Comparison of Multiple Displacement Amplification (MDA) and Multiple Annealing and Looping-Based Amplification Cycles (MALBAC) in Limited DNA Sequencing Based on Tube and Droplet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Device Fabrication
2.2. Whole Genome Amplification
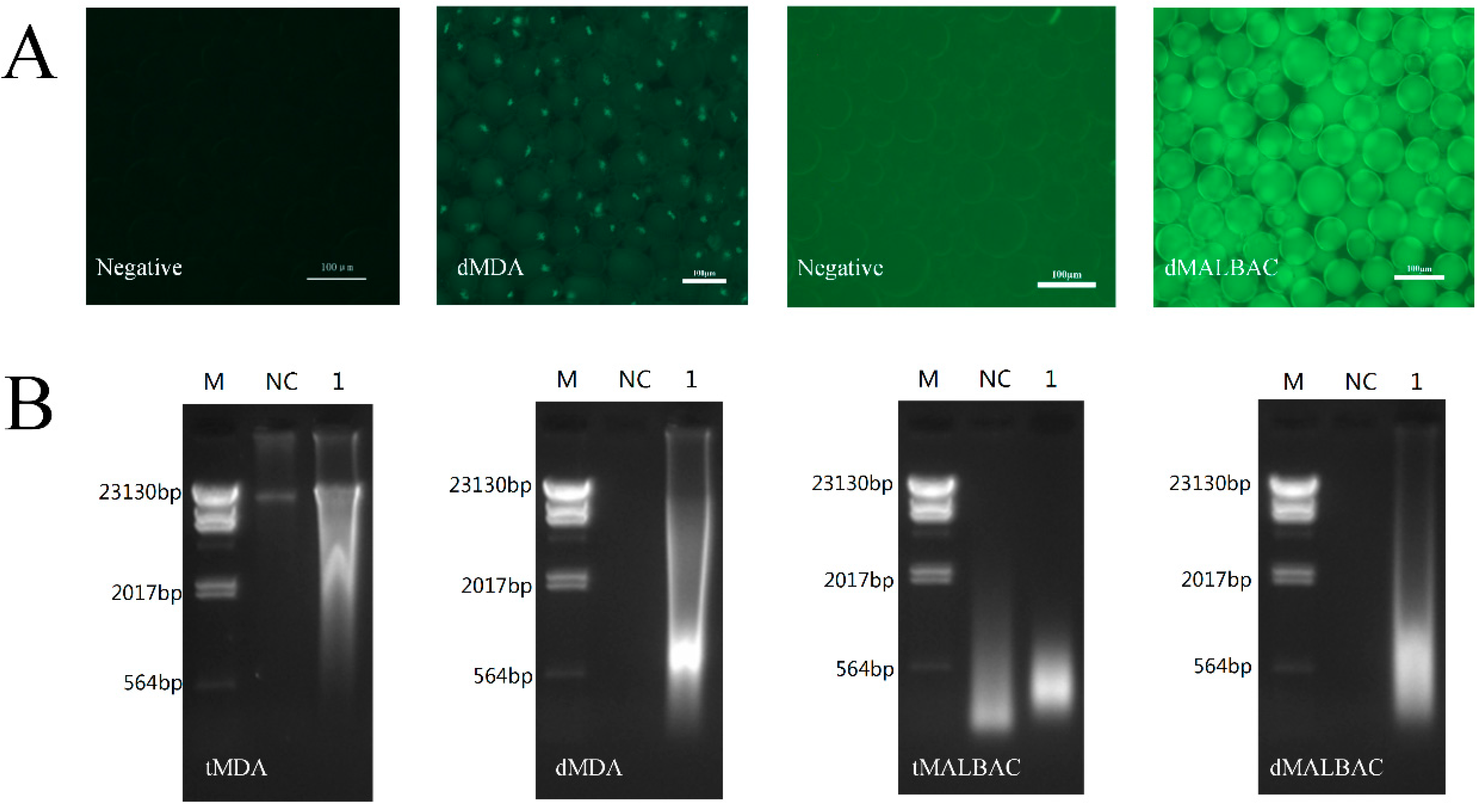
2.3. Quality Assay Based on Fluorescence and Electrophoresis
2.4. Library Preparation and Whole Genome Sequencing
2.5. Sequencing Analysis
3. Results and Discussion
3.1. Overview of the Method
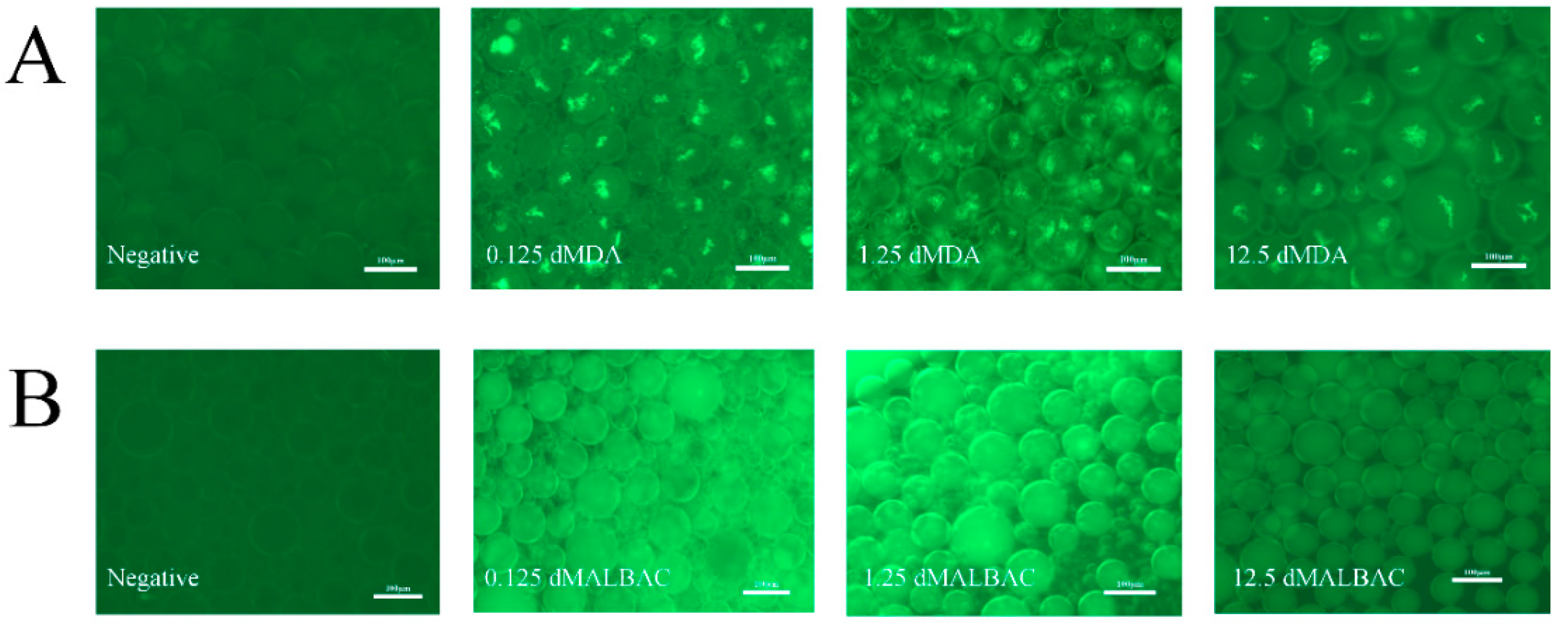
3.2. Whole Genome Amplification in Droplets
3.3. Coverage Breadth and Genome Recovery
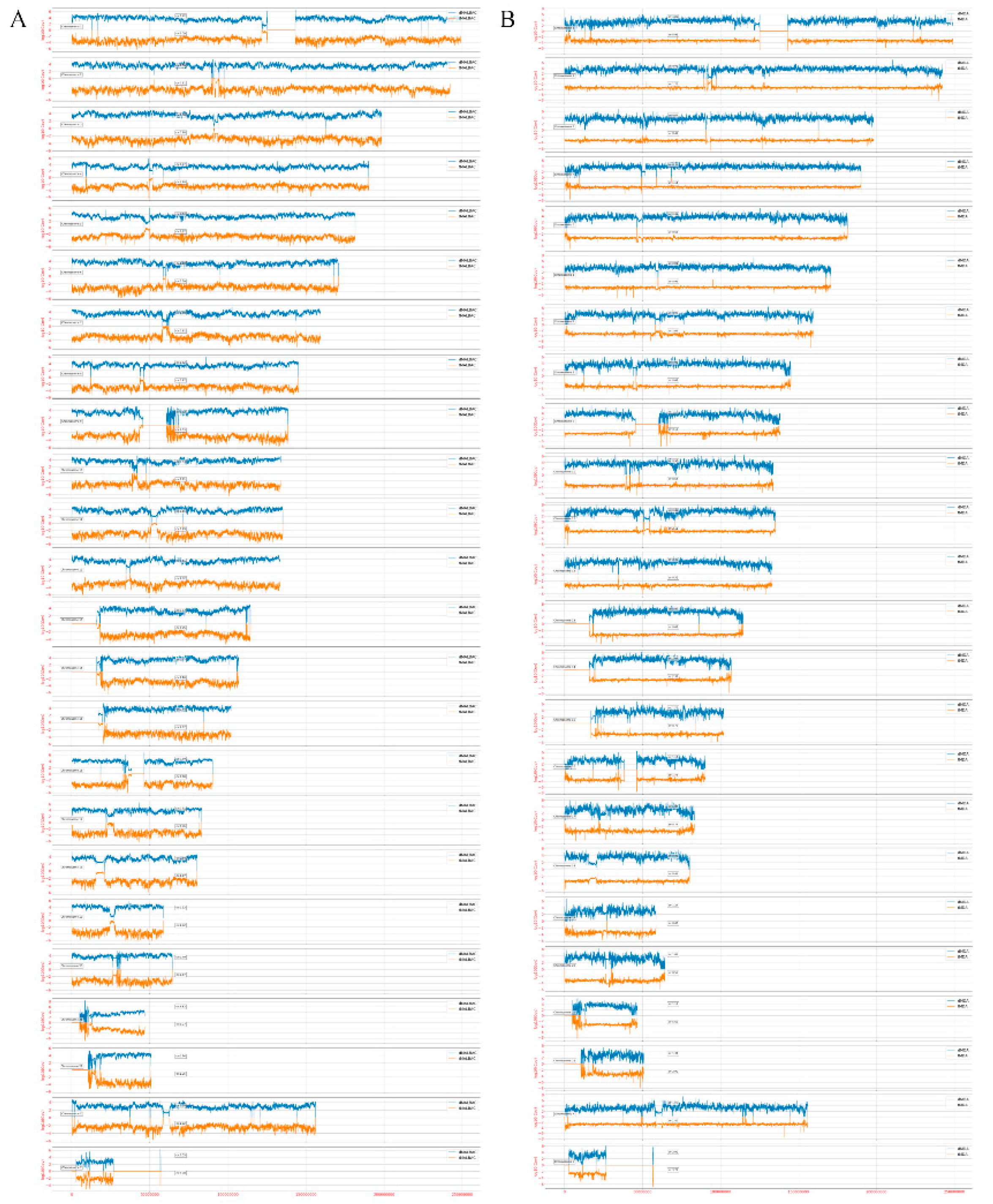
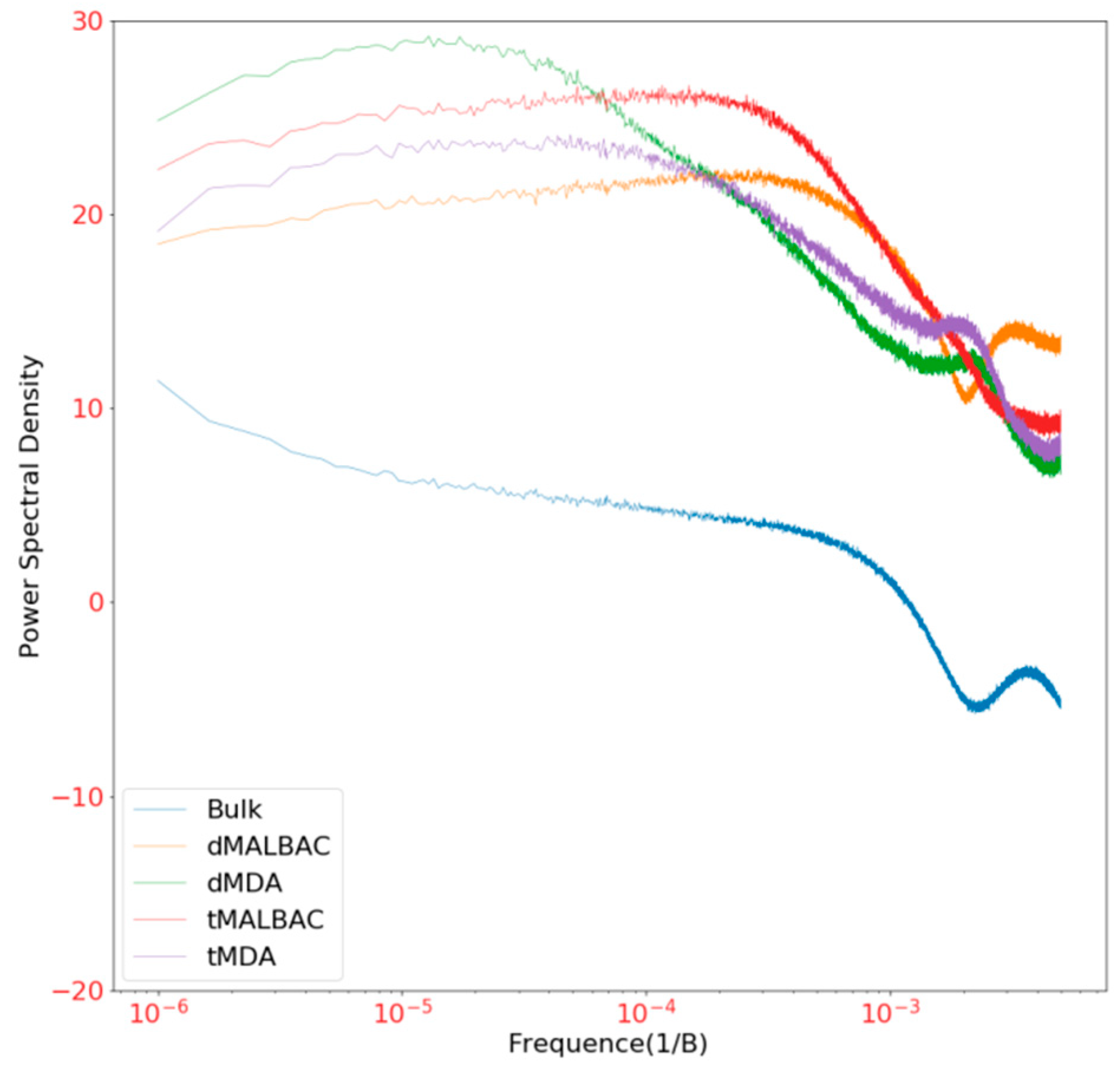
3.4. Amplification Uniformity and SNV Detection
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Riaz, N.; Havel, J.J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martin-Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949.e16. [Google Scholar] [CrossRef] [Green Version]
- Jain, C.; Rodriguez, R.L.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [Green Version]
- Lord, J.; McMullan, D.J.; Eberhardt, R.Y.; Rinck, G.; Hamilton, S.J.; Quinlan-Jones, E.; Prigmore, E.; Keelagher, R.; Best, S.K.; Carey, G.K.; et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): A cohort study. Lancet 2019, 393, 747–757. [Google Scholar] [CrossRef] [Green Version]
- Ishaque, N.; Abba, M.L.; Hauser, C.; Patil, N.; Paramasivam, N.; Huebschmann, D.; Leupold, J.H.; Balasubramanian, G.P.; Kleinheinz, K.; Toprak, U.H.; et al. Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer. Nat. Commun. 2018, 9, 4782. [Google Scholar] [CrossRef] [Green Version]
- Rusch, M.; Nakitandwe, J.; Shurtleff, S.; Newman, S.; Zhang, Z.; Edmonson, M.N.; Parker, M.; Jiao, Y.; Ma, X.; Liu, Y.; et al. Clinical cancer genomic profiling by three-Platform sequencing of whole genome, whole exome and transcriptome. Nat. Commun. 2018, 9, 3962. [Google Scholar] [CrossRef] [Green Version]
