A 3D Microfluidic ELISA for the Detection of Severe Dengue: Sensitivity Improvement and Vroman Effect Amelioration by EDC–NHS Surface Modification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Instruments
2.2. Recruitment of Patients and Study Approval
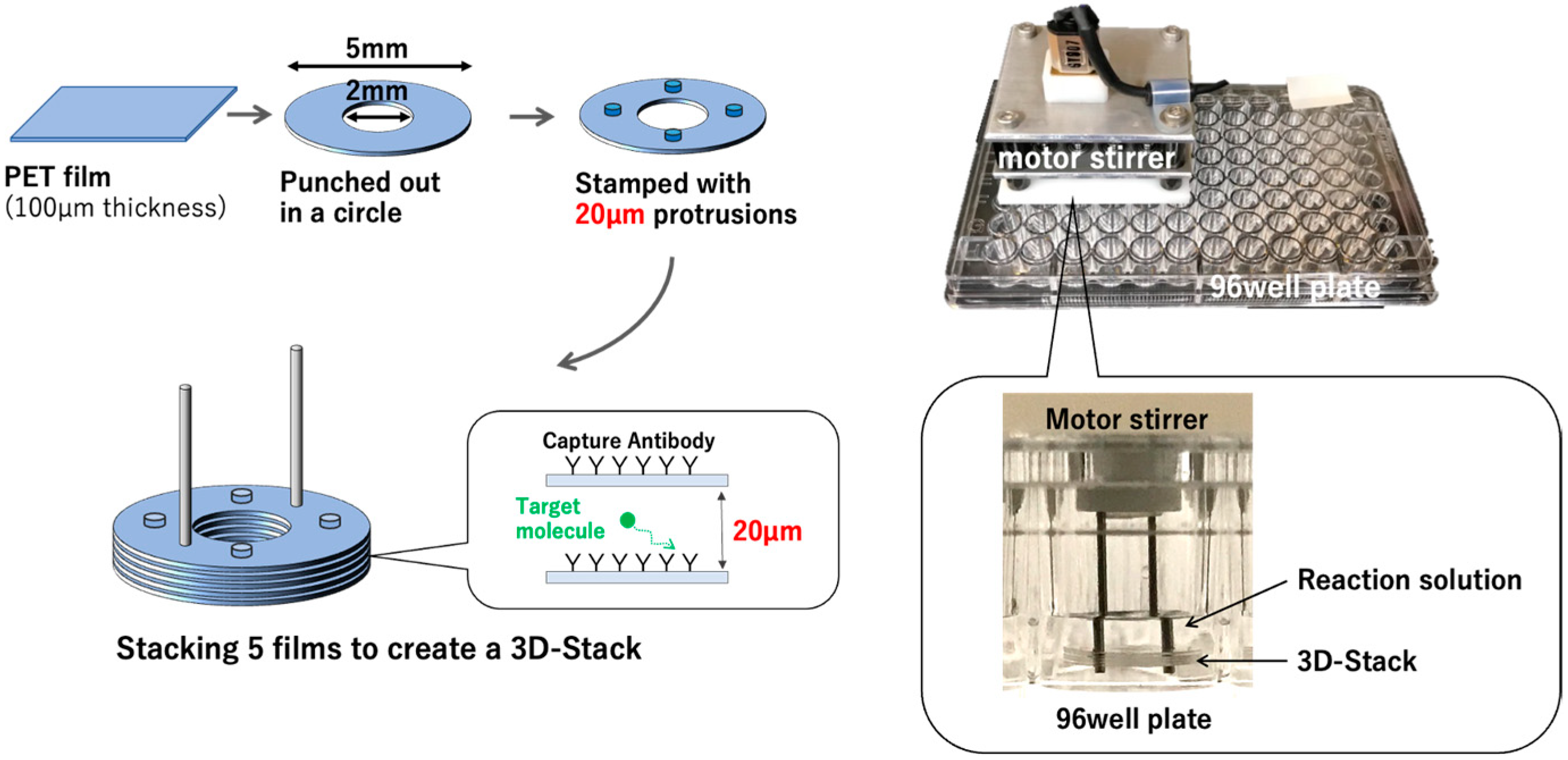
2.3. Hydrolysis of 3D Stack
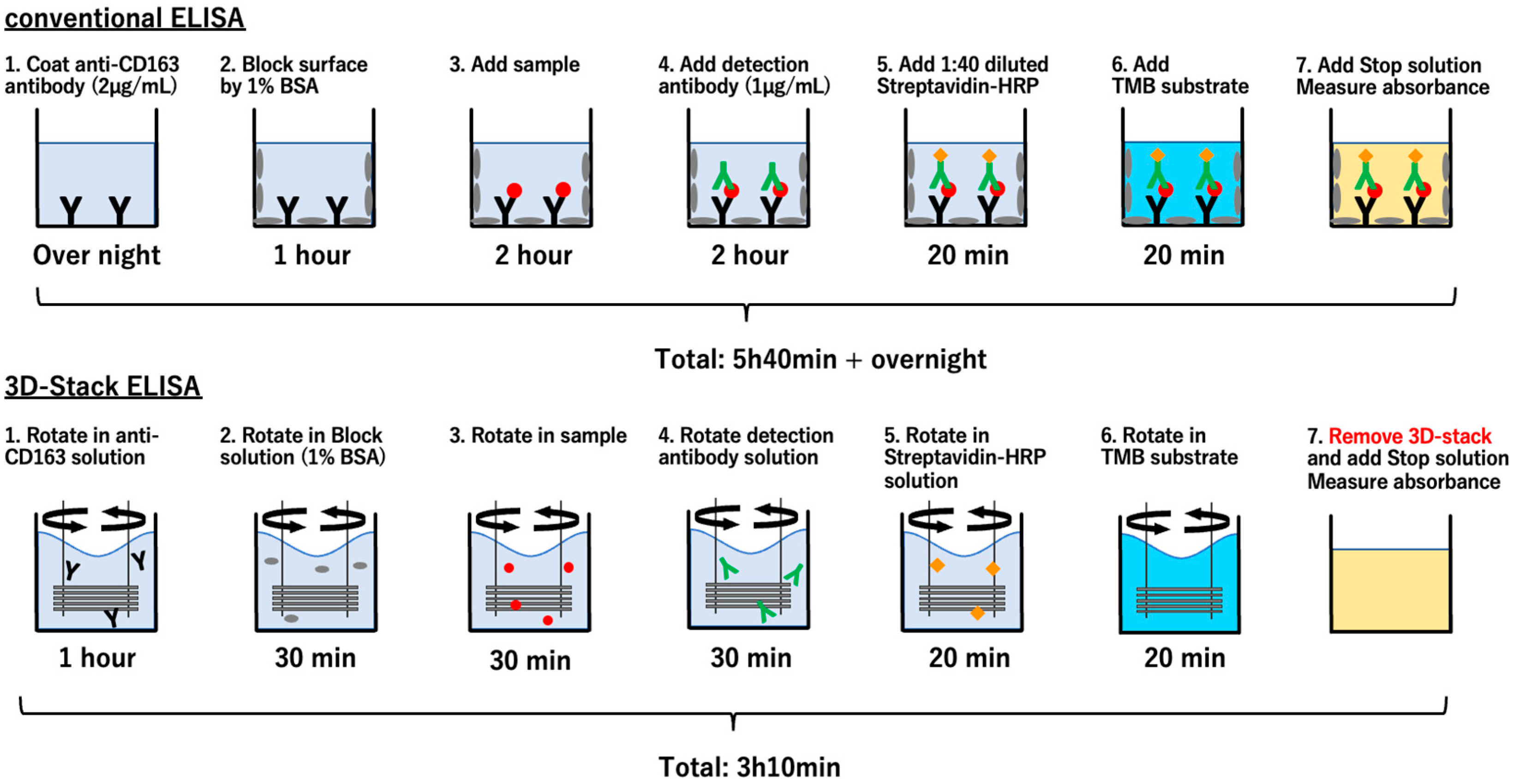
2.4. Sandwich ELISA Using 3D Stack and Conventional Method
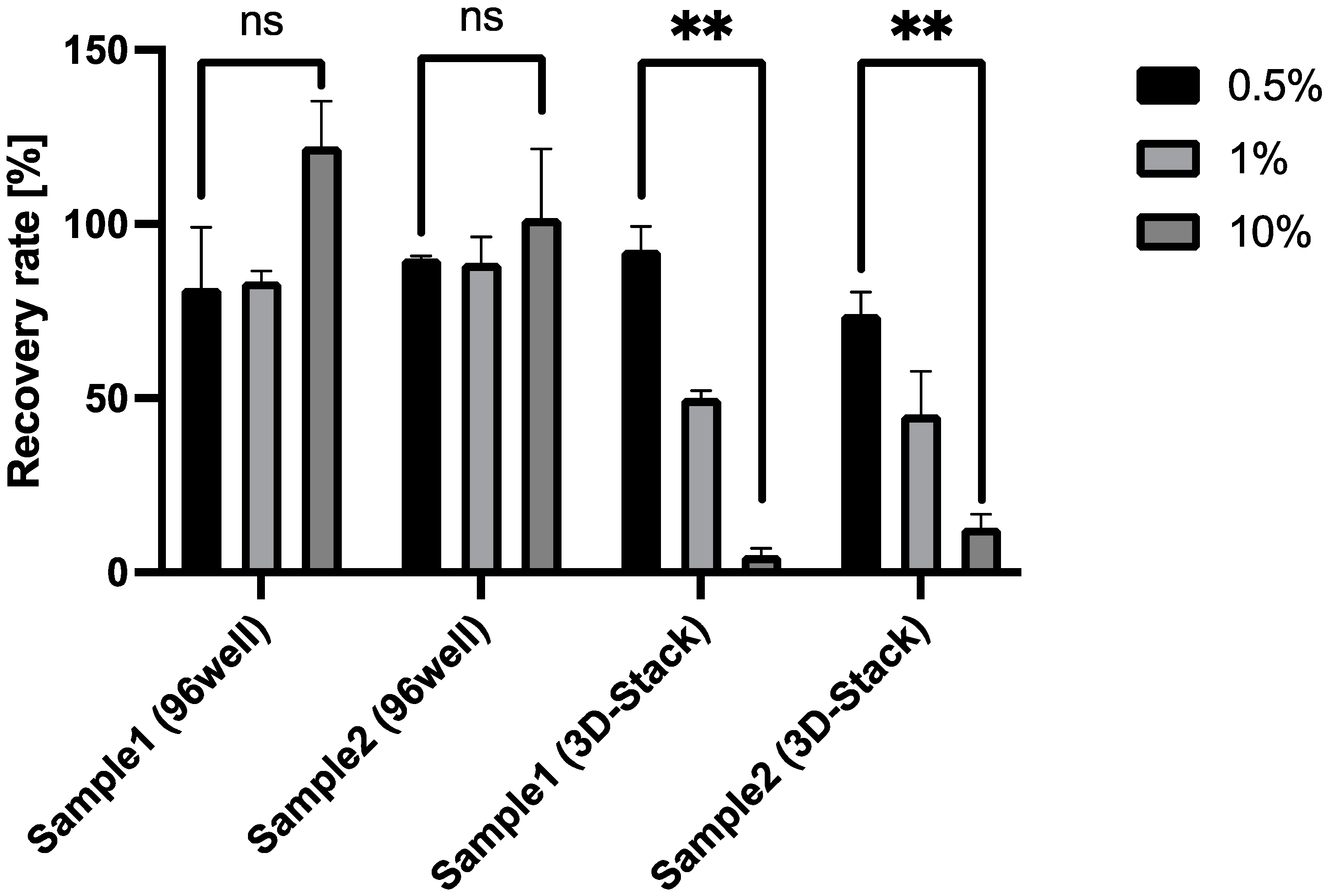
2.5. Spike-and-Recovery Assay
2.6. Detection of the Remaining Coated Antibodies on the Surface of 3D Stack
2.7. EDC–NHS Coupling of Antibody
2.8. Statistical Analysis
3. Results
3.1. Detection Sensitivity of 3D Stack from Standard Sample
3.2. Detection from Serum Samples
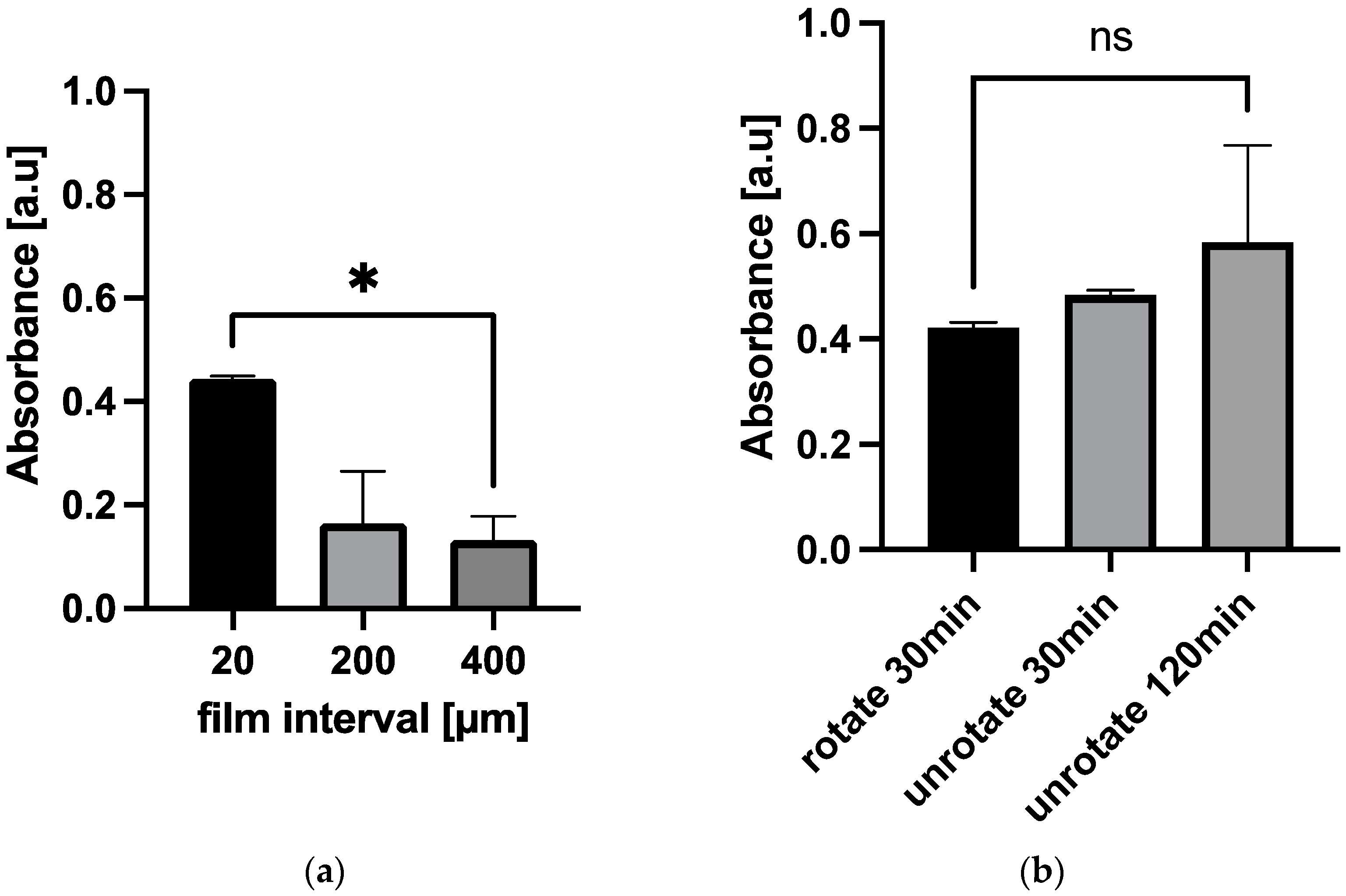
3.3. The Effect of 3D Stack Characteristics on the Detection of sCD163 from Serum
3.4. The Influence of Vroman Effect on the Detection of sCD163 from Serum
3.5. Antibody Immobilization by EDC–NHS Coupling
3.6. Detection of sCD163 from Dengue Patients’ Sera
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Engvall, E.; Perlmann, P. Enzyme-Linked immunosorbent assay (elisa) quantitative assay of immunoglobulin-G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Vanweeme, B.K.; Schuurs, A.H.W. Immunoassay using antigen-enzyme conjugates. FEBS Lett. 1971, 15, 232–236. [Google Scholar] [CrossRef]
