Recent Progress in Nanomaterials Modified Electrochemical Biosensors for the Detection of MicroRNA
Abstract
1. Introduction
2. Amplification Strategies in miRNA Biosensor
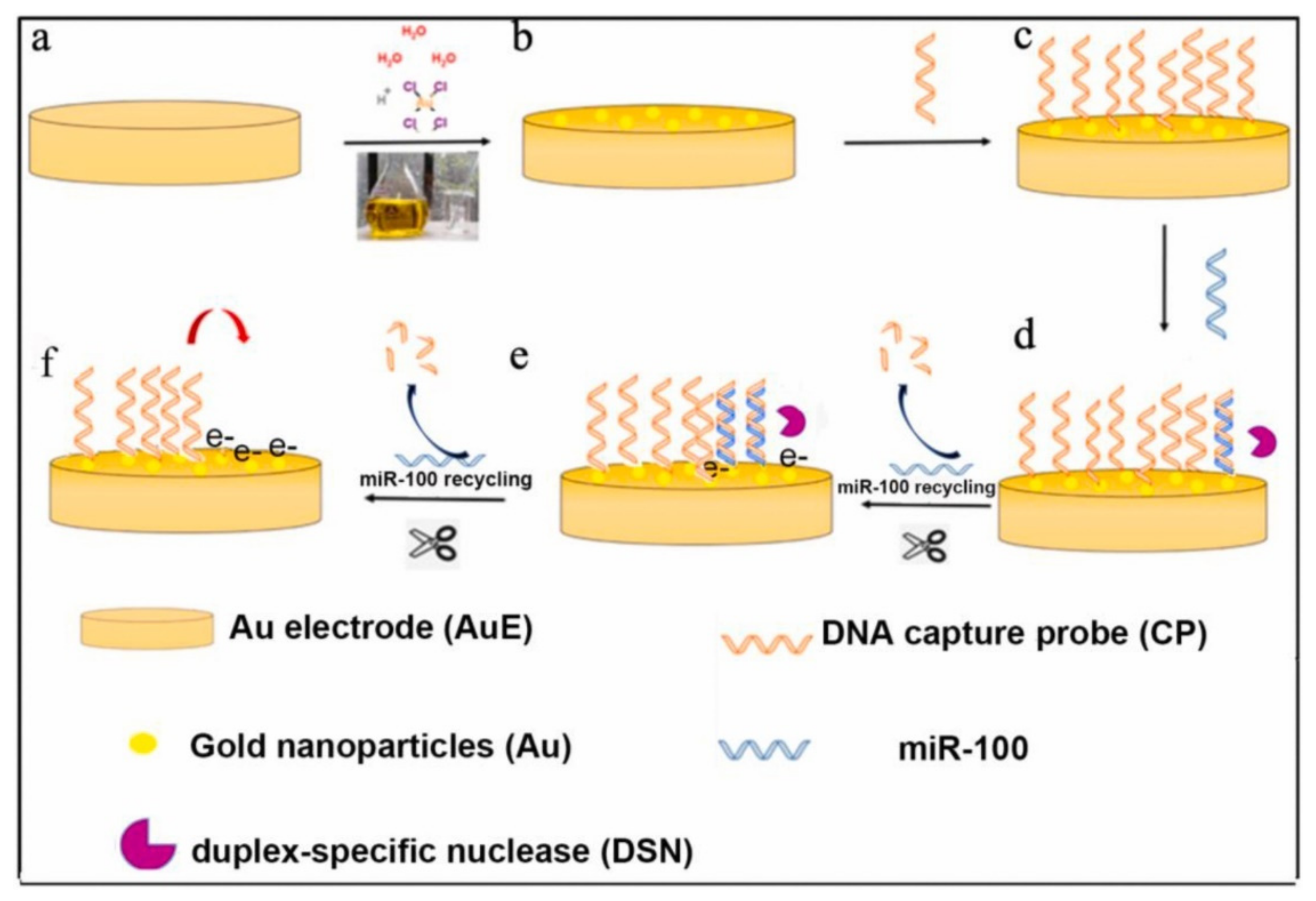
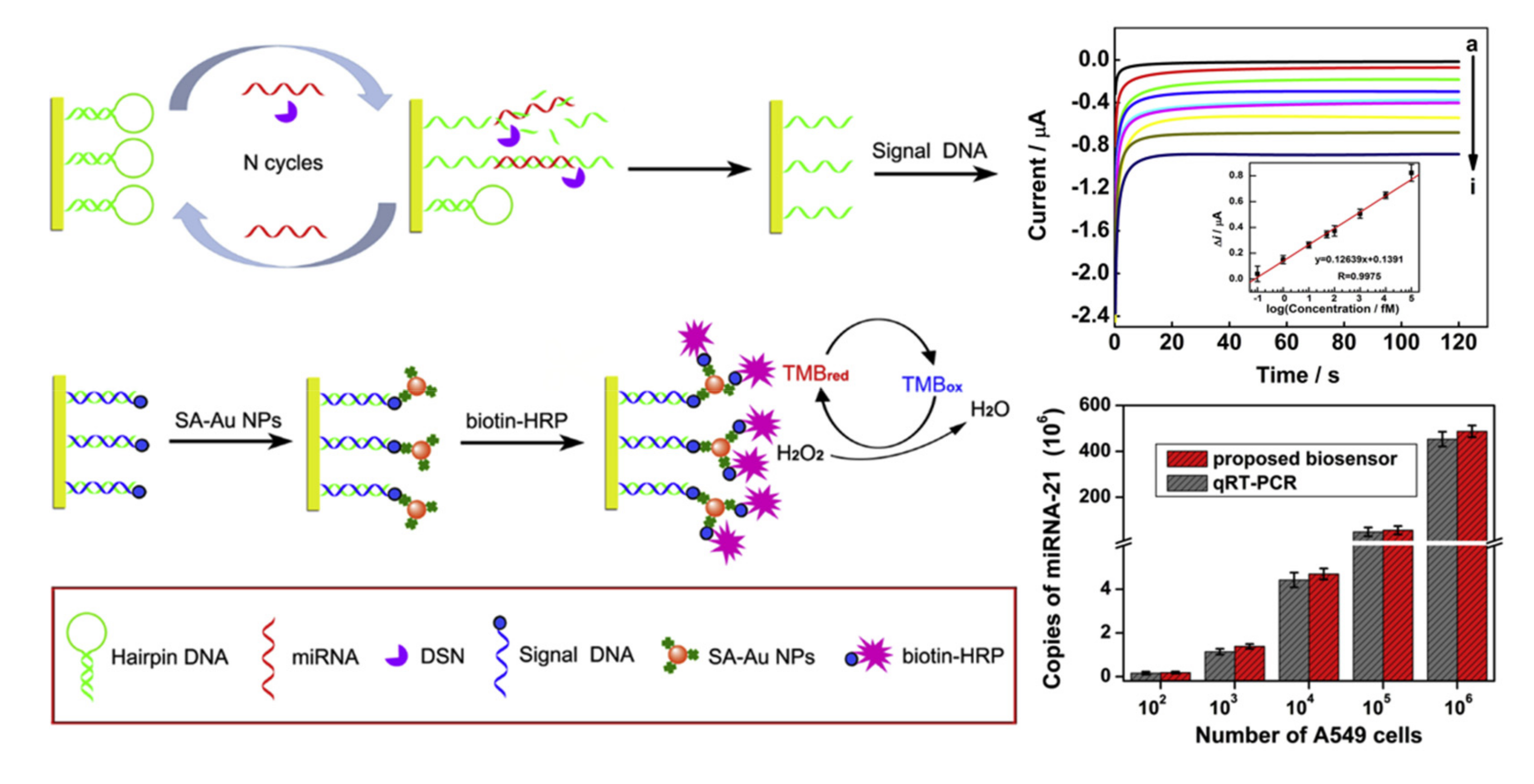
2.1. Cyclic Enzymatic Amplification Method (CEAM) Based on Nuclease
2.2. Rolling Circle Amplification (RCA)
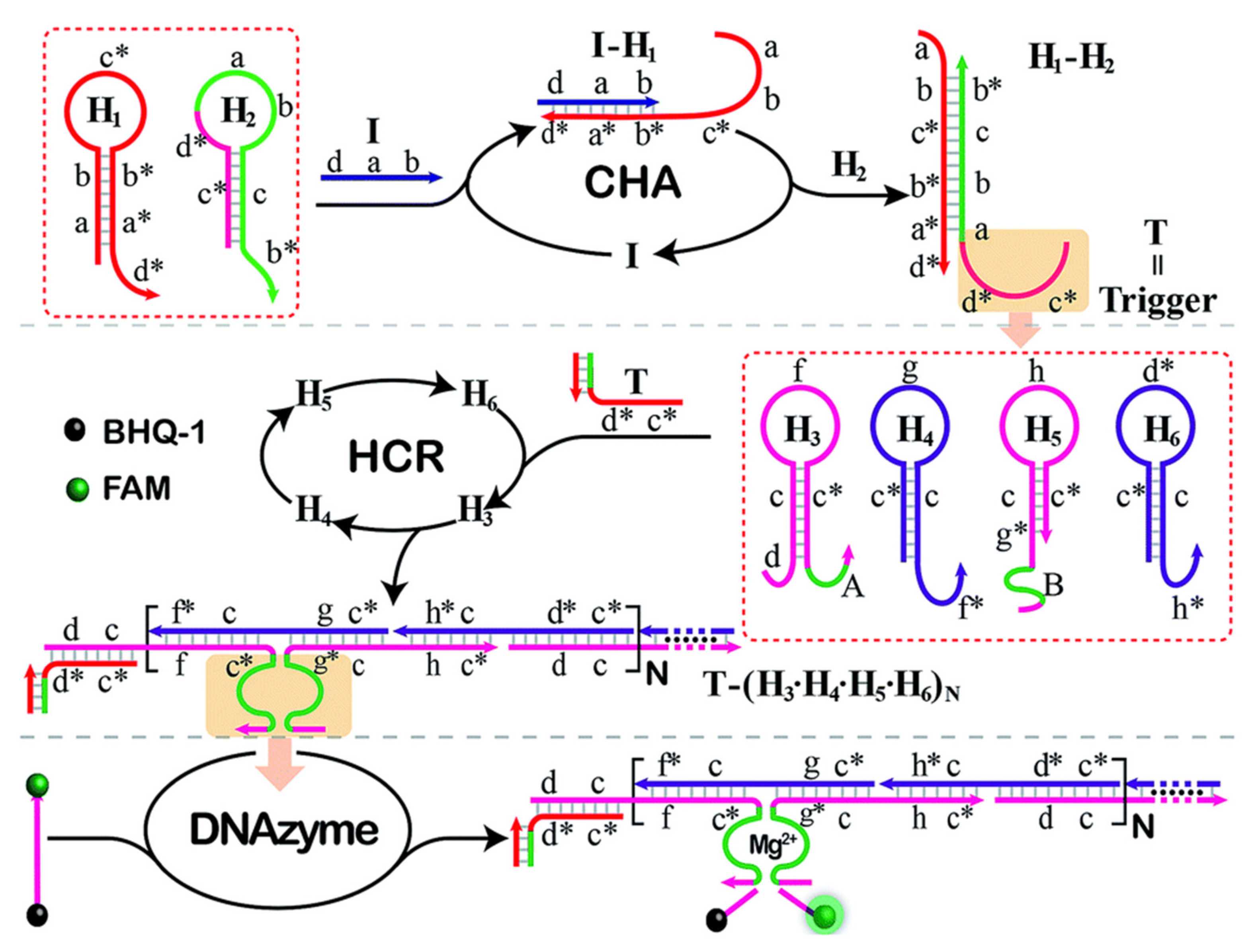
2.3. Enzyme-Free Amplification Strategy
2.4. Intercalation of Redox Mediator
3. Nanomaterial-Based Electrochemical Biosensors for miRNA Detection
3.1. Detection of miR-21
3.2. Detection of miR-144 and miR-200a
3.3. Detection of Other miRNAs
3.4. Simultaneous Detection of miRNAs
4. Conclusions and Future Perspectives
- (i)
- Coupling of novel nanomaterials with recognition element—The progress in nanomaterial synthesis will lead to the construction of novel nanomaterials with desired properties that are suitable for application in electrochemical biosensor. Therefore, the coupling of novel nanomaterials with recognition element for miRNA biosensor, which is normally complementary DNA probe will lead to the fabrication of biosensor with high sensitivity and low detection limit. In addition, novel nanomaterials could also be applied as nanolabels that bind specifically to duplexes, generating enhanced electrochemical signal output and multiplex detection capability.
- (ii)
- Reproducibility of nanomaterial-based electrochemical biosensor—The modification of biosensor with nanomaterials could improve the sensing performance of the biosensor, but there might be variation among each biosensor that was modified with the same nanomaterial. This could be due to the variation in the conformation or topology of modified nanomaterial on the biosensor surface, which is associated with the increased complexity of modified surface, giving rise to the reproducibility issue. Therefore, it is possible to perform statistical sampling on a batch of fabricated sensor and apply the testing and calibration to the entire batch.
