar] [CrossRef]
- Folch, A. Introduction to BioMEMS; CRC Press: Boca Raton, FL, USA, 2013; ISBN 9781439818398. [Google Scholar]
- Bhattacharya, S.; Jang, J.; Yang, L.; Akin, D.; Bashir, R. BioMEMS and Nanotechnology-based Approaches for Rapid Detection of Biological Entities. J. Rapid Methods Autom. Microbiol. 2007, 15, 1–32. [Google Scholar] [CrossRef]
- Franssila, S.; Davis, C.E.; LeVasseur, M.K.; Cao, Z.; Yobas, L. Microfluidics and bioMEMS in silicon. In Handbook of Silicon Based MEMS Materials and Technologies; Tilli, M., Paulasto-Kröckel, M., Petzold, M., Theuss, H., Motooka, T., Lindroos, V., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 547–563. ISBN 9780128177860. [Google Scholar]
- Xu, Y.; Hu, X.; Kundu, S.; Nag, A.; Afsarimanesh, N.; Sapra, S.; Mukhopadhyay, S.C.; Han, T. Silicon-Based Sensors for Biomedical Applications. Sensors 2019, 19, 2908. [Google Scholar] [CrossRef] [Green Version]
- Caldorera-Moore, M.; Peppas, N.A. Micro- and Nanotechnologies for Intelligent and Responsive Biomaterial-Based Medical Systems. Adv. Drug Deliv. Rev. 2009, 61, 1391–1401. [Google Scholar] [CrossRef] [Green Version]
- Sia, S.K.; Whitesides, G.M. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef] [PubMed]
- Nordström, M.; Keller, S.; Lillemose, M.; Johansson, A.; Dohn, S.; Haefliger, D.; Blagoi, G.; Havsteen-Jakobsen, M.; Boisen, A. SU-8 cantilevers for bio/chemical sensing; fabrication, characterisation and development of novel read-out methods. Sensors 2008, 8, 1595–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsao, C.W. Polymer microfluidics: Simple, low-cost fabrication process bridging academic lab research to commercialized production. Micromachines 2016, 7, 225. [Google Scholar] [CrossRef] [Green Version]
- Becker, H.; Gärtner, C. Polymer microfabrication methods for microfluidic analytical applications. Electrophoresis 2000, 21, 12–26. [Google Scholar] [CrossRef]
- Soper, S.A.; Henry, A.C.; Vaidya, B.; Galloway, M.; Wabuyele, M.; McCarley, R.L. Surface modification of polymer-based microfluidic devices. Anal. Chim. Acta 2002, 470, 87–99. [Google Scholar] [CrossRef]
- Nuxoll, E. BioMEMS in drug delivery. Adv. Drug Deliv. Rev. 2013, 65, 1611–1625. [Google Scholar] [CrossRef] [PubMed]
- Coffel, J.; Nuxoll, E. BioMEMS for biosensors and closed-loop drug delivery. Int. J. Pharm. 2018, 544, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Ceylan Koydemir, H.; Külah, H.; Özgen, C.; Alp, A.; Hasçelik, G. MEMS biosensors for detection of methicillin resistant Staphylococcus aureus. Biosens. Bioelectron. 2011, 29, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Caulfield, B.; Reginatto, B.; Slevin, P. Not all sensors are created equal: A framework for evaluating human performance measurement technologies. NPJ Digit. Med. 2019, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thevenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical Biosensors: Recommended Definitions and Classification. Pure Appl. Chem. 1999, 71, 2333–2348. [Google Scholar] [CrossRef] [Green Version]
- Lowe, C.R. Overview of Biosensor and Bioarray Technologies. In Handbook of Biosensors and Biochips; Marks, R.S., Cullen, D.C., Karube, I., Lowe, C.R., Weetall, H.H., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2008; ISBN 9780470019054. [Google Scholar]
- Perumal, V.; Hashim, U. Advances in biosensors: Principle, architecture and applications. J. Appl. Biomed. 2014, 12, 1–15. [Google Scholar] [CrossRef]
- Zhu, C.; Yang, G.; Li, H.; Du, D.; Lin, Y. Electrochemical sensors and biosensors based on nanomaterials and nanostructures. Anal. Chem. 2015, 87, 230–249. [Google Scholar] [CrossRef]
- Vigneshvar, S.; Sudhakumari, C.C.; Senthilkumaran, B.; Prakash, H. Recent advances in biosensor technology for potential applications—an overview. Front. Bioeng. Biotechnol. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Qian, L.; Li, Q.; Baryeh, K.; Qiu, W.; Li, K.; Zhang, J.; Yu, Q.; Xu, D.; Liu, W.; Brand, R.E.; et al. Biosensors for early diagnosis of pancreatic cancer: A review. Transl. Res. 2019, 213, 67–89. [Google Scholar] [CrossRef]
- Derkus, B. Applying the miniaturization technologies for biosensor design. Biosens. Bioelectron. 2016, 79, 901–913. [Google Scholar] [CrossRef]
- Kumar, S.; Tripathy, S.; Jyoti, A.; Singh, S.G. Recent advances in biosensors for diagnosis and detection of sepsis: A comprehensive review. Biosens. Bioelectron. 2019, 124–125, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Mohankumar, P.; Ajayan, J.; Mohanraj, T.; Yasodharan, R. Recent developments in biosensors for healthcare and biomedical applications: A review. Measurement 2020, 167, 108293. [Google Scholar] [CrossRef]
- Jayanthi, V.S.P.K.S.A.; Das, A.B.; Saxena, U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Metkar, S.K.; Girigoswami, K. Diagnostic biosensors in medicine—A review. Biocatal. Agric. Biotechnol. 2019, 17, 271–283. [Google Scholar] [CrossRef]
- Narayan, R.J. (Ed.) Medical Biosensors for Point of Care (POC) Applications; Elsevier: Woodhead Publishing: Duxford, UK, 2017; ISBN 9780081000786. [Google Scholar]
- Gubala, V.; Harris, L.F.; Ricco, A.J.; Tan, M.X.; Williams, D.E. Point of care diagnostics: Status and future. Anal. Chem. 2012, 84, 487–515. [Google Scholar] [CrossRef]
- Sandbhor Gaikwad, P.; Banerjee, R. Advances in point-of-care diagnostic devices in cancers. Analyst 2018, 143, 1326–1348. [Google Scholar] [CrossRef]
- Jung, W.; Han, J.; Choi, J.W.; Ahn, C.H. Point-of-care testing (POCT) diagnostic systems using microfluidic lab-on-a-chip technologies. Microelectron. Eng. 2015, 132, 46–57. [Google Scholar] [CrossRef]
- Choi, S.; Goryll, M.; Sin, L.Y.M.; Wong, P.K.; Chae, J. Microfluidic-based biosensors toward point-of-care detection of nucleic acids and proteins. Microfluid. Nanofluidics 2011, 10, 231–247. [Google Scholar] [CrossRef]
- Zhu, H.; Fohlerová, Z.; Pekárek, J.; Basova, E.; Neužil, P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020, 153. [Google Scholar] [CrossRef]
- Lafleur, J.P.; Jönsson, A.; Senkbeil, S.; Kutter, J.P. Recent advances in lab-on-a-chip for biosensing applications. Biosens. Bioelectron. 2016, 76, 213–233. [Google Scholar] [CrossRef]
- Isiksacan, Z.; Guler, M.T.; Kalantarifard, A.; Asghari, M.; Elbuken, C. Lab-on-a-Chip Platforms for Disease Detection and Diagnosis. In Biosensors and Nanotechnology: Applications in Health Care Diagnostics; Altıntas, Z., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018. [Google Scholar]
- Wu, J.; Dong, M.; Santos, S.; Rigatto, C.; Liu, Y.; Lin, F. Lab-on-a-chip platforms for detection of cardiovascular disease and cancer biomarkers. Sensors 2017, 17, 2934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mark, D.; Haeberle, S.; Roth, G.; Stetten, F.V.; Zengerle, R. Microfluidic lab-on-a-chip platforms: Requirements, characteristics and applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.K.; Wu, R.G.; Chuang, Y.J.; Khoo, H.S.; Huang, S.H.; Tseng, F.G. Microfluidic systems for biosensing. Sensors 2010, 10, 6623–6661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samiei, E.; Tabrizian, M.; Hoorfar, M. A review of digital microfluidics as portable platforms for lab-on a-chip applications. Lab Chip 2016, 16, 2376–2396. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Luka, G.; Ahmadi, A.; Najjaran, H.; Alocilja, E.; Derosa, M.; Wolthers, K.; Malki, A.; Aziz, H.; Althani, A.; Hoorfar, M. Microfluidics integrated biosensors: A leading technology towards lab-on-A-chip and sensing applications. Sensors 2015, 15, 30011–30031. [Google Scholar] [CrossRef] [Green Version]