- Fong, K.; Tremblay, D.M.; Delaquis, P.; Goodridge, L.; Levesque, R.C.; Moineau, S.; Suttle, C.A.; Wang, S. Diversity and Host Specificity Revealed by Biological Characterization and Whole Genome Sequencing of Bacteriophages Infecting Salmonella enterica. Viruses 2019, 11, 854. [Google Scholar] [CrossRef] [Green Version]
- Russell, J.S.; Caly, L.; Kostecki, R.; McGuinness, S.L.; Carter, G.; Bulach, D.; Seemann, T.; Stinear, T.P.; Baird, R.; Catton, M.; et al. The First Isolation and Whole Genome Sequencing of Murray Valley Encephalitis Virus from Cerebrospinal Fluid of a Patient with Encephalitis. Viruses 2018, 10, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob-Hirsch, J.; Eyal, E.; Knisbacher, B.A.; Roth, J.; Cesarkas, K.; Dor, C.; Farage-Barhom, S.; Kunik, V.; Simon, A.J.; Gal, M.; et al. Whole-Genome sequencing reveals principles of brain retrotransposition in neurodevelopmental disorders. Cell Res. 2018, 28, 187–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motley, S.T.; Picuri, J.M.; Crowder, C.D.; Minich, J.J.; Hofstadler, S.A.; Eshoo, M.W. Improved multiple displacement amplification (iMDA) and ultraclean reagents. BMC Genom. 2014, 15, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, M.; Light, Y.K.; Meagher, R.J.; Singh, A.K. Digital Droplet Multiple Displacement Amplification (ddMDA) for Whole Genome Sequencing of Limited DNA Samples. PLoS ONE 2016, 11, e0153699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gole, J.; Gore, A.; Richards, A.; Chiu, Y.J.; Fung, H.L.; Bushman, D.; Chiang, H.I.; Chun, J.; Lo, Y.H.; Zhang, K. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat. Biotechnol. 2013, 31, 1126–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigue, S.; Malmstrom, R.R.; Berlin, A.M.; Birren, B.W.; Henn, M.R.; Chisholm, S.W. Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE 2009, 4, e6864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jan, Y.J.; Chen, J.F.; Zhu, Y.; Lu, Y.T.; Chen, S.H.; Chung, H.; Smalley, M.; Huang, Y.W.; Dong, J.; Chen, L.C.; et al. NanoVelcro rare-Cell assays for detection and characterization of circulating tumor cells. Adv. Drug Deliv. Rev. 2018, 125, 78–93. [Google Scholar] [CrossRef]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-Chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [Green Version]
- Lao, K.; Xu, N.L.; Straus, N.A. Whole genome amplification using single-Primer PCR. Biotechnol. J. 2008, 3, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Dean, F.B.; Nelson, J.R.; Giesler, T.L.; Lasken, R.S. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-Primed rolling circle amplification. Genome Res. 2001, 11, 1095–1099. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Li, C.; Lu, S.; Zhou, W.; Tang, F.; Xie, X.S.; Huang, Y. Uniform and accurate single-Cell sequencing based on emulsion whole-Genome amplification. Proc. Natl. Acad. Sci. USA 2015, 112, 11923–11928. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, M.; Nishikawa, Y.; Kogawa, M.; Takeyama, H. Massively parallel whole genome amplification for single-Cell sequencing using droplet microfluidics. Sci. Rep. 2017, 7, 5199. [Google Scholar] [CrossRef] [Green Version]
- Zong, C.; Lu, S.; Chapman, A.R.; Xie, X.S. Genome-Wide detection of single-Nucleotide and copy-Number variations of a single human cell. Science 2012, 338, 1622–1626. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Xing, D.; Tan, L.; Li, H.; Zhou, G.; Huang, L.; Xie, X.S. Single-Cell whole-Genome analyses by Linear Amplification via Transposon Insertion (LIANTI). Science 2017, 356, 189–194. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Lu, S.; Huang, Y. Microfluidic whole genome amplification device for single cell sequencing. Anal. Chem. 2014, 86, 9386–9390. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, J.; Wan, J.; Lu, Y.; Zhang, Z.; Xu, Z. A low cost and input tailing method of quality control on multiple annealing, and looping-Based amplification cycles-Based whole-Genome amplification products. J. Clin. Lab. Anal. 2019, 33, e22697. [Google Scholar] [CrossRef] [PubMed]