- (iii)
- Validation of miRNA biosensor via real sample detection—It is crucial that the miRNA biosensor could function in clinical sample, providing accurate and reliable results as diagnostic tool. As miRNAs are present in various bodily fluids, for example saliva, plasma, tear, interstitial fluid, serum, urine, and others, the clinical samples obtained will be complex matrices, which will interfere the detection and recovery ratio. In addition, real sample often contains many species that might affect the electrochemical process or non-specific adsorption to sensing surface. Therefore, innovative materials and methods need to be developed to create boundary or functional linkage for the specific adhesion of target analyte to the sensor surface, ensuring the accuracy and recovery of biosensor.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Basson, M.; Liu, Z.; Ambros, V.; Horvitz, H.R.; Ruvkun, G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 2000, 5, 659–669. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of microRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Chugh, P.; Dittmer, D.P. Potential pitfalls in microRNA profiling. Wiley Interdiscip. Rev. RNA 2012, 3, 601–616. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research advances in the detection of miRNA. J. Pharm. Anal. 2019, 9, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Krepelkova, I.; Mrackova, T.; Izakova, J.; Dvorakova, B.; Chalupova, L.; Mikulik, R.; Slaby, O.; Bartos, M.; Ruzicka, V. Evaluation of miRNA detection methods for the analytical characteristic necessary for clinical utilization. Biotechniques 2019, 66, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, Q. The role of nanomaterials in electroanalytical biosensors: A mini review. J. Electroanal. Chem. 2016, 781, 401–409. [Google Scholar] [CrossRef]
- Yoon, J.; Shin, M.; Lee, T.; Choi, J.-W. Highly Sensitive Biosensors Based on Biomolecules and Functional Nanomaterials Depending on the Types of Nanomaterials: A Perspective Review. Materials 2020, 13, 299. [Google Scholar] [CrossRef]
- Holzinger, M.; Le Goff, A.; Cosnier, S. Nanomaterials for biosensing applications: A review. Front. Chem. 2014, 2, 63. [Google Scholar] [CrossRef] [PubMed]
- Alsaiari, N.S.; Katubi, K.M.; Alzahrani, F.M.; Siddeeg, S.M.; Tahoon, M.A. The Application of Nanomaterials for the Electrochemical Detection of Antibiotics: A Review. Micromachines 2021, 12, 308. [Google Scholar] [CrossRef]
- Farzin, L.; Shamsipur, M.; Samandari, L.; Sheibani, S. Advances in the design of nanomaterial-based electrochemical affinity and enzymatic biosensors for metabolic biomarkers: A review. Microchim. Acta 2018, 185, 276. [Google Scholar] [CrossRef]
- Wang, B.; Akiba, U.; Anzai, J. Recent Progress in Nanomaterial-Based Electrochemical Biosensors for Cancer Biomarkers: A Review. Molecules 2017, 22, 1048. [Google Scholar] [CrossRef]
- Wang, Y.-H.; He, L.-L.; Huang, K.-J.; Chen, Y.-X.; Wang, S.-Y.; Liu, Z.-H.; Li, D. Recent advances in nanomaterial-based electrochemical and optical sensing platforms for microRNA assays. Analyst 2019, 144, 2849–2866. [Google Scholar] [CrossRef] [PubMed]
- Low, S.S.; Loh, H.-S.; Boey, J.S.; Khiew, P.S.; Chiu, W.S.; Tan, M.T.T. Sensitivity enhancement of graphene/zinc oxide nanocomposite-based electrochemical impedance genosensor for single stranded RNA detection. Biosens. Bioelectron. 2017, 94, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Low, S.S.; Tan, M.T.T.; Loh, H.-S.; Khiew, P.S.; Chiu, W.S. Facile hydrothermal growth graphene/ZnO nanocomposite for development of enhanced biosensor. Anal. Chim. Acta 2016, 903, 131–141. [Google Scholar] [CrossRef]
- Gerasimova, Y.V.; Kolpashchikov, D.M. Enzyme-assisted target recycling (EATR) for nucleic acid detection. Chem. Soc. Rev. 2014, 43, 6405–6438. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, H.; Yammouri, G.; Amine, A. Current advances in electrochemical genosensors for detecting microRNA cancer markers. Curr. Opin. Electrochem. 2019, 16, 96–105. [Google Scholar] [CrossRef]
- Chen, Y.-X.; Huang, K.-J.; Niu, K.-X. Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens. Bioelectron. 2018, 99, 612–624. [Google Scholar] [CrossRef]
- Shagin, D.A.; Rebrikov, D.V.; Kozhemyako, V.B.; Altshuler, I.M.; Shcheglov, A.S.; Zhulidov, P.A.; Bogdanova, E.A.; Staroverov, D.B.; Rasskazov, V.A.; Lukyanov, S. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 2002, 12, 1935–1942. [Google Scholar] [CrossRef]