- Keçili, R.; Büyüktiryaki, S.; Hussain, C.M. Micro total analysis systems with nanomaterials. In Handbook of Nanomaterials in Analytical Chemistry: Modern Trends in Analysis; Elsevier: Amsterdam, The Netherlands, 2020; pp. 185–198. ISBN 9780128166994. [Google Scholar]
- Manz, A.; Grabner, N.; Widmer, H.M. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sensors Actuators B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Guo, L.; Feng, J.; Fang, Z.; Xu, J.; Lu, X. Application of microfluidic “lab-on-a-chip” for the detection of mycotoxins in foods. Trends Food Sci. Technol. 2015, 46, 252–263. [Google Scholar] [CrossRef]
- Kovarik, M.L.; Ornoff, D.M.; Melvin, A.T.; Dobes, N.C.; Wang, Y.; Dickinson, A.J.; Gach, P.C.; Shah, P.K.; Allbritton, N.L. Micro total analysis systems: Fundamental advances and applications in the laboratory, clinic, and field. Anal. Chem. 2013, 85, 451–472. [Google Scholar] [CrossRef] [Green Version]
- Dittrich, P.S.; Tachikawa, K.; Manz, A. Micro total analysis systems. Latest advancements and trends. Anal. Chem. 2006, 78, 3887–3907. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Simone, G.; Salieb-Beugelaar, G.B.; Kim, J.T.; Manz, A. Latest developments in micro total analysis systems. Anal. Chem. 2010, 82, 4830–4847. [Google Scholar] [CrossRef] [PubMed]
- Saliterman, S. Fundamentals of BioMEMS and Medical Microdevices; SPIE Press: Bellingham, WA, USA, 2006. [Google Scholar]
- Sajeesh, P.; Sen, A.K. Particle separation and sorting in microfluidic devices: A review. Microfluid. Nanofluidics 2014, 17, 1–52. [Google Scholar] [CrossRef]
- Çetin, B.; Özer, M.B.; Solmaz, M.E. Microfluidic bio-particle manipulation for biotechnology. Biochem. Eng. J. 2014, 92, 63–82. [Google Scholar] [CrossRef]
- Dalili, A.; Samiei, E.; Hoorfar, M. A review of sorting, separation and isolation of cells and microbeads for biomedical applications: Microfluidic approaches. Analyst 2019, 144, 87–113. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, S.; Yuan, D.; Alici, G.; Nguyen, N.T.; Ebrahimi Warkiani, M.; Li, W. Fundamentals and applications of inertial microfluidics: A review. Lab Chip 2016, 16, 10–34. [Google Scholar] [CrossRef] [Green Version]
- Di Carlo, D. Inertial microfluidics. Lab Chip 2009, 9, 3038–3046. [Google Scholar] [CrossRef]
- Liu, C.; Hu, G. High-throughput particle manipulation based on hydrodynamic effects in microchannels. Micromachines 2017, 8, 73. [Google Scholar] [CrossRef] [Green Version]
- Bayareh, M. An updated review on particle separation in passive microfluidic devices. Chem. Eng. Process. Process Intensif. 2020, 153. [Google Scholar] [CrossRef]
- Pamme, N. Magnetism and microfluidics. Lab Chip 2006, 6, 24–38. [Google Scholar] [CrossRef]
- Van Reenen, A.; De Jong, A.M.; Den Toonder, J.M.J.; Prins, M.W.J. Integrated lab-on-chip biosensing systems based on magnetic particle actuation-a comprehensive review. Lab Chip 2014, 14, 1966–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hejazian, M.; Li, W.; Nguyen, N.T. Lab on a chip for continuous-flow magnetic cell separation. Lab Chip 2015, 15, 959–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarn, M.D.; Lopez-Martinez, M.J.; Pamme, N. On-chip processing of particles and cells via multilaminar flow streams. Anal. Bioanal. Chem. 2014, 406, 139–161. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.K.; Koomson, E.; Zou, H.; Yi, C.; Li, C.W.; Xu, T.; Yang, M. Recent advances in microfluidic technology for manipulation and analysis of biological cells (2007–2017). Anal. Chim. Acta 2018, 1044, 29–65. [Google Scholar] [CrossRef]
- Dienerowitz, M.; Mazilu, M.; Dholakia, K. Optical manipulation of nanoparticles: A review. J. Nanophotonics 2008, 2, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Mohanty, S. Optically-actuated translational and rotational motion at the microscale for microfluidic manipulation and characterization. Lab Chip 2012, 12, 3624–3636. [Google Scholar] [CrossRef]
- Huang, N.T.; Zhang, H.L.; Chung, M.T.; Seo, J.H.; Kurabayashi, K. Recent advancements in optofluidics-based single-cell analysis: Optical on-chip cellular manipulation, treatment, and property detection. Lab Chip 2014, 14, 1230–1245. [Google Scholar] [CrossRef]
- Wang, X.; Chen, S.; Kong, M.; Wang, Z.; Costa, K.D.; Li, R.A.; Sun, D. Enhanced cell sorting and manipulation with combined optical tweezer and microfluidic chip technologies. Lab Chip 2011, 11, 3656–3662. [Google Scholar] [CrossRef]
- Nasiri, R.; Shamloo, A.; Ahadian, S.; Amirifar, L.; Akbari, J.; Goudie, M.J.; Lee, K.J.; Ashammakhi, N.; Dokmeci, M.R.; Di Carlo, D.; et al. Microfluidic-Based Approaches in Targeted Cell/Particle Separation Based on Physical Properties: Fundamentals and Applications. Small 2020, 16, 1–27. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Onck, P.; den Toonder, J. A concise review of microfluidic particle manipulation methods. Microfluid. Nanofluidics 2020, 24, 1–20. [Google Scholar] [CrossRef]
- Lenshof, A.; Magnusson, C.; Laurell, T. Acoustofluidics 8: Applications of acoustophoresis in continuous flow microsystems. Lab Chip 2012, 12, 1210–1223. [Google Scholar] [CrossRef] [PubMed]
- Guldiken, R.; Jo, M.C.; Gallant, N.D.; Demirci, U.; Zhe, J. Sheathless size-based acoustic particle separation. Sensors 2012, 12, 905–922. [Google Scholar] [CrossRef]
- Qiu, Y.; Wang, H.; Demore, C.E.M.; Hughes, D.A.; Glynne-Jones, P.; Gebhardt, S.; Bolhovitins, A.; Poltarjonoks, R.; Weijer, K.; Schönecker, A.; et al. Acoustic devices for particle and cell manipulation and sensing. Sensors 2014, 14, 14806–14838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkes, J.J.; Barber, R.W.; Emerson, D.R.; Coakley, W.T. Continuous cell washing and mixing driven by an ultrasound standing wave within a microfluidic channel. Lab Chip 2004, 4, 446–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurell, T.; Petersson, F.; Nilsson, A. Chip integrated strategies for acoustic separation and manipulation of cells and particles. Chem. Soc. Rev. 2007, 36, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Wyatt Shields, C., IV; Reyes, C.D.; López, G.P. Microfluidic cell sorting: A review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 2015, 15, 1230–1249. [Google Scholar] [CrossRef] [Green Version]
- Pohl, H.A.; Crane, J.S. Dielectrophoresis of Cells. Biophys. J. 1971, 11, 711–727. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Chang, H.; Neuzil, P. DEP-on-a-Chip: Dielectrophoresis Applied to Microfluidic Platforms. Micromachines 2019, 10, 423. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Zhu, G.; Zhao, T.; Takei, M. Microfluidic device embedding electrodes for dielectrophoretic manipulation of cells-A review. Electrophoresis 2019, 40, 1166–1177. [Google Scholar] [CrossRef]