- Thibault, D.; Jensen, P.A.; Wood, S.; Qabar, C.; Clark, S.; Shainheit, M.G.; Isberg, R.R.; van Opijnen, T. Droplet Tn-Seq combines microfluidics with Tn-Seq for identifying complex single-cell phenotypes. Nat. Commun. 2019, 10, 5729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haliburton, J.R.; Shao, W.; Deutschbauer, A.; Arkin, A.; Abate, A.R. Genetic interaction mapping with microfluidic-Based single cell sequencing. PLoS ONE 2017, 12, e0171302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.; He, Y.; Zhu, Z.; Guo, J.; Wang, G.; Deng, C.; Zhong, Z. Small, Traceable, Endosome-Disrupting, and Bioresponsive Click Nanogels Fabricated via Microfluidics for CD44-Targeted Cytoplasmic Delivery of Therapeutic Proteins. ACS Appl. Mater. Interfaces 2019, 11, 22171–22180. [Google Scholar] [CrossRef] [PubMed]
- Leipert, J.; Tholey, A. Miniaturized sample preparation on a digital microfluidics device for sensitive bottom-Up microproteomics of mammalian cells using magnetic beads and mass spectrometry-Compatible surfactants. Lab Chip 2019, 19, 3490–3498. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Dong, H.; Zhao, W.; Zhang, X.; Duan, X.; Zhang, H.; Liu, S.; Sui, G. An Air-Liquid Interface Organ-Level Lung Microfluidics Platform for Analysis on Molecular Mechanisms of Cytotoxicity Induced by Cancer-Causing Fine Particles. ACS Sens. 2019, 4, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Wimmers, F.; Subedi, N.; van Buuringen, N.; Heister, D.; Vivie, J.; Beeren-Reinieren, I.; Woestenenk, R.; Dolstra, H.; Piruska, A.; Jacobs, J.F.M.; et al. Single-Cell analysis reveals that stochasticity and paracrine signaling control interferon-Alpha production by plasmacytoid dendritic cells. Nat. Commun. 2018, 9, 3317. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Lu, N.; Shi, X.; Qiao, Y.; Chen, L.; Duan, M.; Hou, Y.; Ge, Q.; Tao, Y.; Tu, J.; et al. 1D-Reactor Decentralized MDA for Uniform and Accurate Whole Genome Amplification. Anal. Chem. 2017, 89, 10147–10152. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Jia, F.; Zhang, W.; Shi, F.; Fang, Z.; Zhao, H.; Hu, Z.; Wei, Z. Device for whole genome sequencing single circulating tumor cells from whole blood. Lab Chip 2019, 19, 3168–3178. [Google Scholar] [CrossRef]
- Nishikawa, Y.; Hosokawa, M.; Maruyama, T.; Yamagishi, K.; Mori, T.; Takeyama, H. Monodisperse Picoliter Droplets for Low-Bias and Contamination-Free Reactions in Single-Cell Whole Genome Amplification. PLoS ONE 2015, 10, e0138733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azizi, M.; Zaferani, M.; Cheong, S.H.; Abbaspourrad, A. Pathogenic Bacteria Detection Using RNA-Based Loop-Mediated Isothermal-Amplification-Assisted Nucleic Acid Amplification via Droplet Microfluidics. ACS Sens. 2019, 4, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.; Sciambi, A.; Treusch, S.; Durruthy-Durruthy, R.; Gokhale, K.; Jacob, J.; Chen, T.X.; Geis, J.A.; Oldham, W.; Matthews, J.; et al. High-Throughput single-Cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res. 2018, 28, 1345–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidore, A.M.; Lan, F.; Lim, S.W.; Abate, A.R. Enhanced sequencing coverage with digital droplet multiple displacement amplification. Nucleic Acids Res. 2016, 44, e66. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Whitesides, G.M. Soft Lithography. Angew. Chem. 1998, 37, 550–575. [Google Scholar] [CrossRef]
- Husi, H.; Skipworth, R.J.; Fearon, K.C.; Ross, J.A. LSCluster, a large-Scale sequence clustering and aligning software for use in partial identity mapping and splice-Variant analysis. J. Proteom. 2013, 84, 185–189. [Google Scholar] [CrossRef]