- Yi, H.; Cho, Y.-J.; Won, S.; Lee, J.-E.; Jin Yu, H.; Kim, S.; Schroth, G.P.; Luo, S.; Chun, J. Duplex-specific nuclease efficiently removes rRNA for prokaryotic RNA-seq. Nucleic Acids Res. 2011, 39, e140. [Google Scholar] [CrossRef]
- Qiu, X.; Zhang, H.; Yu, H.; Jiang, T.; Luo, Y. Duplex-specific nuclease-mediated bioanalysis. Trends Biotechnol. 2015, 33, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Wan, H.; Zhang, X. Electrochemical detection of miRNA-100 in the sera of gastric cancer patients based on DSN-assisted amplification. Talanta 2021, 225, 121981. [Google Scholar] [CrossRef]
- Grossman, L. Enzymes Involved in the Repair of DNA. In Advances in Radiation Biology; Lett, J.T., Adler, H., Zelle, M.B.T., Eds.; Elsevier: Amsterdam, The Netherlands, 1974; Volume 4, pp. 77–129. ISBN 0065-3292. [Google Scholar]
- Eun, H.-M. Nucleases. In Enzymology Primer for Recombinant DNA Technology; Eun, H.-M., Ed.; Academic Press: San Diego, CA, USA, 1996; pp. 145–232. ISBN 978-0-12-243740-3. [Google Scholar]
- Shinozaki, K.; Tuneko, O. T7 gene 6 exonuclease has an RNase H activity. Nucleic Acids Res. 1978, 5, 4245–4262. [Google Scholar] [CrossRef][Green Version]
- Yang, X.; Feng, M.; Xia, J.; Zhang, F.; Wang, Z. An electrochemical biosensor based on AuNPs/Ti3C2 MXene three-dimensional nanocomposite for microRNA-155 detection by exonuclease III-aided cascade target recycling. J. Electroanal. Chem. 2020, 878, 114669. [Google Scholar] [CrossRef]
- Yan, X.-M.; Wang, Y.-Q.; Chen, Y.; Chen, Z.-P.; Yu, R.-Q. Detection of microRNAs by the combination of Exonuclease-III assisted target recycling amplification and repeated-fishing strategy. Anal. Chim. Acta 2020, 1131, 1–8. [Google Scholar] [CrossRef]
- Miao, P.; Zhang, T.; Xu, J.; Tang, Y. Electrochemical Detection of miRNA Combining T7 Exonuclease-Assisted Cascade Signal Amplification and DNA-Templated Copper Nanoparticles. Anal. Chem. 2018, 90, 11154–11160. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Fu, Z.; Li, B.; Zhou, Y.; Yin, H.; Ai, S. One-Step, Ultrasensitive, and Electrochemical Assay of microRNAs Based on T7 Exonuclease Assisted Cyclic Enzymatic Amplification. Anal. Chem. 2014, 86, 5606–5610. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.G.; Kool, E.T. The Discovery of Rolling Circle Amplification and Rolling Circle Transcription. Acc. Chem. Res. 2016, 49, 2540–2550. [Google Scholar] [CrossRef]
- Zhao, W.; Ali, M.M.; Brook, M.A.; Li, Y. Rolling Circle Amplification: Applications in Nanotechnology and Biodetection with Functional Nucleic Acids. Angew. Chem. Int. Ed. 2008, 47, 6330–6337. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Jonstrup, S.P.; Koch, J.; Kjems, J. A microRNA detection system based on padlock probes and rolling circle amplification. RNA 2006, 12, 1747–1752. [Google Scholar] [CrossRef] [PubMed]
- Dirks, R.M.; Pierce, N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 2004, 101, 15275–15278. [Google Scholar] [CrossRef]
- Evanko, D. Hybridization chain reaction. Nat. Methods 2004, 1, 186. [Google Scholar] [CrossRef]
- Liang, M.; Pan, M.; Hu, J.; Wang, F.; Liu, X. Electrochemical Biosensor for MicroRNA Detection Based on Cascade Hybridization Chain Reaction. ChemElectroChem 2018, 5, 1380–1386. [Google Scholar] [CrossRef]
- Kim, J.; Shim, J.S.; Han, B.H.; Kim, H.J.; Park, J.; Cho, I.-J.; Kang, S.G.; Kang, J.Y.; Bong, K.W.; Choi, N. Hydrogel-based hybridization chain reaction (HCR) for detection of urinary exosomal miRNAs as a diagnostic tool of prostate cancer. Biosens. Bioelectron. 2021, 192, 113504. [Google Scholar] [CrossRef]
- Li, M.; Cheng, J.; Yuan, Z.; Zhou, H.; Zhang, L.; Dai, Y.; Shen, Q.; Fan, Q. Sensitive electrochemical detection of microRNA based on DNA walkers and hyperbranched HCR-DNAzyme cascade signal amplification strategy. Sens. Actuators B Chem. 2021, 345, 130348. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Xie, H.; Zhao, L.; Zheng, L.; Ye, H. Applications of Catalytic Hairpin Assembly Reaction in Biosensing. Small 2019, 15, 1902989. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Wu, Q.; Liang, M.; Liu, X.; Wang, F. A DNAzyme-amplified DNA circuit for highly accurate microRNA detection and intracellular imaging. Chem. Sci. 2019, 10, 9597–9604. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Briggs, N.; McLain, J.R.; Ellington, A.D. Stacking nonenzymatic circuits for high signal gain. Proc. Natl. Acad. Sci. USA 2013, 110, 5386–5391. [Google Scholar] [CrossRef] [PubMed]