- Rahman, N.A.; Ibrahim, F.; Yafouz, B. Dielectrophoresis for biomedical sciences applications: A review. Sensors 2017, 17, 449. [Google Scholar] [CrossRef] [Green Version]
- Pysher, M.D.; Hayes, M.A. Electrophoretic and dielectrophoretic field gradient technique for separating bioparticles. Anal. Chem. 2007, 79, 4552–4557. [Google Scholar] [CrossRef] [PubMed]
- Henslee, E.A.; Sano, M.B.; Rojas, A.D.; Schmelz, E.M.; Davalos, R.V. Selective concentration of human cancer cells using contactless dielectrophoresis. Electrophoresis 2011, 32, 2523–2529. [Google Scholar] [CrossRef] [PubMed]
- Alshareef, M.; Metrakos, N.; Juarez Perez, E.; Azer, F.; Yang, F.; Yang, X.; Wang, G. Separation of tumor cells with dielectrophoresis-based microfluidic chip. Biomicrofluidics 2013, 7. [Google Scholar] [CrossRef] [Green Version]
- Cheng, I.F.; Huang, W.L.; Chen, T.Y.; Liu, C.W.; Lin, Y.D.; Su, W.C. Antibody-free isolation of rare cancer cells from blood based on 3D lateral dielectrophoresis. Lab Chip 2015, 15, 2950–2959. [Google Scholar] [CrossRef] [PubMed]
- Khoshmanesh, K.; Baratchi, S.; Tovar-Lopez, F.J.; Nahavandi, S.; Wlodkowic, D.; Mitchell, A.; Kalantar-Zadeh, K. On-chip separation of Lactobacillus bacteria from yeasts using dielectrophoresis. Microfluid. Nanofluidics 2012, 12, 597–606. [Google Scholar] [CrossRef]
- Regtmeier, J.; Eichhorn, R.; Viefhues, M.; Bogunovic, L.; Anselmetti, D. Electrodeless dielectrophoresis for bioanalysis: Theory, devices and applications. Electrophoresis 2011, 32, 2253–2273. [Google Scholar] [CrossRef]
- Pethig, R.; Menachery, A.; Pells, S.; De Sousa, P. Dielectrophoresis: A review of applications for stem cell research. J. Biomed. Biotechnol. 2010. [Google Scholar] [CrossRef]
- Alazzam, A.; Stiharu, I.; Bhat, R.; Meguerditchian, A.N. Interdigitated comb-like electrodes for continuous separation of malignant cells from blood using dielectrophoresis. Electrophoresis 2011, 32, 1327–1336. [Google Scholar] [CrossRef]
- Gupta, V.; Jafferji, I.; Garza, M.; Melnikova, V.O.; Hasegawa, D.K.; Pethig, R.; Davis, D.W. ApoStreamTM, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012, 6. [Google Scholar] [CrossRef] [Green Version]
- Qian, C.; Huang, H.; Chen, L.; Li, X.; Ge, Z.; Chen, T.; Yang, Z.; Sun, L. Dielectrophoresis for bioparticle manipulation. Int. J. Mol. Sci. 2014, 15, 18281–18309. [Google Scholar] [CrossRef] [Green Version]
- Çetin, B.; Li, D. Dielectrophoresis in microfluidics technology. Electrophoresis 2011, 32, 2410–2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adekanmbi, E.O.; Srivastava, S.K. Dielectrophoretic applications for disease diagnostics using lab-on-a-chip platforms. Lab Chip 2016, 16, 2148–2167. [Google Scholar] [CrossRef] [PubMed]
- Broche, L.M.; Bhadal, N.; Lewis, M.P.; Porter, S.; Hughes, M.P.; Labeed, F.H. Early detection of oral cancer—Is dielectrophoresis the answer? Oral Oncol. 2007, 43, 199–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.Y.; Ahmad Kayani, A.B.; Md Ali, M.A.; Kok, C.K.; Yeop Majlis, B.; Hoe, S.L.L.; Marzuki, M.; Khoo, A.S.B.; Ostrikov, K.; Ataur Rahman, M.; et al. Dielectrophoresis-based microfluidic platforms for cancer diagnostics. Biomicrofluidics 2018, 12. [Google Scholar] [CrossRef]
- Mathew, B.; Alazzam, A.; Khashan, S.; Abutayeh, M. Lab-on-chip for liquid biopsy (LoC-LB) based on dielectrophoresis. Talanta 2017, 164, 608–611. [Google Scholar] [CrossRef]
- Szydzik, C.; Khoshmanesh, K.; Mitchell, A.; Karnutsch, C. Microfluidic platform for separation and extraction of plasma from whole blood using dielectrophoresis. Biomicrofluidics 2015, 9. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Daggolu, P.R.; Burgess, S.C.; Minerick, A.R. Dielectrophoretic characterization of erythrocytes: Positive ABO blood types. Electrophoresis 2008, 29, 5033–5046. [Google Scholar] [CrossRef]
- Han, S.I.; Lee, S.M.; Joo, Y.D.; Han, K.H. Lateral dielectrophoretic microseparators to measure the size distribution of blood cells. Lab Chip 2011, 11, 3864–3872. [Google Scholar] [CrossRef]
- Yoshioka, J.; Ohsugi, Y.; Yoshitomi, T.; Yasukawa, T.; Sasaki, N.; Yoshimoto, K. Label-free rapid separation and enrichment of bone marrow-derived mesenchymal stem cells from a heterogeneous cell mixture using a dielectrophoresis device. Sensors 2018, 18, 3007. [Google Scholar] [CrossRef] [Green Version]
- Elitas, M.; Dhar, N.; Schneider, K.; Valero, A.; Braschler, T.; McKinney, J.D.; Renaud, P. Dielectrophoresis as a single cell characterization method for bacteria. Biomed. Phys. Eng. Express 2017, 3, 015005. [Google Scholar] [CrossRef]
- Fernandez, R.E.; Rohani, A.; Farmehini, V.; Swami, N.S. Review: Microbial analysis in dielectrophoretic microfluidic systems. Anal. Chim. Acta 2017, 966, 11–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapizco-Encinas, B.H.; Simmons, B.A.; Cummings, E.B.; Fintschenko, Y. Insulator-based dielectrophoresis for the selective concentration and separation of live bacteria in water. Electrophoresis 2004, 25, 1695–1704. [Google Scholar] [CrossRef] [PubMed]
- Abt, V.; Gringel, F.; Han, A.; Neubauer, P.; Birkholz, M. Separation, characterization, and handling of microalgae by dielectrophoresis. Microorganisms 2020, 8, 540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, M.P.; Morgan, H.; Rixon, F.J.; Burt, J.P.H.; Pethig, R. Manipulation of herpes simplex virus type 1 by dielectrophoresis. Biochim. Biophys. Acta Gen. Subj. 1998, 1425, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Jesús-Pérez, N.M.; Lapizco-Encinas, B.H. Dielectrophoretic monitoring of microorganisms in environmental applications. Electrophoresis 2011, 32, 2331–2357. [Google Scholar] [CrossRef]
- Hayes, M.A. Dielectrophoresis of proteins: Experimental data and evolving theory. Anal. Bioanal. Chem. 2020, 412, 3801–3811. [Google Scholar] [CrossRef]
- Lapizco-Encinas, B.H.; Ozuna-Chacón, S.; Rito-Palomares, M. Protein manipulation with insulator-based dielectrophoresis and direct current electric fields. J. Chromatogr. A 2008, 1206, 45–51. [Google Scholar] [CrossRef]
- Nakano, A.; Ros, A. Protein dielectrophoresis: Advances, challenges, and applications. Electrophoresis 2013, 34, 1085–1096. [Google Scholar] [CrossRef] [Green Version]
- Viefhues, M.; Eichhorn, R. DNA dielectrophoresis: Theory and applications a review. Electrophoresis 2017, 38, 1483–1506. [Google Scholar] [CrossRef]
- Shokouhmand, H.; Abdollahi, A. Detection of cell-free DNA nanoparticles in insulator based dielectrophoresis systems. J. Chromatogr. A 2020, 1626. [Google Scholar] [CrossRef]
- Asbury, C.L.; Diercks, A.H.; Van Den Engh, G. Trapping of DNA by dielectrophoresis. Electrophoresis 2002, 23, 2658–2666. [Google Scholar] [CrossRef]
- Ayala-Mar, S.; Gallo-Villanueva, R.C.; González-Valdez, J. Dielectrophoretic manipulation of exosomes in a multi-section microfluidic device. Mater. Today Proc. 2019, 13, 332–340. [Google Scholar] [CrossRef]