- Ward, C.M.; Thu-Hien, T.; Pederson, S.M. ngsReports: A Bioconductor package for managing FastQC reports and other NGS related log files. Bioinformatics 2019, 36, 2587–2588. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Conesa, A.; Garcia-Alcalde, F. Qualimap 2: Advanced multi-Sample quality control for high-Throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Garvin, T.; Aboukhalil, R.; Kendall, J.; Baslan, T.; Atwal, G.S.; Hicks, J.; Wigler, M.; Schatz, M.C. Interactive analysis and assessment of single-Cell copy-Number variations. Nat. Methods 2015, 12, 1058–1060. [Google Scholar] [CrossRef] [Green Version]
- de Bourcy, C.F.; De Vlaminck, I.; Kanbar, J.N.; Wang, J.; Gawad, C.; Quake, S.R. A quantitative comparison of single-Cell whole genome amplification methods. PLoS ONE 2014, 9, e105585. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Manghwar, H.; Sun, L.; Wang, P.; Wang, G.; Sheng, H.; Zhang, J.; Liu, H.; Qin, L.; Rui, H.; et al. Whole genome sequencing reveals rare off-Target mutations and considerable inherent genetic or/and somaclonal variations in CRISPR/Cas9-Edited cotton plants. Plant Biotechnol. J. 2019, 17, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Haliburton, J.R.; Yuan, A.; Abate, A.R. Droplet barcoding for massively parallel single-molecule deep sequencing. Nat. Commun. 2016, 7, 11784. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Demaree, B.; Ahmed, N.; Abate, A.R. Single-Cell genome sequencing at ultra-High-Throughput with microfluidic droplet barcoding. Nat. Biotechnol. 2017, 35, 640–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Niu, L.; Zhao, F.; Yan, L.; Nong, J.; Wang, C.; Gao, N.; Zhu, X.; Wu, L.; Bo, T.; et al. Identification of Acinetobacter baumannii and its carbapenem-Resistant gene blaOXA-23-Like by multiple cross displacement amplification combined with lateral flow biosensor. Sci. Rep. 2019, 9, 17888. [Google Scholar] [CrossRef]