- El Aamri, M.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical Biosensors for Detection of MicroRNA as a Cancer Biomarker: Pros and Cons. Biosensors 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Pothipor, C.; Aroonyadet, N.; Bamrungsap, S.; Jakmunee, J.; Ounnunkad, K. A highly sensitive electrochemical microRNA-21 biosensor based on intercalating methylene blue signal amplification and a highly dispersed gold nanoparticles/graphene/polypyrrole composite. Analyst 2021, 146, 2679–2688. [Google Scholar] [CrossRef]
- Zhao, L.-L.; Pan, H.-Y.; Zhang, X.-X.; Zhou, Y.-L. Ultrasensitive detection of microRNA based on a homogeneous label-free electrochemical platform using G-triplex/methylene blue as a signal generator. Anal. Chim. Acta 2020, 1116, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Medina, P.P.; Slack, F.J. MicroRNAs and cancer: An overview. Cell Cycle 2008, 7, 2485–2492. [Google Scholar] [CrossRef]
- Sabahi, A.; Salahandish, R.; Ghaffarinejad, A.; Omidinia, E. Electrochemical nano-genosensor for highly sensitive detection of miR-21 biomarker based on SWCNT-grafted dendritic Au nanostructure for early detection of prostate cancer. Talanta 2020, 209, 120595. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Liu, W.; Zhao, D.; Hao, Q.; Li, J.; Huang, J.; Shi, J.; Chao, J.; Su, S.; Wang, L. Label-Free Electrochemical Sensing Platform for MicroRNA-21 Detection Using Thionine and Gold Nanoparticles Co-Functionalized MoS2 Nanosheet. ACS Appl. Mater. Interfaces 2017, 9, 35597–35603. [Google Scholar] [CrossRef]
- Low, S.S.; Pan, Y.; Ji, D.; Li, Y.; Lu, Y.; He, Y.; Chen, Q.; Liu, Q. Smartphone-based portable electrochemical biosensing system for detection of circulating microRNA-21 in saliva as a proof-of-concept. Sens. Actuators B Chem. 2020, 308, 127718. [Google Scholar] [CrossRef]
- Ghazizadeh, E.; Moosavifard, S.E.; Daneshmand, N.; kamari Kaverlavani, S. Impediometric Electrochemical Sensor Based on The Inspiration of Carnation Italian Ringspot Virus Structure to Detect an Attommolar of miR. Sci. Rep. 2020, 10, 9645. [Google Scholar] [CrossRef] [PubMed]
- Ghazizadeh, E.; Naseri, Z.; Jaafari, M.R.; Forozandeh-Moghadam, M.; Hosseinkhani, S. A fires novel report of exosomal electrochemical sensor for sensing micro RNAs by using multi covalent attachment p19 with high sensitivity. Biosens. Bioelectron. 2018, 113, 74–81. [Google Scholar] [CrossRef]
- Deng, K.; Liu, X.; Li, C.; Huang, H. Sensitive electrochemical sensing platform for microRNAs detection based on shortened multi-walled carbon nanotubes with high-loaded thionin. Biosens. Bioelectron. 2018, 117, 168–174. [Google Scholar] [CrossRef]
- Chen, Z.; Xie, Y.; Huang, W.; Qin, C.; Yu, A.; Lai, G. Exonuclease-assisted target recycling for ultrasensitive electrochemical detection of microRNA at vertically aligned carbon nanotubes. Nanoscale 2019, 11, 11262–11269. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, J.; Dong, S.; Zhu, N.; Gyimah, E.; Wang, K.; Li, Y.; Zhang, Z. An ultrasensitive electrochemical biosensor for detection of microRNA-21 based on redox reaction of ascorbic acid/iodine and duplex-specific nuclease assisted target recycling. Biosens. Bioelectron. 2019, 130, 81–87. [Google Scholar] [CrossRef]
- Zhang, H.; Fan, M.; Jiang, J.; Shen, Q.; Cai, C.; Shen, J. Sensitive electrochemical biosensor for MicroRNAs based on duplex-specific nuclease-assisted target recycling followed with gold nanoparticles and enzymatic signal amplification. Anal. Chim. Acta 2019, 1064, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Wang, Z.; Wang, C.; Han, J.; Bu, H. Combining padlock exponential rolling circle amplification with CoFe2O4 magnetic nanoparticles for microRNA detection by nanoelectrocatalysis without a substrate. Anal. Chim. Acta 2017, 962, 24–31. [Google Scholar] [CrossRef]
- Meng, T.; Jia, H.; An, S.; Wang, H.; Yang, X.; Zhang, Y. Pd nanoparticles-DNA layered nanoreticulation biosensor based on target-catalytic hairpin assembly for ultrasensitive and selective biosensing of microRNA-21. Sens. Actuators B Chem. 2020, 323, 128621. [Google Scholar] [CrossRef]
- Meng, T.; Shang, N.; Nsabimana, A.; Ye, H.; Wang, H.; Wang, C.; Zhang, Y. An enzyme-free electrochemical biosensor based on target-catalytic hairpin assembly and Pd@UiO-66 for the ultrasensitive detection of microRNA-21. Anal. Chim. Acta 2020, 1138, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Zhang, W.; Zhang, J.; Zhang, Q.; Sheng, T.; Gu, Y. An electrochemical biosensor for sensitive detection of microRNAs based on target-recycled non-enzymatic amplification. Sens. Actuators B Chem. 2018, 271, 15–23. [Google Scholar] [CrossRef]