- Pohl, H.A. The Motion and Precipitation of Suspensoids in Divergent Electric Fields. J. Appl. Phys. 1951, 22, 869–871. [Google Scholar] [CrossRef]
- Hughes, M.P. Fifty years of dielectrophoretic cell separation technology. Biomicrofluidics 2016, 10, 032801. [Google Scholar] [CrossRef] [Green Version]
- Pethig, R. Dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics 2010, 4, 022811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, T.B. Electromechanics of Particles; Cambridge University Press: Cambridge, UK, 1995; ISBN 9780521431965. [Google Scholar]
- RC, G.P.; Jody, V. Particle separation by dielectrophoresis. Electrophoresis 2002, 23, 1973–1983. [Google Scholar] [CrossRef]
- Pamme, N. Continuous flow separations in microfluidic devices. Lab Chip 2007, 7, 1644–1659. [Google Scholar] [CrossRef]
- Jubery, T.Z.; Srivastava, S.K.; Dutta, P. Dielectrophoretic separation of bioparticles in microdevices: A review. Electrophoresis 2014, 35, 691–713. [Google Scholar] [CrossRef] [PubMed]
- Pethig, R.R. Dielectrophoresis: Theory, Methodology and Biological Applications, 1st ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; ISBN 9781118671436. [Google Scholar]
- Li, M.; Li, W.H.; Zhang, J.; Alici, G.; Wen, W. A review of microfabrication techniques and dielectrophoretic microdevices for particle manipulation and separation. J. Phys. D Appl. Phys. 2014, 47, 063001. [Google Scholar] [CrossRef]
- Khoshmanesh, K.; Nahavandi, S.; Baratchi, S.; Mitchell, A.; Kalantar-zadeh, K. Dielectrophoretic platforms for bio-microfluidic systems. Biosens. Bioelectron. 2011, 26, 1800–1814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Khoshmanesh, K.; Mitchell, A.; Kalantar-Zadeh, K. Dielectrophoresis for manipulation of micro/nano particles in microfluidic systems. Anal. Bioanal. Chem. 2010, 396, 401–420. [Google Scholar] [CrossRef]
- Martinez-Duarte, R. Microfabrication technologies in dielectrophoresis applications-A review. Electrophoresis 2012, 33, 3110–3132. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Morgan, H. Single-cell microfluidic Impedance cytometry: A review. Microfluid. Nanofluidics 2010, 8, 423–443. [Google Scholar] [CrossRef]
- Zheng, Y.; Nguyen, J.; Wei, Y.; Sun, Y. Recent advances in microfluidic techniques for single-cell biophysical characterization. Lab Chip 2013, 13, 2464–2483. [Google Scholar] [CrossRef] [Green Version]
- Han, S.-I.; Joo, Y.-D.; Han, K.-H. An electrorotation technique for measuring the dielectric properties of cells with simultaneous use of negative quadrupolar dielectrophoresis and electrorotation. Analyst 2013, 138, 1529–1537. [Google Scholar] [CrossRef]
- Henslee, E.A. Review: Dielectrophoresis in cell characterization. Electrophoresis 2020. [Google Scholar] [CrossRef]
- Gagnon, Z.R. Cellular dielectrophoresis: Applications to the characterization, manipulation, separation and patterning of cells. Electrophoresis 2011, 32, 2466–2487. [Google Scholar] [CrossRef]
- Fatoyinbo, H.O.; Hoettges, K.F.; Hughes, M.P. Rapid-on-chip determination of dielectric properties of biological cells using imaging techniques in a dielectrophoresis dot microsystem. Electrophoresis 2008, 29, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gascoyne, P.R.C.; Shim, S.; Noshari, J.; Becker, F.F.; Stemke-Hale, K. Correlations between the dielectric properties and exterior morphology of cells revealed by dielectrophoretic field-flow fractionation. Electrophoresis 2013, 34, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Narayanan Unni, H.; Hartono, D.; Yue Lanry Yung, L.; Mah-Lee Ng, M.; Pueh Lee, H.; Cheong Khoo, B.; Lim, K.M. Characterization and separation of Cryptosporidium and Giardia cells using on-chip dielectrophoresis. Biomicrofluidics 2012, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, S.; Suzuki, M.; Arimoto, S.; Korenaga, T.; Yasukawa, T. Determination of membrane capacitance and cytoplasm conductivity by simultaneous electrorotation. Analyst 2020, 145, 4188–4195. [Google Scholar] [CrossRef]
- Arnold, W.M.; Zimmermann, U. Electro-rotation: Development of a technique for dielectric measurements on individual cells and particles. J. Electrostat. 1988, 21, 151–191. [Google Scholar] [CrossRef]
- Yang, J.; Huang, Y.; Wang, X.; Wang, X.B.; Becker, F.F.; Gascoyne, P.R.C. Dielectric properties of human leukocyte subpopulations determined by electrorotation as a cell separation criterion. Biophys. J. 1999, 76, 3307–3314. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Arnold, W.M.; Zimmermann, U. Alterations in the electrical properties of T and B lymphocyte membranes induced by mitogenic stimulation. Activation monitored by electro-rotation of single cells. Biochim. Biophys. Acta 1990, 1021, 191–200. [Google Scholar] [CrossRef]
- Griffith, A.W.; Cooper, J.M. Single-Cell Measurements of Human Neutrophil Activation Using Electrorotation. Anal. Chem. 1998, 70, 2607–2612. [Google Scholar] [CrossRef]
- Gimsa, J.; Müller, T.; Schnelle, T.; Fuhr, G. Dielectric spectroscopy of single human erythrocytes at physiological ionic strength: Dispersion of the cytoplasm. Biophys. J. 1996, 71, 495–506. [Google Scholar] [CrossRef] [Green Version]
- Georgieva, R.; Neu, B.; Shilov, V.M.; Knippel, E.; Budde, A.; Latza, R.; Donath, E.; Kiesewetter, H.; Bäumler, H. Low frequency electrorotation of fixed red blood cells. Biophys. J. 1998, 74, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Egger, M.; Donath, E.; Spangenberg, P.; Bimmler, M.; Glaser, R.; Till, U. Human platelet electrorotation change induced by activation: Inducer specificity and correlation to serotonin release. Biochim. Biophys. Acta 1988, 972, 265–276. [Google Scholar] [CrossRef]
- Berardi, V.; Aiello, C.; Bonincontro, A.; Risuleo, G. Alterations of the plasma membrane caused by murine polyomavirus proliferation: An electrorotation study. J. Membr. Biol. 2009, 229, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Hölzel, R. Electrorotation of single yeast cells at frequencies between 100 hz and 1.6 GHz. Biophys. J. 1997, 73, 1103–1109. [Google Scholar] [CrossRef] [Green Version]
- Becker, F.F.; Wang, X.B.; Huang, Y.; Pethig, R.; Vykoukal, J.; Gascoyne, P.R.C. Separation of human breast cancer cells from blood by differential dielectric affinity. Proc. Natl. Acad. Sci. USA 1995, 92, 860–864. [Google Scholar] [CrossRef] [Green Version]
- Cristofanilli, M.; De Gasperis, G.; Zhang, L.; Hung, M.C.; Gascoyne, P.R.C.; Hortobagyi, G.N. Automated electrorotation to reveal dielectric variations related to HER-2/neu overexpression in MCF-7 sublines. Clin. Cancer Res. 2002, 8, 615–619. [Google Scholar] [PubMed]
- Trainito, C.I.; Sweeney, D.C.; Čemažar, J.; Schmelz, E.M.; Français, O.; Le Pioufle, B.; Davalos, R.V. Characterization of sequentially-staged cancer cells using electrorotation. PLoS ONE 2019, 14, e0222289. [Google Scholar] [CrossRef] [PubMed]
- Hölzel, R. Non-invasive determination of bacterial single cell properties by electrorotation. Biochim. Biophys. Acta Mol. Cell Res. 1999, 1450, 53–60. [Google Scholar] [CrossRef] [Green Version]
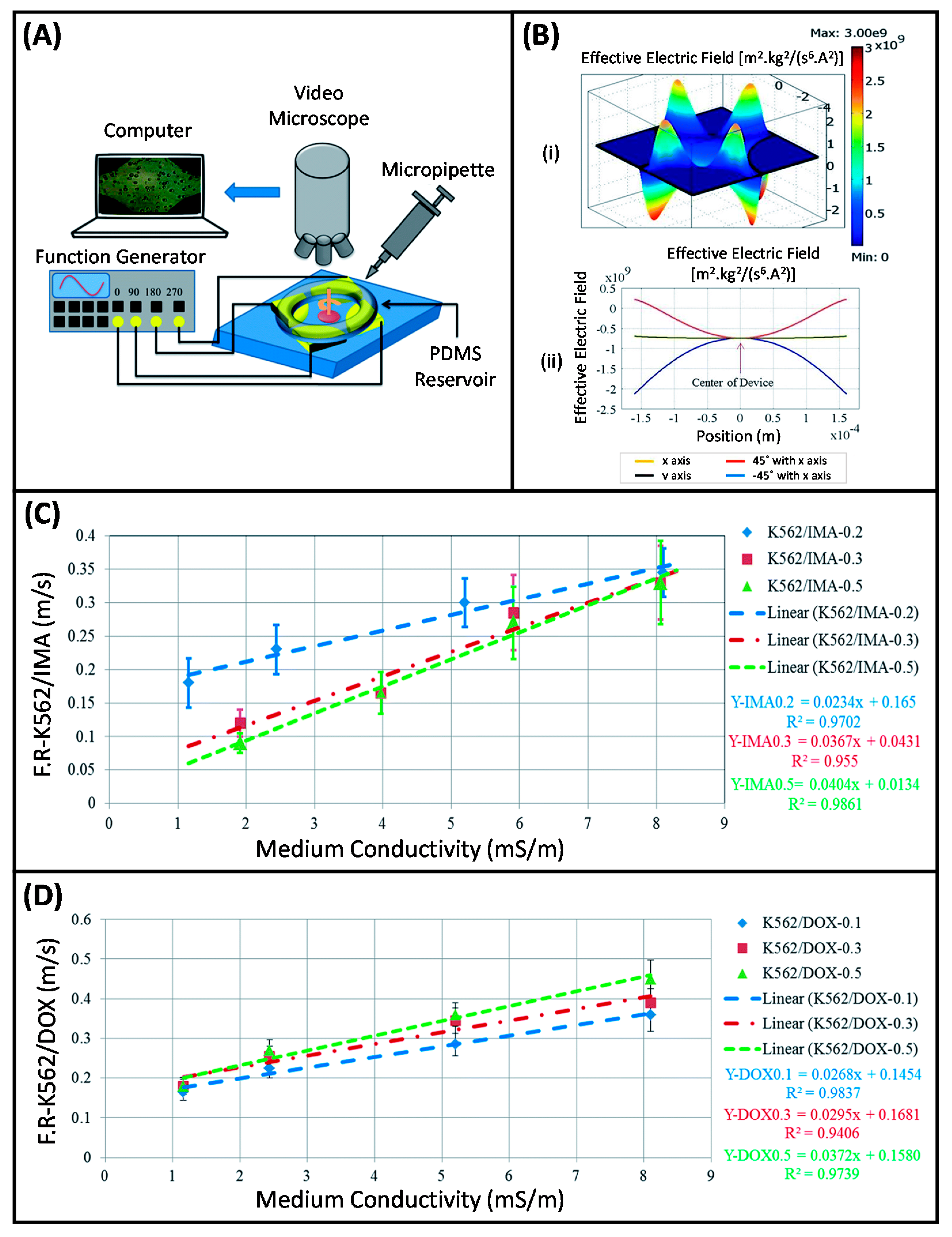