- Niu, L.; Zhao, F.; Chen, J.; Nong, J.; Wang, C.; Wang, J.; Gao, N.; Zhu, X.; Wu, L.; Hu, S. Isothermal amplification and rapid detection of Klebsiella pneumoniae based on the multiple cross displacement amplification (MCDA) and gold nanoparticle lateral flow biosensor (LFB). PLoS ONE 2018, 13, e0204332. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Wang, H.; Xu, J.; Ye, C. A label-Free technique for accurate detection of nucleic acid-Based self-Avoiding molecular recognition systems supplemented multiple cross-displacement amplification and nanoparticles based biosensor. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1671–1684. [Google Scholar] [CrossRef]
- Huang, L.; Ma, F.; Chapman, A.; Lu, S.; Xie, X.S. Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annu. Rev. Genom. Hum. Genet. 2015, 16, 79–102. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zhang, H.; Hu, D.; Lu, S.; Sun, X. The performance of MALBAC and MDA methods in the identification of concurrent mutations and aneuploidy screening to diagnose beta-Thalassaemia disorders at the single- and multiple-Cell levels. J. Clin. Lab. Anal. 2018, 32. [Google Scholar] [CrossRef] [Green Version]



| Sample/Method | Read Mapping Ratio (%) | GC Content (%) | Mean Depth (×) | Genome Coverage (%) |
|---|---|---|---|---|
| bulk | 99.67 | 40.87 | 30.91 | 94.83 |
| tMDA | 99.65 | 41.41 | 30.84 | 94.55 |
| dMDA | 99.69 | 38.76 | 43.62 | 92.94 |
| tMALBAC | 99.61 | 47.13 | 29.96 | 92.82 |
| dMALBAC | 99.45 | 46.13 | 40.11 | 91.11 |
| Parameter | Sample Type | ||||
|---|---|---|---|---|---|
| dMDA | tMDA | dMALBAC | tMALBAC | Bulk | |
| Total SNVs | 24,2393 | 32,5728 | 22,6516 | 21,9911 | 43,4452 |
| Detection rate | 55.79% | 74.97% | 52.14% | 50.62% | N/A |
| Heterozygous SNVs | 124,015 | 153,704 | 111,856 | 122,428 | 165,177 |
| Detection rate | 75.08% | 93.05% | 67.72% | 74.12% | N/A |
| Homozygous SNVs | 118,378 | 172,024 | 114,660 | 97,483 | 269,275 |
| Detection rate | 43.96% | 63.88% | 42.58% | 36.20% | N/A |
| ADO rate | 1.58% | 1.22% | 0.92% | 0.52% | N/A |
| SNV error rate | 0.002% | 0.002% | 0.06% | 0.049% | N/A |
| False-positive rate | 25.13% | 25.03% | 19.96% | 26.05% | N/A |
| Parameter | Heterozygous SNVs | Homozygous SNVs | Total SNVs |