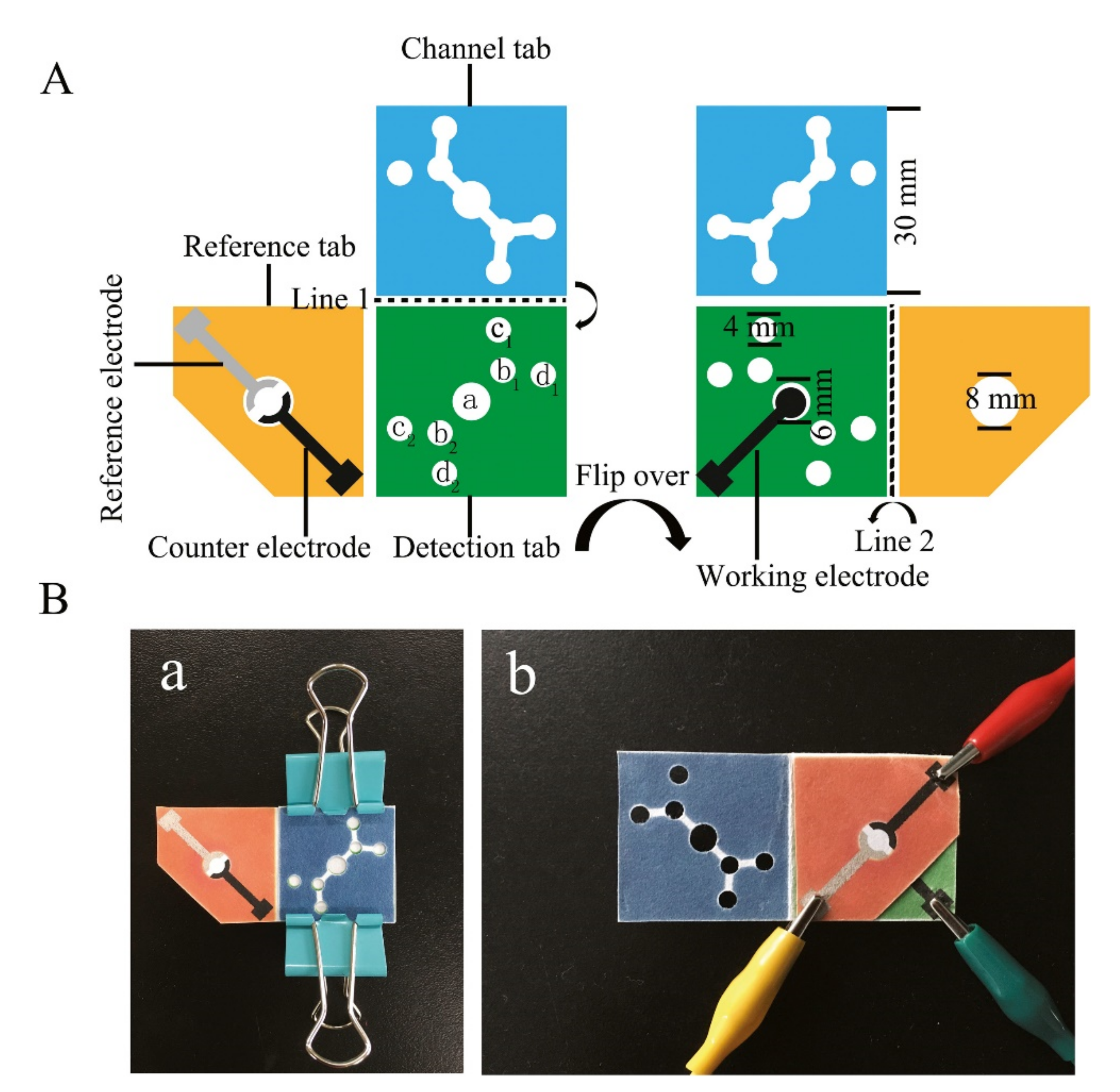
- Liu, X.; Li, X.; Gao, X.; Ge, L.; Sun, X.; Li, F. A Universal Paper-Based Electrochemical Sensor for Zero-Background Assay of Diverse Biomarkers. ACS Appl. Mater. Interfaces 2019, 11, 15381–15388. [Google Scholar] [CrossRef]
- Sun, X.; Wang, H.; Jian, Y.; Lan, F.; Zhang, L.; Liu, H.; Ge, S.; Yu, J. Ultrasensitive microfluidic paper-based electrochemical/visual biosensor based on spherical-like cerium dioxide catalyst for miR-21 detection. Biosens. Bioelectron. 2018, 105, 218–225. [Google Scholar] [CrossRef]
- Zhao, F.; Zhang, H.; Zheng, J. Novel electrochemical biosensing platform for microRNA detection based on G-quadruplex formation in nanochannels. Sens. Actuators B Chem. 2021, 327, 128898. [Google Scholar] [CrossRef]
- Mohammadniaei, M.; Go, A.; Chavan, S.G.; Koyappayil, A.; Kim, S.-E.; Yoo, H.J.; Min, J.; Lee, M.-H. Relay-race RNA/barcode gold nanoflower hybrid for wide and sensitive detection of microRNA in total patient serum. Biosens. Bioelectron. 2019, 141, 111468. [Google Scholar] [CrossRef]
- Gao, Y.; Feng, B.; Han, S.; Zhang, K.; Chen, J.; Li, C.; Wang, R.; Chen, L. The Roles of MicroRNA-141 in Human Cancers: From Diagnosis to Treatment. Cell. Physiol. Biochem. 2016, 38, 427–448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Chai, H.; Meng, F.; Guo, Z.; Jiang, Y.; Miao, P. DNA-Functionalized Porous Fe3O4 Nanoparticles for the Construction of Self-Powered miRNA Biosensor with Target Recycling Amplification. ACS Appl. Mater. Interfaces 2018, 10, 36796–36804. [Google Scholar] [CrossRef]
- Low, S.S.; Chen, Z.; Li, Y.; Lu, Y.; Liu, Q. Design Principle in Biosensing: Critical Analysis based on Graphitic Carbon Nitride (G-C3N4) Photoelectrochemical Biosensor. TrAC Trends Anal. Chem. 2021, 116454. [Google Scholar] [CrossRef]
- Yuan, Y.-H.; Chi, B.-Z.; Wen, S.-H.; Liang, R.-P.; Li, Z.-M.; Qiu, J.-D. Ratiometric electrochemical assay for sensitive detecting microRNA based on dual-amplification mechanism of duplex-specific nuclease and hybridization chain reaction. Biosens. Bioelectron. 2018, 102, 211–216. [Google Scholar] [CrossRef]
- Moazampour, M.; Zare, H.R.; Shekari, Z. Femtomolar determination of an ovarian cancer biomarker (miR-200a) in blood plasma using a label free electrochemical biosensor based on l-cysteine functionalized ZnS quantum dots. Anal. Methods 2021, 13, 2021–2029. [Google Scholar] [CrossRef] [PubMed]
- Jou, A.F.-J.; Chen, Y.-J.; Li, Y.; Chang, Y.-F.; Lee, J.; Liao, A.T.; Ho, J.A. Target-Triggered, Dual Amplification Strategy for Sensitive Electrochemical Detection of a Lymphoma-associated MicroRNA. Electrochim. Acta 2017, 236, 190–197. [Google Scholar] [CrossRef]
- Hakimian, F.; Ghourchian, H. Ultrasensitive electrochemical biosensor for detection of microRNA-155 as a breast cancer risk factor. Anal. Chim. Acta 2020, 1136, 1–8. [Google Scholar] [CrossRef]
- Liang, Z.; Ou, D.; Sun, D.; Tong, Y.; Luo, H.; Chen, Z. Ultrasensitive biosensor for microRNA-155 using synergistically catalytic nanoprobe coupled with improved cascade strand displacement reaction. Biosens. Bioelectron. 2019, 146, 111744. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.V.; Nguyen, N.D.; Piro, B.; Tran, L.T. Fabrication of a quinone containing layer on gold nanoparticles directed to a label-free and reagentless electrochemical miRNA sensor. Anal. Methods 2017, 9, 2696–2702. [Google Scholar] [CrossRef]
- Asadzadeh-Firouzabadi, A.; Zare, H.R. Application of cysteamine-capped gold nanoparticles for early detection of lung cancer-specific miRNA (miR-25) in human blood plasma. Anal. Methods 2017, 9, 3852–3861. [Google Scholar] [CrossRef]
- Akbarnia, A.; Zare, H.R. A voltammetric assay for microRNA-25 based on the use of amino-functionalized graphene quantum dots and ss- and ds-DNAs as gene probes. Microchim. Acta 2018, 185, 503. [Google Scholar] [CrossRef] [PubMed]
- Isin, D.; Eksin, E.; Erdem, A. Graphene oxide modified single-use electrodes and their application for voltammetric miRNA analysis. Mater. Sci. Eng. C 2017, 75, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Congur, G.; Eksin, E.; Erdem, A. Impedimetric detection of miRNA-34a using graphene oxide modified chemically activated graphite electrodes. Sens. Actuators A Phys. 2018, 279, 493–500. [Google Scholar] [CrossRef]