- Bahrieh, G.; Erdem, M.; Özgür, E.; Gündüz, U.; Külah, H. Dielectric analysis of changes in electric properties of doxorubicin resitant K562 leukemic cells through electrorotation with 3-D electrodes. In Proceedings of the 17th International Conference on Miniaturized Systems for Chemistry and Life Sciences, MicroTAS 2013, Freiburg, Germany, 27–31 October 2013; pp. 1502–1504. [Google Scholar]
- Bahrieh, G.; Koydemir, H.C.; Erdem, M.; Ozgur, E.; Gunduz, U.; Kulah, H. Dielectric characterization of Imatinib resistant K562 leukemia cells through electrorotation with 3-D electrodes. In Proceedings of the SENSORS, 2013 IEEE, Baltimore, MD, USA, 3–6 November 2013; pp. 1–4. [Google Scholar]
- Bahrieh, G.; Erdem, M.; Özgür, E.; Gündüz, U.; Külah, H. Assessment of effects of multi drug resistance on dielectric properties of K562 leukemic cells using electrorotation. RSC Adv. 2014, 4, 44879–44887. [Google Scholar] [CrossRef]
- Labeed, F.H.; Coley, H.M.; Thomas, H.; Hughes, M.P. Assessment of multidrug resistance reversal using dielectrophoresis and flow cytometry. Biophys. J. 2003, 85, 2028–2034. [Google Scholar] [CrossRef] [Green Version]
- Bahrieh, G.; Özgür, E.; Koyuncuoğlu, A.; Erdem, M.; Gündüz, U.; Külah, H. Characterization of the distribution of rotational torque on electrorotation chips with 3D electrodes. Electrophoresis 2015, 36, 1785–1794. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, X.B.; Becker, F.F.; Gascoyne, P.R.C. Membrane changes associated with the temperature-sensitive P85gag-mos-dependent transformation of rat kidney cells as determined by dielectrophoresis and electrorotation. Biochim. Biophys. Acta Biomembr. 1996, 1282, 76–84. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Joo, S.; Duhon, M.; Heller, M.; Wallace, B.; Xu, X. Dielectrophoretic cell separation and gene expression profiling on microelectronic chip arrays. Anal. Chem. 2002, 74, 3362–3371. [Google Scholar] [CrossRef]
- Gascoyne, P.; Pethig, R.; Satayavivad, J.; Becker, F.F.; Ruchirawat, M. Dielectrophoretic detection of changes in erythrocyte membranes following malarial infection. Biochim. Biophys. Acta Biomembr. 1997, 1323, 240–252. [Google Scholar] [CrossRef] [Green Version]
- Castellarnau, M.; Errachid, A.; Madrid, C.; Juárez, A.; Samitier, J. Dielectrophoresis as a tool to characterize and differentiate isogenic mutants of Escherichia coli. Biophys. J. 2006, 91, 3937–3945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vykoukal, D.M.; Gascoyne, P.R.C.; Vykoukal, J. Dielectric characterization of complete mononuclear and polymorphonuclear blood cell subpopulations for label-free discrimination. Integr. Biol. 2009, 1, 477–484. [Google Scholar] [CrossRef]
- Ismail, A.; Hughes, M.; Mulhall, H.; Oreffo, R.; Labeed, F. Characterization of human skeletal stem and bone cell populations using dielectrophoresis. J. Tissue Eng. Regen. Med. 2015, 9, 162–168. [Google Scholar] [CrossRef] [Green Version]
- Bunthawin, S.; Wanichapichart, P.; Tuantranont, A.; Coster, H.G.L. Dielectrophoretic spectra of translational velocity and critical frequency for a spheroid in traveling electric field. Biomicrofluidics 2010, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaler, K.V.; Jones, T.B. Dielectrophoretic spectra of single cells determined by feedback-controlled levitation. Biophys. J. 1990, 57, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, B.G.; Huang, C.; Arasanipalai, S.; Kirby, B.J. Automated dielectrophoretic characterization of Mycobacterium smegmatis. Anal. Chem. 2011, 83, 3507–3515. [Google Scholar] [CrossRef]
- Mulhall, H.J.; Labeed, F.H.; Kazmi, B.; Costea, D.E.; Hughes, M.P.; Lewis, M.P. Cancer, pre-cancer and normal oral cells distinguished by dielectrophoresis. Anal. Bioanal. Chem. 2011, 401, 2455–2463. [Google Scholar] [CrossRef] [Green Version]
- Sanchis, A.; Brown, A.P.; Sancho, M.; Martínez, G.; Sebastián, J.L.; Muñoz, S.; Miranda, J.M. Dielectric characterization of bacterial cells using dielectrophoresis. Bioelectromagnetics 2007, 28, 393–401. [Google Scholar] [CrossRef]
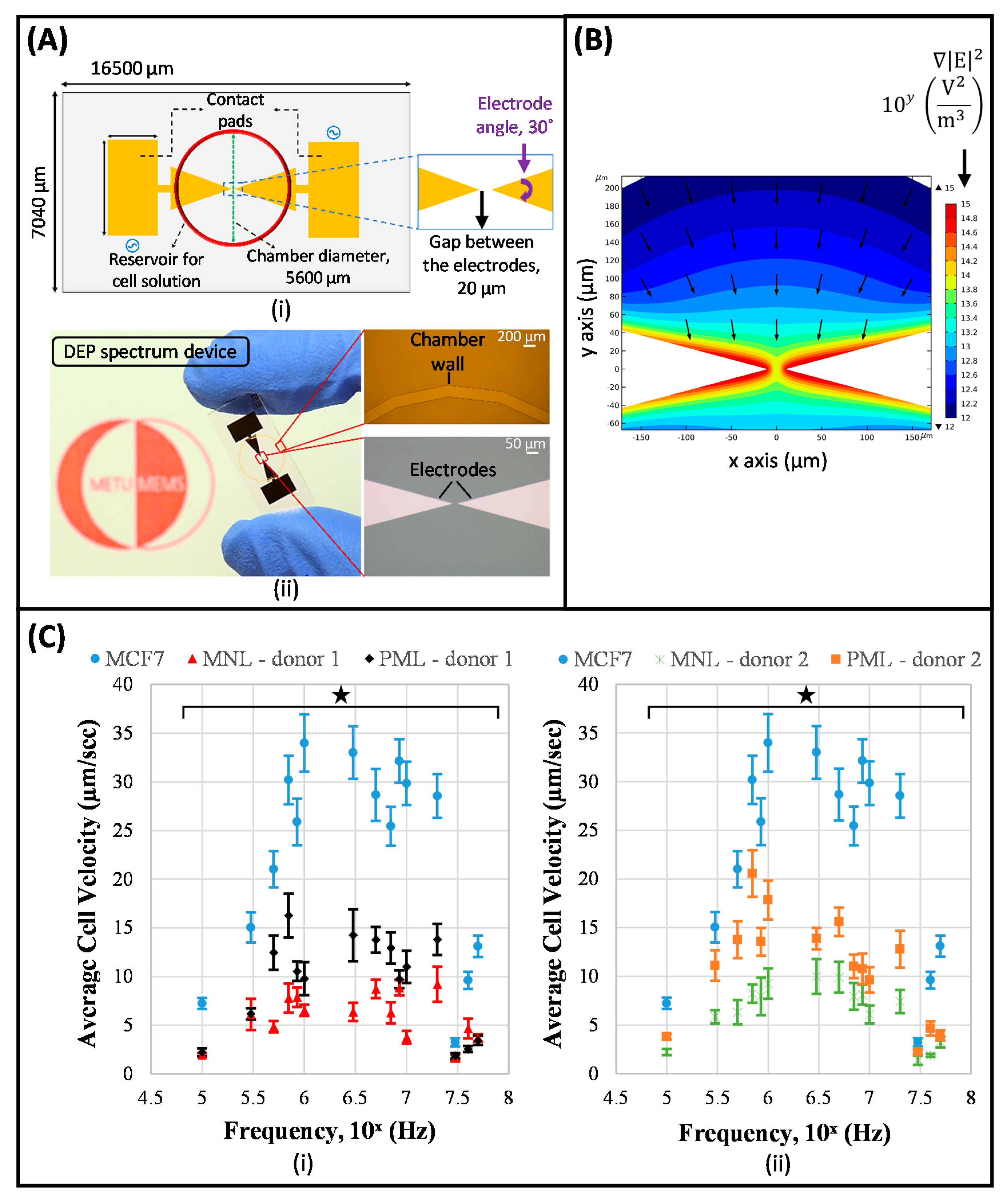
- Çağlayan, Z.; Sel, K.; Yalçın, Y.D.; Sukas, Ö.Ş.; Külah, H. Analysis of the Dielectrohporetic (DEP) Spectra of Biological Cells. In Proceedings of the 2017 19th International Conference on Solid-State Sensors, Actuators and Microsystems (TRANSDUCERS), Kaohsiung, Taiwan, 18–22 June 2017; pp. 1644–1647. [Google Scholar]
- Çağlayan, Z.; Demircan Yalçın, Y.; Külah, H. Examination of the dielectrophoretic spectra of MCF7 breast cancer cells and leukocytes. Electrophoresis 2020, 41, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Majidinia, M.; Mirza-Aghazadeh-Attari, M.; Rahimi, M.; Mihanfar, A.; Karimian, A.; Safa, A.; Yousefi, B. Overcoming multidrug resistance in cancer: Recent progress in nanotechnology and new horizons. Int. Union Biochem. Mol. Biol. 2020, 72, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Bar-Zeev, M.; Livney, Y.D.; Assaraf, Y.G. Targeted nanomedicine for cancer therapeutics: Towards precision medicine overcoming drug resistance. Drug Resist. Updat. 2017, 31, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J. (Ed.) Multi-Drug Resistance in Cancer; Humana Press: New York, NY, USA, 2010. [Google Scholar]
- Demircan, Y.; Özgür, E.; Külah, H. Dielectrophoresis: Applications and future outlook in point of care. Electrophoresis 2013, 34, 1008–1027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiang, H.; Sun, F.; Wang, H.; Zhao, J.; Chen, B.; Wang, X. Rapid diagnosis of multidrug resistance in cancer by electrochemical sensor based on carbon nanotubes–drug supramolecular nanocomposites. Biosens. Bioelectron. 2011, 26, 3361–3366. [Google Scholar] [CrossRef]
- Yalçın, Y.D.; Sukas, S.; Töral, T.B.; Gündüz, U.; Külah, H. Exploring the relationship between cytoplasmic ion content variation and multidrug resistance in cancer cells via ion-release based impedance spectroscopy. Sensors Actuators B Chem. 2019, 290, 180–187. [Google Scholar] [CrossRef]
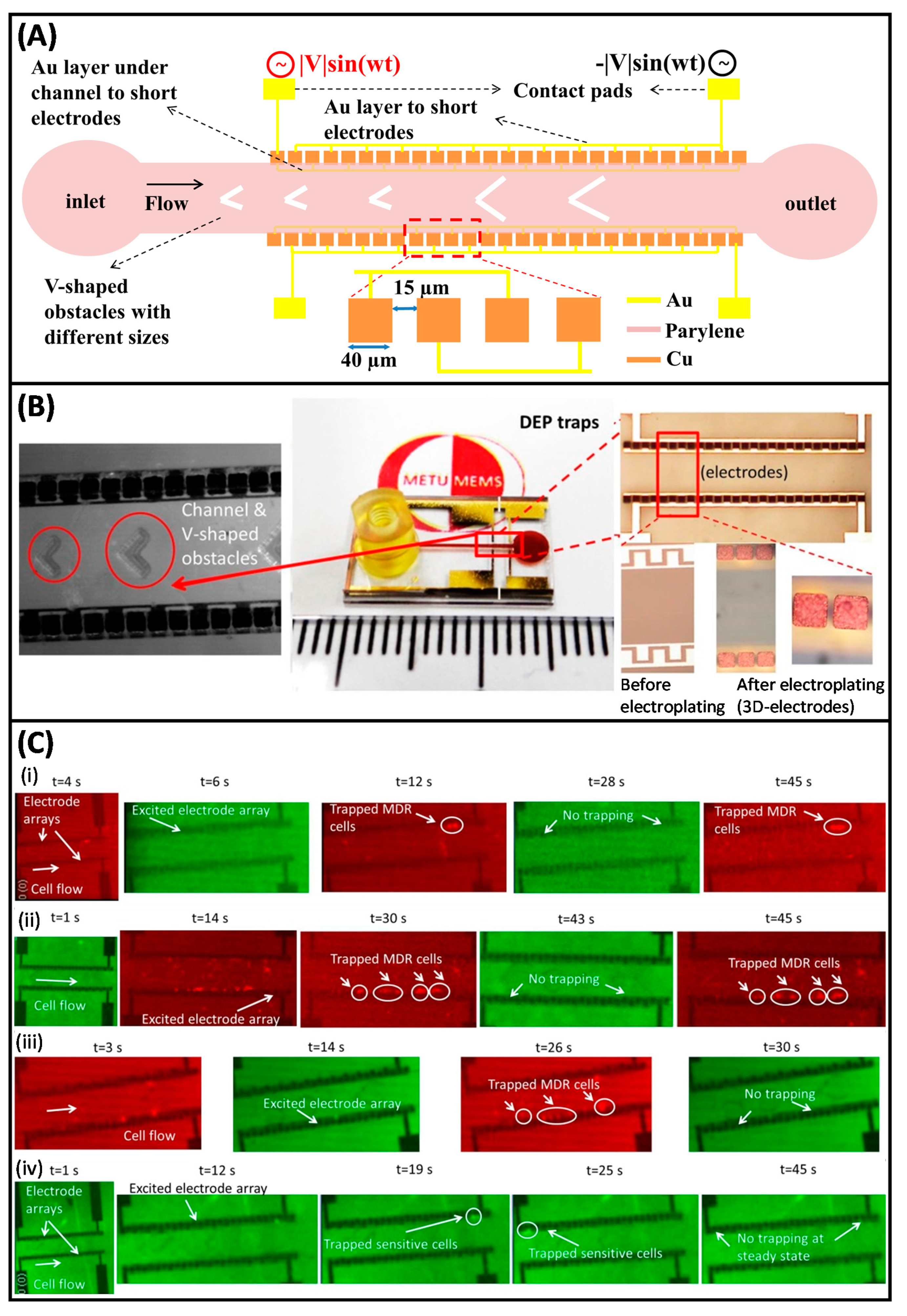
- Demircan, Y.; Koyuncuoğlu, A.; Erdem, M.; Özgür, E.; Gündüz, U.; Külah, H. Label-free detection of multidrug resistance in K562 cells through isolated 3D-electrode dielectrophoresis. Electrophoresis 2015, 36, 1149–1157. [Google Scholar] [CrossRef]
- Duncan, L.; Shelmerdine, H.; Hughes, M.P.; Coley, H.M.; Hübner, Y.; Labeed, F.H. Dielectrophoretic analysis of changes in cytoplasmic ion levels due to ion channel blocker action reveals underlying differences between drug-sensitive and multidrug-resistant leukaemic cells. Phys. Med. Biol. 2008, 53, N1–N7. [Google Scholar] [CrossRef]
- Labeed, F.H.; Coley, H.M.; Hughes, M.P. Differences in the biophysical properties of membrane and cytoplasm of apoptotic cells revealed using dielectrophoresis. Biochim. Biophys. Acta 2006, 1760, 922–929. [Google Scholar] [CrossRef]
- Coley, H.M.; Labeed, F.H.; Thomas, H.; Hughes, M.P. Biophysical characterization of MDR breast cancer cell lines reveals the cytoplasm is critical in determining drug sensitivity. Biochim. Biophys. Acta Gen. Subj. 2007, 1770, 601–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demircan, Y. Detection of Imatinib and Doxorubicin Resistance in K562 Leukemia Cells By 3D-Electrode Contactless Dielectrophoresis. In Proceedings of the TRANSDUCERS 2013, Barcelona, Spain, 16–20 June 2013; pp. 2086–2089. [Google Scholar]