- Lequin, R.M. Enzyme Immunoassay (EIA)/Enzyme-Linked Immunosorbent Assay (ELISA). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Vázquez-Villegas, P.; Rito-Palomares, M.; Martinez-Chapa, S.O. Advantages, disadvantages and modifications of conventional ELISA. In Enzyme-Linked Immunosorbent Assay (ELISA); Springer: Berlin/Heidelberg, Germany, 2018; pp. 67–115. [Google Scholar]
- Lilyanna, S.; Ng, E.M.W.; Moriguchi, S.; Chan, S.P.; Kokawa, R.; Huynh, S.H.; Chong, P.C.J.; Ng, Y.X.; Richards, A.M.; Ng, T.W.; et al. Variability in Microplate Surface Properties and Its Impact on ELISA. J. Appl. Lab. Med. 2018, 2, 687–699. [Google Scholar] [CrossRef]
- Kusnezow, W.; Syagailo, Y.V.; Goychuk, I.; Hoheisel, J.D.; Wild, D.G. Antibody microarrays: The crucial impact of mass transport on assay kinetics and sensitivity. Expert Rev. Mol. Diagn. 2006, 6, 111–124. [Google Scholar] [CrossRef]
- Squires, T.M.; Quake, S.R. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005, 77, 977. [Google Scholar] [CrossRef]
- Zheng, C.; Wang, J.; Pang, Y.; Wang, J.; Li, W.; Ge, Z.; Huang, Y. High-throughput immunoassay through in-channel microfluidic patterning. Lab Chip 2012, 12, 2487–2490. [Google Scholar] [CrossRef]