- Azimzadeh, M.; Nasirizadeh, N.; Rahaie, M.; Naderi-Manesh, H. Early detection of Alzheimer’s disease using a biosensor based on electrochemically-reduced graphene oxide and gold nanowires for the quantification of serum microRNA-137. RSC Adv. 2017, 7, 55709–55719. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Nikokar, I.; Zokaei, M.; Bozorgzadeh, E. Design, development and evaluation of microRNA-199a-5p detecting electrochemical nanobiosensor with diagnostic application in Triple Negative Breast Cancer. Talanta 2018, 189, 592–598. [Google Scholar] [CrossRef]
- Sun, Y.; Jin, H.; Jiang, X.; Gui, R. Black phosphorus nanosheets adhering to thionine-doped 2D MOF as a smart aptasensor enabling accurate capture and ratiometric electrochemical detection of target microRNA. Sens. Actuators B Chem. 2020, 309, 127777. [Google Scholar] [CrossRef]
- Zuo, J.; Yuan, Y.; Zhao, M.; Wang, J.; Chen, Y.; Zhu, Q.; Bai, L. An efficient electrochemical assay for miR-3675-3p in human serum based on the nanohybrid of functionalized fullerene and metal-organic framework. Anal. Chim. Acta 2020, 1140, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh-Firouzabadi, A.; Zare, H.R. Preparation and application of AgNPs/SWCNTs nanohybrid as an electroactive label for sensitive detection of miRNA related to lung cancer. Sens. Actuators B Chem. 2018, 260, 824–831. [Google Scholar] [CrossRef]
- Tao, Y.; Yin, D.; Jin, M.; Fang, J.; Dai, T.; Li, Y.; Li, Y.; Pu, Q.; Xie, G. Double-loop hairpin probe and doxorubicin-loaded gold nanoparticles for the ultrasensitive electrochemical sensing of microRNA. Biosens. Bioelectron. 2017, 96, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Li, Y.; Bai, J. Hierarchical assembled nanomaterial paper based analytical devices for simultaneously electrochemical detection of microRNAs. Anal. Chim. Acta 2019, 1058, 89–96. [Google Scholar] [CrossRef]
- Mohammadniaei, M.; Koyappayil, A.; Sun, Y.; Min, J.; Lee, M.-H. Gold nanoparticle/MXene for multiple and sensitive detection of oncomiRs based on synergetic signal amplification. Biosens. Bioelectron. 2020, 159, 112208. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.-H.; Wu, Y.-D.; Chi, B.-Z.; Wen, S.-H.; Liang, R.-P.; Qiu, J.-D. Simultaneously electrochemical detection of microRNAs based on multifunctional magnetic nanoparticles probe coupling with hybridization chain reaction. Biosens. Bioelectron. 2017, 97, 325–331. [Google Scholar] [CrossRef]
- Chang, J.; Wang, X.; Wang, J.; Li, H.; Li, F. Nucleic Acid-Functionalized Metal–Organic Framework-Based Homogeneous Electrochemical Biosensor for Simultaneous Detection of Multiple Tumor Biomarkers. Anal. Chem. 2019, 91, 3604–3610. [Google Scholar] [CrossRef]
- Azzouzi, S.; Fredj, Z.; Turner, A.P.F.; Ali, M.B.; Mak, W.C. Generic Neutravidin Biosensor for Simultaneous Multiplex Detection of MicroRNAs via Electrochemically Encoded Responsive Nanolabels. ACS Sens. 2019, 4, 326–334. [Google Scholar] [CrossRef]
- Wang, L.-L.; Shao, H.-H.; Wang, W.-J.; Zhang, J.-R.; Zhu, J.-J. Nitrogen-doped hollow carbon nanospheres for high-energy-density biofuel cells and self-powered sensing of microRNA-21 and microRNA-141. Nano Energy 2018, 44, 95–102. [Google Scholar] [CrossRef]

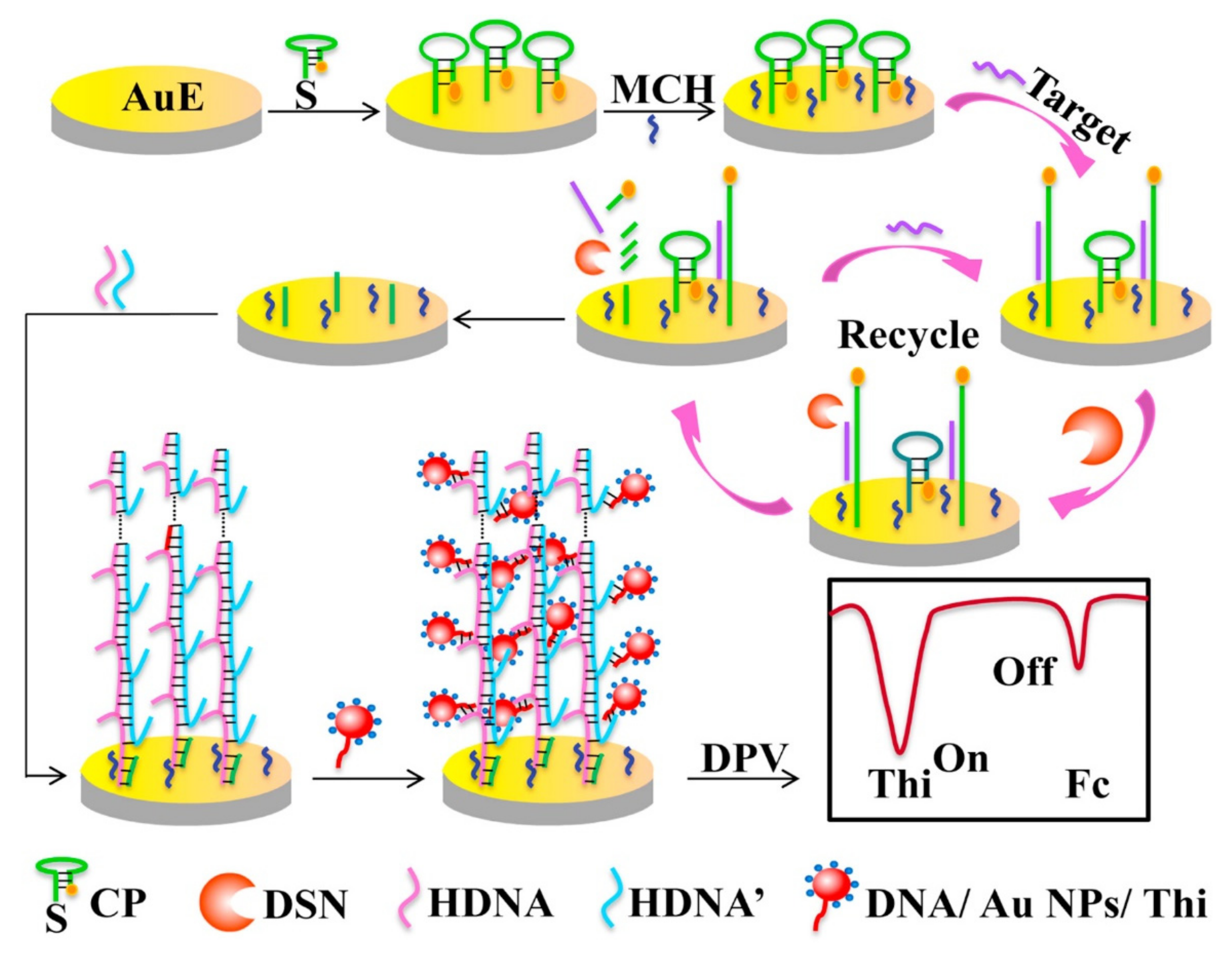
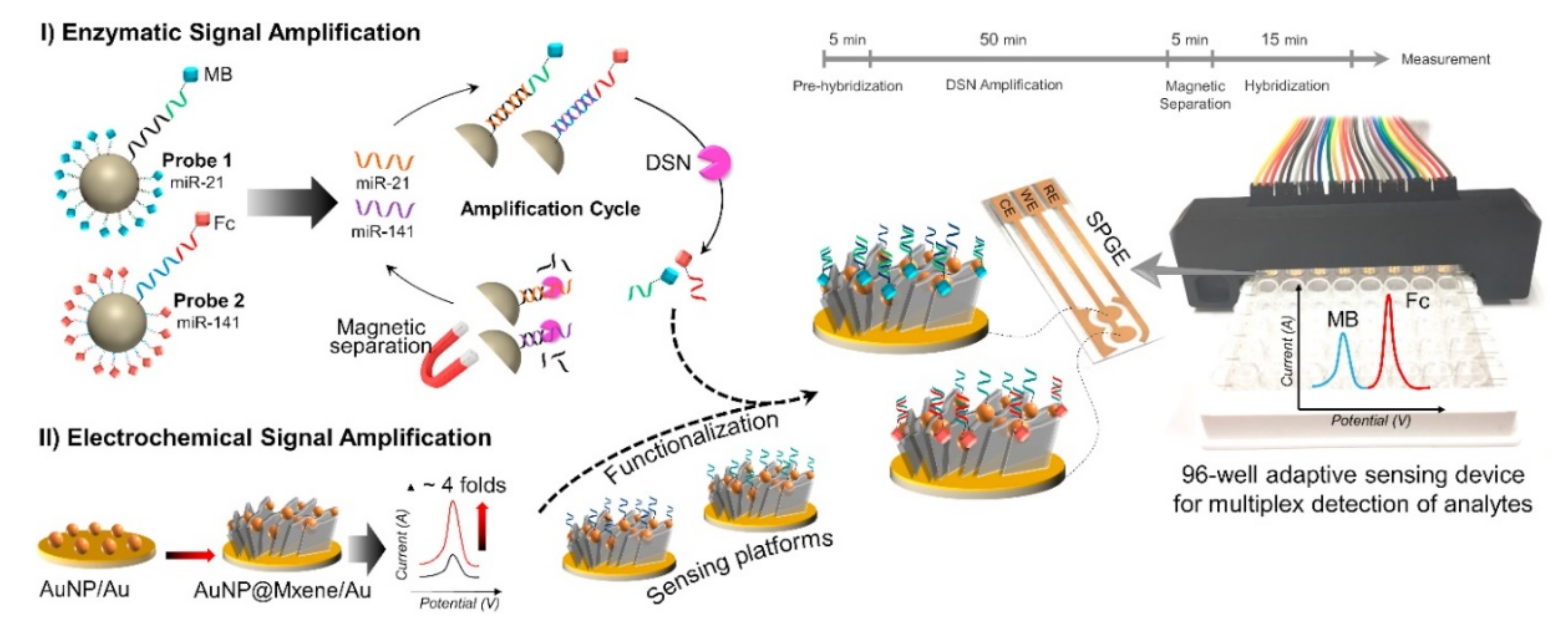
| miRNA | Nanomaterial Used | Linear Range | Limit of Detection (LOD) | Electrochemical Technique | Remark | Ref |
|---|---|---|---|---|---|---|
| miR-21 | Pd@UiO-66 | 20 fM to 600 pM | 0.713 fM | DPV | CHA amplification | [63] |
| SWCNT | 0.01 to 100 pM | 3.5 fM | DPV | T7 Exonuclease-Assisted Cascade Signal Amplification | [58] | |
| AuNPs | 0.1 fM to 100 pM | 43.3 aM | Amperometry | Triple amplification via DSN-assisted target recycling combined with gold nanoparticles, and horseradish peroxidase (HRP) enzymatic catalysis | [60] | |
| CNTs | 1 fM to 1 μM | - | DPV | Target-induced synthesis of Mg2+-dependent DNAzyme | [65] | |
| GO; Pd NPs | 1 fM to 50 pM | 63.1 aM | DPV | CHA amplification | [62] | |
| AuNPs; CoFe2O4 MNPs | 1 fM to 2 nM | 0.3 fM | SWV | Padlock exponential rolling circle amplification (P-ERCA) | [61] | |
| GNF@Pt | 1 μM to 500 aM | 135 aM | DPV | Relay-race RNA/barcode gold nanoflower hybrid | [68] | |
| AuNPs | 1 aM to 500 pM | 1 aM | DPV | Exosomal electrochemical properties as electrochemical amplifier bed | [56] | |
| AuNRs; CeO2-Au@GOx | 1 fM to 1000 fM | 0.434 fM | DPV | - | [66] | |