- Demircan, Y.; Erdem, M.; Ozgur, E.; Gunduz, U.; Kulah, H. Determination of multidrug resistance level in K562 Leukemia cells by 3D-electrode contactless dielectrophoresis. In Proceedings of the IEEE International Conference on Micro Electro Mechanical Systems (MEMS), San Francisco, CA, USA, 26–30 January 2014; pp. 837–840. [Google Scholar]
- Yalçın, Y.D.; Özkayar, G.; Özgür, E.; Gündüz, U.; Külah, H. A DEP-based lab-on-a-chip system for the detection of multidrug resistance in K562 leukemia cells. In Proceedings of the 2016 Solid-State Sensors, Actuators and Microsystems Workshop, Hilton Head 2016, Hilton Head Island, South Caroline, 5–9 June 2016; Volume 2, pp. 316–319. [Google Scholar]
- Vykoukal, J.; Vykoukal, D.M.; Freyberg, S.; Alt, E.U.; Gascoyne, P.R.C. Enrichment of putative stem cells from adipose tissue using dielectrophoretic field-flow fractionation. Lab Chip 2008, 8, 1386–1393. [Google Scholar] [CrossRef] [Green Version]
- Adams, T.N.G.; Jiang, A.Y.L.; Vyas, P.D.; Flanagan, L.A. Separation of neural stem cells by whole cell membrane capacitance using dielectrophoresis. Methods 2018, 133, 91–103. [Google Scholar] [CrossRef]
- Song, H.; Rosano, J.M.; Wang, Y.; Garson, C.J.; Prabhakarpandian, B.; Pant, K.; Klarmann, G.J.; Perantoni, A.; Alvarez, L.M.; Lai, E. Continuous-flow sorting of stem cells and differentiation products based on dielectrophoresis. Lab Chip 2015, 15, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.; Stemke-Hale, K.; Tsimberidou, A.M.; Noshari, J.; Anderson, T.E.; Gascoyne, P.R.C. Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis. Biomicrofluidics 2013, 7. [Google Scholar] [CrossRef] [Green Version]
- Cheng, I.F.; Froude, V.E.; Zhu, Y.; Chang, H.C.; Chang, H.C. A continuous high-throughput bioparticle sorter based on 3D traveling-wave dielectrophoresis. Lab Chip 2009, 9, 3193–3201. [Google Scholar] [CrossRef] [PubMed]
- Borgatti, M.; Altomare, L.; Baruffa, M.; Fabbri, E.; Breveglieri, G.; Feriotto, G.; Manaresi, N.; Medoro, G.; Romani, A.; Tartagni, M.; et al. Separation of white blood cells from erythrocytes on a dielectrophoresis (DEP) based “Lab-on-a-chip” device. Int. J. Mol. Med. 2005, 15, 913–920. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, Y.; Cui, X.; Fan, Y.; Xue, Y.; Miao, H.; Li, G. Extraction of Cell-Free Whole Blood Plasma Using a Dielectrophoresis-Based Microfluidic Device. Biotechnol. J. 2019, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Bashir, R. Dielectrophoretic separation and manipulation of live and heat-treated cells of Listeria on microfabricated devices with interdigitated electrodes. Sensors Actuators B Chem. 2002, 86, 215–221. [Google Scholar] [CrossRef]
- Fatoyinbo, H.O.; McDonnell, M.C.; Hughes, M.P. Dielectrophoretic sample preparation for environmental monitoring of microorganisms: Soil particle removal. Biomicrofluidics 2014, 8. [Google Scholar] [CrossRef] [Green Version]
- Elitas, M.; Martinez-Duarte, R.; Dhar, N.; McKinney, J.D.; Renaud, P. Dielectrophoresis-based purification of antibiotic-treated bacterial subpopulations. Lab Chip 2014, 14, 1850–1857. [Google Scholar] [CrossRef]
- Sonnenberg, A.; Marciniak, J.Y.; Mccanna, J.; Krishnan, R.; Rassenti, L.; Kipps, T.J.; Heller, M.J. Dielectrophoretic isolation and detection of cfc-DNA nanoparticulate biomarkers and virus from blood. Electrophoresis 2013, 34, 1076–1084. [Google Scholar] [CrossRef]
- Masuda, T.; Maruyama, H.; Honda, A.; Arai, F. Virus enrichment for single virus infection by using 3D insulator based dielectrophoresis. PLoS ONE 2014, 9, e94083. [Google Scholar] [CrossRef] [Green Version]
- Sonnenberg, A.; Marciniak, J.Y.; Skowronski, E.A.; Manouchehri, S.; Rassenti, L.; Ghia, E.M.; Widhopf, G.F.; Kipps, T.J.; Heller, M.J. Dielectrophoretic isolation and detection of cancer-related circulating cell-free DNA biomarkers from blood and plasma. Electrophoresis 2014, 35, 1828–1836. [Google Scholar] [CrossRef] [Green Version]
- Manouchehri, S.; Ibsen, S.; Wright, J.; Rassenti, L.; Ghia, E.M.; Widhopf, G.F.; Kipps, T.J.; Heller, M.J. Dielectrophoretic recovery of DNA from plasma for the identification of chronic lymphocytic leukemia point mutations. Int. J. Hematol. Oncol. 2016, 5, 27–35. [Google Scholar] [CrossRef]
- Jones, P.V.; Salmon, G.L.; Ros, A. Continuous Separation of DNA Molecules by Size Using Insulator-Based Dielectrophoresis. Anal. Chem. 2017, 89, 1531–1539. [Google Scholar] [CrossRef]
- Ciftlik, A.T.; Kulah, H. A direct injection method for blood cells into microchannels from pure blood droplets with switchable in-situ distillation of erythrocytes. In Proceedings of the 2008 PhD Research in Microelectronics and Electronics, İstanbul, Turkey, 25 April–22 June 2008; pp. 29–32. [Google Scholar]
- Gao, D.; Jin, F.; Zhou, M.; Jiang, Y. Recent advances in single cell manipulation and biochemical analysis on microfluidics. Analyst 2019, 144, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Mohr, S.; Liew, C.C. The peripheral-blood transcriptome: New insights into disease and risk assessment. Trends Mol. Med. 2007, 13, 422–432. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Shim, S. Isolation of circulating tumor cells by dielectrophoresis. Cancers 2014, 6, 545–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gascoyne, P.R.C.; Wang, X.-B.; Huang, Y.; Becker, F.F. Dielectrophoretic Separation of Cancer Cells from Blood. IEEE Trans. Ind. Appl. 1997, 33, 670–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shim, S.; Stemke-Hale, K.; Noshari, J.; Becker, F.F.; Gascoyne, P.R.C. Dielectrophoresis has broad applicability to marker-free isolation of tumor cells from blood by microfluidic systems. Biomicrofluidics 2013, 7, 011808. [Google Scholar] [CrossRef] [Green Version]
- Hong, B.; Zu, Y. Detecting circulating tumor cells: Current challenges and new trends. Theranostics 2013, 3, 377–394. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Rao, C.; Bui, T.; Connelly, M.; Doyle, G.; Karydis, I.; Middleton, M.R.; Clack, G.; Malone, M.; Coumans, F.A.W.; Terstappen, L.W.M.M. Circulating melanoma cells and survival in metastatic melanoma. Int. J. Oncol. 2011, 38, 755–760. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol. Adv. 2018, 36, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Low, W.S.; Wan Abas, W.A.B. Benchtop Technologies for Circulating Tumor Cells Separation Based on Biophysical Properties. Biomed Res. Int. 2015, 2015, 239362. [Google Scholar] [CrossRef]
- Dobrzynska, I.; Skrzydlewska, E.; Figaszewski, Z.A. Changes in Electric Properties of Human Breast Cancer Cells. J. Membr. Biol. 2013, 246, 161–166. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.H.; Huang, C.T.; Wu, H.H.; Zamay, T.N.; Zamay, A.S.; Jen, C.P. Isolating and concentrating rare cancerous cells in large sample volumes of blood by using dielectrophoresis and stepping electric fields. Biochip. J. 2014, 8, 67–74. [Google Scholar] [CrossRef]