- Liao, T.; Yuan, F.; Yu, H.; Li, Z. An ultrasensitive ELISA method for the detection of procalcitonin based on magnetic beads and enzyme-antibody labeled gold nanoparticles. Anal. Methods 2016, 8, 1577–1585. [Google Scholar] [CrossRef]
- Lee, K.K.; Ahn, C.H. A new on-chip whole blood/plasma separator driven by asymmetric capillary forces. Lab Chip 2013, 13, 3261–3267. [Google Scholar] [CrossRef]
- Zhao, Y.; Zeng, D.; Yan, C.; Chen, W.; Ren, J.; Jiang, Y.; Jiang, L.; Xue, F.; Ji, D.; Tang, F. Rapid and accurate detection of Escherichia coli O157: H7 in beef using microfluidic wax-printed paper-based ELISA. Analyst 2020, 145, 3106–3115. [Google Scholar] [CrossRef] [PubMed]
- Kai, J.H.; Puntambekar, A.; Santiago, N.; Lee, S.H.; Sehy, D.W.; Moore, V.; Han, J.; Ahn, C.H. A novel microfluidic microplate as the next generation assay platform for enzyme linked immunoassays (ELISA). Lab Chip 2012, 12, 4257–4262. [Google Scholar] [CrossRef]
- Su, W.; Cook, B.S.; Fang, Y.; Tentzeris, M.M. Fully inkjet-printed microfluidics: A solution to low-cost rapid three-dimensional microfluidics fabrication with numerous electrical and sensing applications. Sci. Rep. 2016, 6, 35111. [Google Scholar] [CrossRef]
- Li, H.; Fan, Y.; Kodzius, R.; Foulds, I.G. Fabrication of polystyrene microfluidic devices using a pulsed CO2 laser system. Microsyst. Technol. 2012, 18, 373–379. [Google Scholar] [CrossRef]