|---|---|---|---|
| Bulk (30×) | |||
| SNVs | 165,177 | 269,275 | 434,452 |
| Bulk (10×) | |||
| SNVs | 153,212 | 165,070 | 318,282 |
| Detection rate | 92.76% | 61.30% | 73.26% |
| dMDA (10×) | |||
| SNVs | 52,325 | 15,005 | 67,330 |
| Detection rate | 91.135% | 69.525% | 80.330% |
| tMDA (10×) | |||
| SNVs | 103,535 | 101,481 | 205,016 |
| Detection rate | 62.68% | 37.69% | 47.19% |
| dMALBAC (10×) | |||
| SNVs | 69,417 | 66,793 | 136,210 |
| Detection rate | 42.02% | 24.80% | 31.35% |
| tMALBAC (10×) | |||
| SNVs | 67,410 | 47,705 | 115,115 |
| Detection rate | 40.81% | 17.72% | 26.50% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Xu, Y.; Zhu, L.; Su, Z.; Han, X.; Zhang, Z.; Huang, Y.; Liu, Q. Comparison of Multiple Displacement Amplification (MDA) and Multiple Annealing and Looping-Based Amplification Cycles (MALBAC) in Limited DNA Sequencing Based on Tube and Droplet. Micromachines 2020, 11, 645. https://doi.org/10.3390/mi11070645
Zhou X, Xu Y, Zhu L, Su Z, Han X, Zhang Z, Huang Y, Liu Q. Comparison of Multiple Displacement Amplification (MDA) and Multiple Annealing and Looping-Based Amplification Cycles (MALBAC) in Limited DNA Sequencing Based on Tube and Droplet. Micromachines. 2020; 11(7):645. https://doi.org/10.3390/mi11070645
Chicago/Turabian StyleZhou, Xiaoxiang, Ying Xu, Libo Zhu, Zhen Su, Xiaoming Han, Zhen Zhang, Yan Huang, and Quanjun Liu. 2020. "Comparison of Multiple Displacement Amplification (MDA) and Multiple Annealing and Looping-Based Amplification Cycles (MALBAC) in Limited DNA Sequencing Based on Tube and Droplet" Micromachines 11, no. 7: 645. https://doi.org/10.3390/mi11070645
APA StyleZhou, X., Xu, Y., Zhu, L., Su, Z., Han, X., Zhang, Z., Huang, Y., & Liu, Q. (2020). Comparison of Multiple Displacement Amplification (MDA) and Multiple Annealing and Looping-Based Amplification Cycles (MALBAC) in Limited DNA Sequencing Based on Tube and Droplet. Micromachines, 11(7), 645. https://doi.org/10.3390/mi11070645