- Moon, H.S.; Kwon, K.; Kim, S.I.; Han, H.; Sohn, J.; Lee, S.; Jung, H.I. Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP). Lab Chip 2011, 11, 1118–1125. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Noshari, J.; Anderson, T.J.; Becker, F.F. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis 2009, 30, 1388–1398. [Google Scholar] [CrossRef] [Green Version]
- Waheed, W.; Alazzam, A.; Mathew, B.; Christoforou, N.; Abu-Nada, E. Lateral fluid flow fractionation using dielectrophoresis (LFFF-DEP) for size-independent, label-free isolation of circulating tumor cells. J. Chromatogr. B 2018, 1087–1088, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Le Du, F.; Fujii, T.; Kida, K.; Davis, D.W.; Park, M.; Liu, D.D.; Wu, W.; Chavez-MacGregor, M.; Barcenas, C.H.; Valero, V.; et al. EpCAM-independent isolation of circulating tumor cells with epithelial-to-mesenchymal transition and cancer stem cell phenotypes using ApoStream® in patients with breast cancer treated with primary systemic therapy. PLoS ONE 2020, 15, e0229903. [Google Scholar] [CrossRef] [PubMed]
- Rugo, H.S.; Cortes, J.; Awada, A.; O’Shaughnessy, J.; Twelves, C.; Im, S.A.; Hannah, A.; Lu, L.; Sy, S.; Caygill, K.; et al. Change in topoisomerase 1–positive circulating tumor cells affects overall survival in patients with advanced breast cancer after treatment with etirinotecan pegol. Clin. Cancer Res. 2018, 24, 3348–3357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yılmaz, G.; Çiftlik, A.T.; Külah, H. A Dielectrophoretic Cell/Particle Separator Fabricated by Spiral Channels and COncentric Gold Electrodes. In Proceedings of the Transducers 2009, Denver, CO, USA, 21–25 June 2009; pp. 73–76. [Google Scholar]
- Özkayar, G.; Yalçın, Y.D.; Özgür, E.; Gündüz, U.; Külah, H. A High-throughput Microfluidic Rare Cell Enrichment System Based on Dielectrophoresis and Filtering. Procedia Technol. 2017, 27, 177–178. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Vykoukal, J.V.; Schwartz, J.A.; Anderson, T.J.; Vykoukal, D.M.; Current, K.W.; McConaghy, C.; Becker, F.F.; Andrews, C. Dielectrophoresis-based programmable fluidic processors. Lab Chip 2004, 4, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Han, S.I.; Soo Kim, H.; Han, A. In-droplet cell concentration using dielectrophoresis. Biosens. Bioelectron. 2017, 97, 41–45. [Google Scholar] [CrossRef]
- Agarwal, T.; Maiti, T.K. Dielectrophoresis-based devices for cell patterning. In Bioelectronics and Medical Devices; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; pp. 493–511. ISBN 9780081024201. [Google Scholar]
- Lin, R.Z.; Ho, C.T.; Liu, C.H.; Chang, H.Y. Dielectrophoresis based-cell patterning for tissue engineering. Biotechnol. J. 2006, 1, 949–957. [Google Scholar] [CrossRef]
- Suzuki, M.; Yasukawa, T.; Shiku, H.; Matsue, T. Negative dielectrophoretic patterning with different cell types. Biosens. Bioelectron. 2008, 24, 1043–1047. [Google Scholar] [CrossRef]
- Suehiro, J.; Ikeda, N.; Ohtsubo, A.; Imasaka, K. Fabrication of bio/nano interfaces between biological cells and carbon nanotubes using dielectrophoresis. Microfluid. Nanofluidics 2008, 5, 741–747. [Google Scholar] [CrossRef]
- Yahya, W.N.W.; Kadri, N.A.; Ibrahim, F. Cell patterning for liver tissue engineering via dielectrophoretic mechanisms. Sensors 2014, 14, 11714–11734. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.; Vykoukal, J.; Vykoukal, D.M.; Schwartz, J.A.; Shi, L.; Gascoyne, P.R.C. A three-dimensional dielectrophoretic particle focusing channel for microcytometry applications. J. Microelectromechanical Syst. 2005, 14, 480–487. [Google Scholar] [CrossRef]
- Lin, C.H.; Lee, G.B.; Fu, L.M.; Hwey, B.H. Vertical focusing device utilizing dielectrophoretic force and its application on microflow cytometer. J. Microelectromechanical Syst. 2004, 13, 923–932. [Google Scholar] [CrossRef]
- Nguyen, N.V.; Jen, C.P. Impedance detection integrated with dielectrophoresis enrichment platform for lung circulating tumor cells in a microfluidic channel. Biosens. Bioelectron. 2018, 121, 10–18. [Google Scholar] [CrossRef]
- Barik, A.; Zhang, Y.; Grassi, R.; Nadappuram, B.P.; Edel, J.B.; Low, T.; Koester, S.J.; Oh, S.H. Graphene-edge dielectrophoretic tweezers for trapping of biomolecules. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Ettehad, H.M.; Yadav, R.K.; Guha, S.; Wenger, C. Towards CMOS integrated microfluidics using dielectrophoretic immobilization. Biosensors 2019, 9, 77. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, C.; Iizuka, R.; Ohki, Y.; Fujimaki, M. Development of a dielectrophoresis-assisted surface plasmon resonance fluorescence biosensor for detection of bacteria. Jpn. J. Appl. Phys. 2018, 57, 057001. [Google Scholar] [CrossRef]
- Páez-Avilés, C.; Juanola-Feliu, E.; Punter-Villagrasa, J.; Del Moral Zamora, B.; Homs-Corbera, A.; Colomer-Farrarons, J.; Miribel-Català, P.L.; Samitier, J. Combined dielectrophoresis and impedance systems for bacteria analysis in microfluidic on-chip platforms. Sensors 2016, 16, 1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velmanickam, L.; Fondakowski, M.; Nawarathna, D. Integrated dielectrophoresis and fluorescence-based platform for biomarker detection from serum samples. Biomed. Phys. Eng. Express 2018, 4. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Caplin, J.D.; Granados, N.G.; James, M.R.; Montazami, R.; Hashemi, N. Microfluidic Organ-on-a-Chip Technology for Advancement of Drug Development and Toxicology. Adv. Healthc. Mater. 2015, 4, 1426–1450. [Google Scholar] [CrossRef] [Green Version]
- Tian, C.; Tu, Q.; Liu, W.; Wang, J. Recent advances in microfluidic technologies for organ-on-a-chip. Trends Anal. Chem. 2019, 117, 146–156. [Google Scholar] [CrossRef]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Ahadian, S.; Civitarese, R.; Bannerman, D.; Mohammadi, M.H.; Lu, R.; Wang, E.; Davenport-Huyer, L.; Lai, B.; Zhang, B.; Zhao, Y.; et al. Organ-On-A-Chip Platforms: A Convergence of Advanced Materials, Cells, and Microscale Technologies. Adv. Healthc. Mater. 2018, 7, 1–53. [Google Scholar] [CrossRef]
- Chan, C.Y.; Huang, P.H.; Guo, F.; Ding, X.; Kapur, V.; Mai, J.D.; Yuen, P.K.; Huang, T.J. Accelerating drug discovery via organs-on-chips. Lab Chip 2013, 13, 4697–4710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, C.T.; Lin, R.Z.; Chen, R.J.; Chin, C.K.; Gong, S.E.; Chang, H.Y.; Peng, H.L.; Hsu, L.; Yew, T.R.; Chang, S.F.; et al. Liver-cell patterning Lab Chip: Mimicking the morphology of liver lobule tissue. Lab Chip 2013, 13, 3578–3587. [Google Scholar] [CrossRef] [Green Version]
- Schütte, J.; Hagmeyer, B.; Holzner, F.; Kubon, M.; Werner, S.; Freudigmann, C.; Benz, K.; Böttger, J.; Gebhardt, R.; Becker, H.; et al. “Artificial micro organs”—A microfluidic device for dielectrophoretic assembly of liver sinusoids. Biomed. Microdevices 2011, 13, 493–501. [Google Scholar] [CrossRef]
- Yang, L. Dielectrophoresis assisted immuno-capture and detection of foodborne pathogenic bacteria in biochips. Talanta 2009, 80, 551–558. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 21 October 2020).