- Singh, H.; Morita, T.; Suzuki, Y.; Shimojima, M.; Van, A.L.; Sugamata, M.; Yang, M. High sensitivity, high surface area Enzyme-linked Immunosorbent Assay (ELISA). Bio-Med. Mater. Eng. 2015, 26, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Morioka, K.; Ohata, S.; Shimizu, T.; Nakajima, H.; Uchiyama, K.; Yang, M. Rapid ELISA Using a Film-Stack Reaction Field with Micropillar Arrays. Sensors 2017, 17, 1608. [Google Scholar] [CrossRef]
- Suzuki, Y.; Morioka, K.; Shimizu, T.; Nakajima, H.; Uchiyama, K.; Yang, M. Influence of structural dimensions of micro-pillar array in reaction field on sensitivity of enzyme-linked immunosorbent assay (ELISA). Biotechnol. Biotechnol. Equip. 2018, 32, 520–529. [Google Scholar] [CrossRef]
- Tate, J.; Ward, G. Interferences in immunoassay. Clin. Biochem. Rev. 2004, 25, 105. [Google Scholar]
- Kragstrup, T.W.; Vorup-Jensen, T.; Deleuran, B.; Hvid, M. A simple set of validation steps identifies and removes false results in a sandwich enzyme-linked immunosorbent assay caused by anti-animal IgG antibodies in plasma from arthritis patients. Springerplus 2013, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, M.; Hou, X. Competitive Protein Adsorption Multilayer Adsorption and Surface Induced Protein Aggregation. Langmuir 2009, 25, 2081–2089. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, M.; Hou, X. Fibrinogen adsorption on blocked surface of albumin. Colloids Surf. B Biointerfaces 2011, 84, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, M.; Stibius, K.B.; Larsen, N.B.; Hou, X. Competitive protein adsorption to polymer surfaces from human serum. J. Mater. Sci. Mater. Med. 2008, 19, 2179–2185. [Google Scholar] [CrossRef] [PubMed]
- Vroman, L.; Adams, A.L. Findings with the recording ellipsometer suggesting rapid exchange of specific plasma proteins at liquid/solid interfaces. Surf. Sci. 1969, 16, 438–446. [Google Scholar] [CrossRef]
- Noh, H.; Vogler, E.A. Volumetric interpretation of protein adsorption: Competition from mixtures and the Vroman effect. Biomaterials 2007, 28, 405–422. [Google Scholar] [CrossRef] [PubMed]
- Bartali, R.; Lorenzelli, L.; Scarpa, M.; Morganti, E.; Collini, C.; Micheli, V.; Gottardi, G.; Gambetti, A.; Gambetti, G.; Coser, G.; et al. Super-Hydrophilic PDMS and PET Surfaces for Microfluidic Devices. Adv. Sci. Technol. 2013, 81, 96–100. [Google Scholar] [CrossRef]
- Ruben, B.; Elisa, M.; Leandro, L.; Victor, M.; Gloria, G.; Marina, S.; Pandiyan, R.; Nadhira, L. Oxygen plasma treatments of polydimethylsiloxane surfaces: Effect of the atomic oxygen on capillary flow in the microchannels. Micro Nano Lett. 2017, 12, 754–757. [Google Scholar] [CrossRef]
- Choi, S.; Chae, J. Methods of reducing non-specific adsorption in microfluidic biosensors. J. Micromech. Microeng. 2010, 20, 075015. [Google Scholar] [CrossRef]