| SWCNTs/dendritic Au | 0.01 fM to 1 μM | 0.01 fM | DPV | - | [52] | |
| MWCNTs-COOH | 0.1 fmol to 5 pmol | 56.7 amol | DPV | Target-recycled non-enzymatic amplification | [64] | |
| MoS2-Thionine-AuNPs | 1.0 pM to 10.0 nM | 0.26 pM | SWV | - | [53] | |
| rGO/Au | 0.1 mM to 1 pM | 1 pM | DPV | Smartphone-based portable electrochemical biosensing system | [54] | |
| Carbon nanofibers | 1 aM to 10 pM | 0.5 aM | DPV | Label-free sensing based on guanine-quadruplex (G-quadruplex) formation | [67] | |
| AuNPs; MWCNTs | 0.1 to 12000 pM | 0.032 pM | DPV | - | [57] | |
| CuCo2O4 | 100 fM to1 aM | 1 aM | DPV | Virus-like hollow structure of CuCo2O4 filled with p19 protein | [55] | |
| MWCNTs@GONRs/AuNPs | 0.1 nM to 0.1 fM | 0.034 fM | DPV | DSN amplification | [59] | |
| miR-141 | AuNPs | 0.1 fM to 100 pM | 11 aM | DPV | Dual-amplification: DSN, HCR | [72] |
| CuNPs | 0.1 pM to 0.1 fM | 0.45 aM | DPV | T7 Exonuclease-Assisted Cascade Signal Amplification | [34] | |
| GO/AuNPs/Gox; Fe3O4 NPs | 10 aM to 10 fM | 1.4 aM | EIS | Self-powered system with DSN amplification | [70] | |
| miR-155 | AuNPs | - | 3.57 fM | SWV | Dual amplification via DSN amplification and strand displacement reaction | [74] |
| Ag-PEI NPs | 2 × 10−20 to 2 × 10−12 mol | 20 zmol | CV | - | [75] | |
| Cu-NMOF@PtNPs | 0.50 to 1.0 × 105 fM | 0.13 fM | SWV | Synergistically catalytic nanoprobe coupled with improved cascade strand displacement reaction | [76] | |
| AuNPs/Ti3C2 Mxene | 10 nM to 1 fM | 0.35 fM | DPV | Exonuclease III-aided cascade target recycling | [32] | |
| miR-103 | AuNPs | 100 fM to 5 nM | 100 fM | SWV | Label-free and reagentless detection | [77] |
| miR-25 | Cysteamine-AuNPs | 1 pM to 0.1 nM; 0.1 nM to 1 µM | 0.25 pM | EIS | - | [78] |
| AgNPs/SWCNTs | 1 pM to 0.1 nM; 0.1 nM to 0.1 10 nM | 0.313 pM | DPV | - | [86] | |
| Amino-functionalized GQDs | 0.3 nM to 1.0 μM | 95.0 pM | DPV | Accumulation of p-Biphenol | [79] | |
| miR-34a | GO | 5 to 35 μg/mL | 7.52 μg/mL | DPV | - | [80] |
| GO | 0 to 10 µg/mL | 261.7 nM | EIS | - | [81] | |
| miR-137 | ERGO + AuNWs | 5 to 750 fM | 1.7 fM | DPV | - | [82] |
| miR-200a | L-cysteine functionalized ZnS QDs | 1 µM to 10 fM | 8.4 fM | EIS | - | [73] |
| miR-199a-5p | GO-AuNRs | 15 fM to 148 pM | 4.5 fM | EIS | - | [83] |
| miR-3123 | BPNSs/Thionine/Cu-MOF | 2 pM to 2 μM | 0.3 pM | SWV | - | [84] |
| miR-3675-3p | C60@PAMAM-MOF; Au@PtNPs | 10 fM to 10 nM | 2.99 fM | DPV | - | [85] |
| Let-7d | AuNPs@Doxorucibin | 1 pM to 10 nM | 0.17 pM | SWV | Double-loop hairpin probe | [87] |
| Simultaneous detection: miR-141 and miR-21 | Fe3O4 NPs | 1 nM to 1 fM | 0.44 fM (miR-141) 0.46 fM (miR-21) | DPV | HCR amplification | [90] |
| MoS2/AuNPs/AgNW | 1 nM to 1 fM | 0.1 fM | SWV | - | [88] | |
| AuNPs; AgNPs | 50 to 1000 pM (miR-141); 0.5 to 1000 pM (miR-21) | 10 pM (miR-141); 0.3 pM (miR-21) | SSWV | Neutravidin—biotin affinity | [92] | |
| AuNPs/Mxene | 500 aM to 50 nM | 138 aM (miR-141); 204 aM (miR-21) | DPV | DSN amplification | [89] | |
| Simultaneous detection: miR-21 and let-7a | pNHCSs | 0.1 nM to 3.16 fM | 4.0 fM (miR-141); 0.1 fM (miR-21) | EIS | High-energy-density biofuel cells for self-powered sensing | [93] |
| UIO-66-NH2 | 0.01 to 100 pM | 8.2 fM (miR-21); 3.6 fM (let-7a) | DPV | - | [91] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Low, S.S.; Ji, D.; Chai, W.S.; Liu, J.; Khoo, K.S.; Salmanpour, S.; Karimi, F.; Deepanraj, B.; Show, P.L. Recent Progress in Nanomaterials Modified Electrochemical Biosensors for the Detection of MicroRNA. Micromachines 2021, 12, 1409. https://doi.org/10.3390/mi12111409
Low SS, Ji D, Chai WS, Liu J, Khoo KS, Salmanpour S, Karimi F, Deepanraj B, Show PL. Recent Progress in Nanomaterials Modified Electrochemical Biosensors for the Detection of MicroRNA. Micromachines. 2021; 12(11):1409. https://doi.org/10.3390/mi12111409
Chicago/Turabian StyleLow, Sze Shin, Daizong Ji, Wai Siong Chai, Jingjing Liu, Kuan Shiong Khoo, Sadegh Salmanpour, Fatemeh Karimi, Balakrishnan Deepanraj, and Pau Loke Show. 2021. "Recent Progress in Nanomaterials Modified Electrochemical Biosensors for the Detection of MicroRNA" Micromachines 12, no. 11: 1409. https://doi.org/10.3390/mi12111409
APA StyleLow, S. S., Ji, D., Chai, W. S., Liu, J., Khoo, K. S., Salmanpour, S., Karimi, F., Deepanraj, B., & Show, P. L. (2021). Recent Progress in Nanomaterials Modified Electrochemical Biosensors for the Detection of MicroRNA. Micromachines, 12(11), 1409. https://doi.org/10.3390/mi12111409