- Aguilar, J.B.; Faust, J.S.; Westafer, L.M.; Gutierrez, J.B. A Model Describing COVID-19 Community Transmission Taking into Account Asymptomatic Carriers and Risk Mitigation. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Virgin, H.W. The virome in mammalian physiology and disease. Cell 2014, 157, 142–150. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, H.; Kotani, K.; Masuda, T.; Honda, A.; Takahata, T.; Arai, F. Nanomanipulation of single influenza virus using dielectrophoretic concentration and optical tweezers for single virus infection to a specific cell on a microfluidic chip. Microfluid. Nanofluidics 2011, 10, 1109–1117. [Google Scholar] [CrossRef]
- Nakano, M.; Ding, Z.; Suehiro, J. Dielectrophoresis and dielectrophoretic impedance detection of adenovirus and rotavirus. Jpn. J. Appl. Phys. 2016, 55. [Google Scholar] [CrossRef]
- Iswardy, E.; Tsai, T.C.; Cheng, I.F.; Ho, T.C.; Perng, G.C.; Chang, H.C. A bead-based immunofluorescence-assay on a microfluidic dielectrophoresis platform for rapid dengue virus detection. Biosens. Bioelectron. 2017, 95, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Han, C.H.; Woo, S.Y.; Bhardwaj, J.; Sharma, A.; Jang, J. Rapid and selective concentration of bacteria, viruses, and proteins using alternating current signal superimposition on two coplanar electrodes. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Lawrence, R.M.; Jones, P.V.; Hogue, B.G.; Hayes, M.A. Concentration of Sindbis virus with optimized gradient insulator-based dielectrophoresis. Analyst 2016, 141, 1997–2008. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, G.; Çiftlik, A.T.; Külah, H. A MEMS-based spiral channel dielectrophoretic chromatography system for cytometry applications. Biotechnol. J. 2011, 6, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Demircan, Y.; Orguc, S.; Musayev, J.; Ozgur, E.; Erdem, M.; Gunduz, U.; Eminoglu, S.; Kulah, H.; Akin, T. Label-free detection of leukemia cells with a lab-on-a-chip system integrating dielectrophoresis and CMOS imaging. In Proceedings of the 2015 Transducers-2015 18th International Conference on Solid-State Sensors, Actuators and Microsystems, TRANSDUCERS 2015, Anchorage, AK, USA, 21–25 June 2015; pp. 1589–1592. [Google Scholar]
- Aslan, M.K.; Kulah, H. Android based portable cell counting system for label free quantification of dep manipulated cancer cells. In Proceedings of the TRANSDUCERS 2017-19th International Conference on Solid-State Sensors, Actuators and Microsystems, Kaohsiung, Taiwan, 18–22 June 2017; pp. 556–559. [Google Scholar]
- Li, M.; Anand, R.K. Cellular dielectrophoresis coupled with single-cell analysis. Anal. Bioanal. Chem. 2018, 410, 2499–2515. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lu, J.; Marchenko, S.A.; Monuki, E.S.; Flanagan, L.A.; Lee, A.P. Dual frequency dielectrophoresis with interdigitated sidewall electrodes for microfluidic flow-through separation of beads and cells. Electrophoresis 2009, 30, 782–791. [Google Scholar] [CrossRef]
- Islam, M.; Natu, R.; Larraga-Martinez, M.F.; Martinez-Duarte, R. Enrichment of diluted cell populations from large sample volumes using 3D carbon-electrode dielectrophoresis. Biomicrofluidics 2016, 10. [Google Scholar] [CrossRef] [Green Version]
- Cheng, I.F.; Chang, H.C.; Hou, D.; Chang, H.C. An integrated dielectrophoretic chip for continuous bioparticle filtering, focusing, sorting, trapping, and detecting. Biomicrofluidics 2007, 1. [Google Scholar] [CrossRef] [Green Version]
- Čemažar, J.; Douglas, T.A.; Schmelz, E.M.; Davalos, R.V. Enhanced contactless dielectrophoresis enrichment and isolation platform via cell-scale microstructures. Biomicrofluidics 2016, 10. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.Y.; Zhu, J.; Sivan, V.; Gol, B.; Soffe, R.; Zhang, W.; Mitchell, A.; Khoshmanesh, K. Creation of Liquid Metal 3D Microstructures Using Dielectrophoresis. Adv. Funct. Mater. 2015, 25, 4445–4452. [Google Scholar] [CrossRef]
- Jackson, J.M.; Witek, M.A.; Kamande, J.W.; Soper, S.A. Materials and microfluidics: Enabling the efficient isolation and analysis of circulating tumour cells. Chem. Soc. Rev. 2017, 46, 4245–4280. [Google Scholar] [CrossRef]
- Agarwal, A.; Balic, M.; El-Ashry, D.; Cote, R.J. Circulating Tumor Cells: Strategies for Capture, Analyses, and Propagation. Cancer J. 2018, 24, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Wu, A.; Chen, X. Current detection technologies for circulating tumor cells. Chem. Soc. Rev. 2017, 46, 2038–2056. [Google Scholar] [CrossRef]
- Pommer, M.S.; Zhang, Y.; Keerthi, N.; Chen, D.; Thomson, J.A.; Meinhart, C.D.; Soh, H.T. Dielectrophoretic separation of platelets from diluted whole blood in microfluidic channels. Electrophoresis 2008, 29, 1213–1218. [Google Scholar] [CrossRef]
- Braff, W.A.; Pignier, A.; Buie, C.R. High sensitivity three-dimensional insulator-based dielectrophoresis. Lab Chip 2012, 12, 1327–1331. [Google Scholar] [CrossRef]
- Voldman, J. Electrical Forces for Microscale Cell Manipulation. Annu. Rev. Biomed. Eng. 2006, 8, 425–454. [Google Scholar] [CrossRef] [Green Version]
- Mittal, N.; Rosenthal, A.; Voldman, J. nDEP microwells for single-cell patterning in physiological media. Lab Chip 2007, 7, 1146–1153. [Google Scholar] [CrossRef]
- Sabuncu, A.C.; Asmar, A.J.; Stacey, M.W.; Beskok, A. Differential dielectric responses of chondrocyte and Jurkat cells in electromanipulation buffers. Electrophoresis 2015, 36, 1499–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puttaswamy, S.V.; Sivashankar, S.; Chen, R.J.; Chin, C.K.; Chang, H.Y.; Liu, C.H. Enhanced cell viability and cell adhesion using low conductivity medium for negative dielectrophoretic cell patterning. Biotechnol. J. 2010, 5, 1005–1015. [Google Scholar] [CrossRef]
- Park, S.; Zhang, Y.; Wang, T.H.; Yang, S. Continuous dielectrophoretic bacterial separation and concentration from physiological media of high conductivity. Lab Chip 2011, 11, 2893–2900. [Google Scholar] [CrossRef]
- Sin, M.L.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovarik, M.L.; Gach, P.C.; Ornoff, D.M.; Wang, Y.; Balowski, J.; Farrag, L.; Allbritton, N.L. Micro total analysis systems for cell biology and biochemical assays. Anal. Chem. 2012, 84, 516–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Quiñones, J.; Moncada-Hernandez, H.; Rossetto, O.; Martinez-Duarte, R.; Lapizco-Encinas, B.H.; Madou, M.; Martinez-Chapa, S.O. An application specific multi-channel stimulator for electrokinetically-driven microfluidic devices. In Proceedings of the 2011 IEEE 9th International New Circuits and Systems Conference, NEWCAS 2011, Bordeaux, France, 26–29 June 2011; pp. 350–353. [Google Scholar]
- Qiao, W.; Cho, G.; Lo, Y.H. Wirelessly powered microfluidic dielectrophoresis devices using printable RF circuits. Lab Chip 2011, 11, 1074–1080. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Çağlayan, Z.; Demircan Yalçın, Y.; Külah, H. A Prominent Cell Manipulation Technique in BioMEMS: Dielectrophoresis. Micromachines 2020, 11, 990. https://doi.org/10.3390/mi11110990
Çağlayan Z, Demircan Yalçın Y, Külah H. A Prominent Cell Manipulation Technique in BioMEMS: Dielectrophoresis. Micromachines. 2020; 11(11):990. https://doi.org/10.3390/mi11110990
Chicago/Turabian StyleÇağlayan, Zeynep, Yağmur Demircan Yalçın, and Haluk Külah. 2020. "A Prominent Cell Manipulation Technique in BioMEMS: Dielectrophoresis" Micromachines 11, no. 11: 990. https://doi.org/10.3390/mi11110990
APA StyleÇağlayan, Z., Demircan Yalçın, Y., & Külah, H. (2020). A Prominent Cell Manipulation Technique in BioMEMS: Dielectrophoresis. Micromachines, 11(11), 990. https://doi.org/10.3390/mi11110990