- Masson, J.F.; Battaglia, T.M.; Cramer, J.; Beaudoin, S.; Sierks, M.; Booksh, K.S. Reduction of nonspecific protein binding on surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2006, 386, 1951–1959. [Google Scholar] [CrossRef]
- Norde, W. My voyage of discovery to proteins in flatland… and beyond. Colloids Surf. B Biointerfaces 2008, 1, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yan, H.; Ren, K.; Dai, W.; Wu, H. Convenient Method for Modifying Poly(dimethylsiloxane) with Poly(ethylene glycol) in Microfluidics. Anal. Chem. 2009, 81, 6627–6632. [Google Scholar] [CrossRef]
- Barbosa, M.A.; Martins, M.C.L.; Barbosa, J.N. 4-Cellular response to the surface chemistry of nanostructured biomaterials. In Cellular Response to Biomaterials; Di Silvio, L., Ed.; Woodhead Publishing: Sawston, UK, 2009; pp. 85–113. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.A.; Depelsenaire, A.C.; Young, P.R. Clinical and Laboratory Diagnosis of Dengue Virus Infection. J. Infect. Dis. 2017, 215, S89–S95. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Manzano, J.; Chia, P.Y.; Yeo, T.W.; Holmes, A.; Georgiou, P.; Yacoub, S. Improving Dengue Diagnostics and Management through Innovative Technology. Curr. Infect. Dis. Rep. 2018, 20, 25. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Jimena, B.; Bekaert, M.; Bakheit, M.; Frischmann, S.; Patel, P.; Simon-Loriere, E.; Lambrechts, L.; Duong, V.; Dussart, P.; Harold, G.; et al. Development and validation of four one-step real-time RT-LAMP assays for specific detection of each dengue virus serotype. PLoS Negl. Trop. Dis. 2018, 12, e0006381. [Google Scholar] [CrossRef] [PubMed]
- Ab-Rahman, H.A.; Rahim, H.; AbuBakar, S.; Wong, P.-F. Macrophage activation syndrome-associated markers in severe dengue. Int. J. Med. Sci. 2016, 13, 179. [Google Scholar] [CrossRef]
- Awaja, F.; Pavel, D. Recycling of PET. Eur. Polym. J. 2005, 41, 1453–1477. [Google Scholar] [CrossRef]
- Angioletti-Uberti, S.; Ballauff, M.; Dzubiella, J. Competitive adsorption of multiple proteins to nanoparticles: The Vroman effect revisited. Mol. Phys. 2018, 116, 3154–3163. [Google Scholar] [CrossRef]
- Choi, S.; Chae, J. A microfluidic biosensor based on competitive protein adsorption for thyroglobulin detection. Biosens. Bioelectron. 2009, 25, 118–123. [Google Scholar] [CrossRef]
- Reth, M. Matching cellular dimensions with molecular sizes. Nat. Immunol. 2013, 14, 765–767. [Google Scholar] [CrossRef]
- Norde, W.; Lyklema, J. Thermodynamics of protein adsorption. Theory with special reference to the adsorption of human plasma albumin and bovine pancreas ribonuclease at polystyrene surfaces. J. Colloid Interface Sci. 1979, 71, 350–366. [Google Scholar] [CrossRef]
- Schmidt, D.R.; Waldeck, H.; Kao, W.J. Protein adsorption to biomaterials. In Biological Interactions on Materials Surfaces; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–18. [Google Scholar]
- Sam, S.; Touahir, L.; Salvador Andresa, J.; Allongue, P.; Chazalviel, J.-N.; Gouget-Laemmel, A.; Henry de Villeneuve, C.; Moraillon, A.; Ozanam, F.; Gabouze, N. Semiquantitative study of the EDC/NHS activation of acid terminal groups at modified porous silicon surfaces. Langmuir 2010, 26, 809–814. [Google Scholar] [CrossRef]
- Tonigold, M.; Simon, J.; Estupiñán, D.; Kokkinopoulou, M.; Reinholz, J.; Kintzel, U.; Kaltbeitzel, A.; Renz, P.; Domogalla, M.P.; Steinbrink, K. Pre-adsorption of antibodies enables targeting of nanocarriers despite a biomolecular corona. Nat. Nanotechnol. 2018, 13, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Welch, N.G.; Scoble, J.A.; Muir, B.W.; Pigram, P.J. Orientation and characterization of immobilized antibodies for improved immunoassays. Biointerphases 2017, 12, 02D301. [Google Scholar] [CrossRef] [PubMed]
- Gervais, T.; Jensen, K.F. Mass transport and surface reactions in microfluidic systems. Chem. Eng. Sci. 2006, 61, 1102–1121. [Google Scholar] [CrossRef]


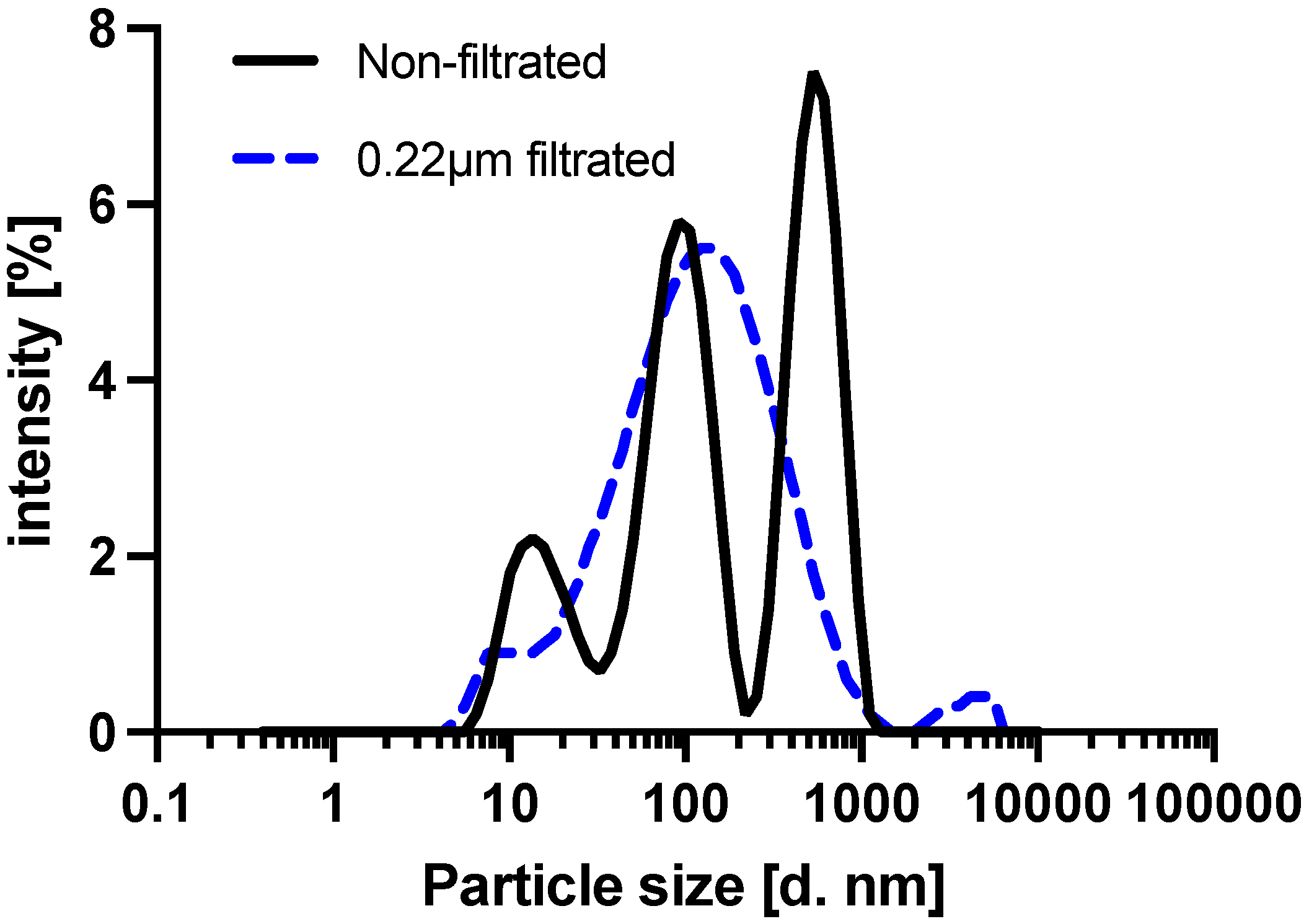
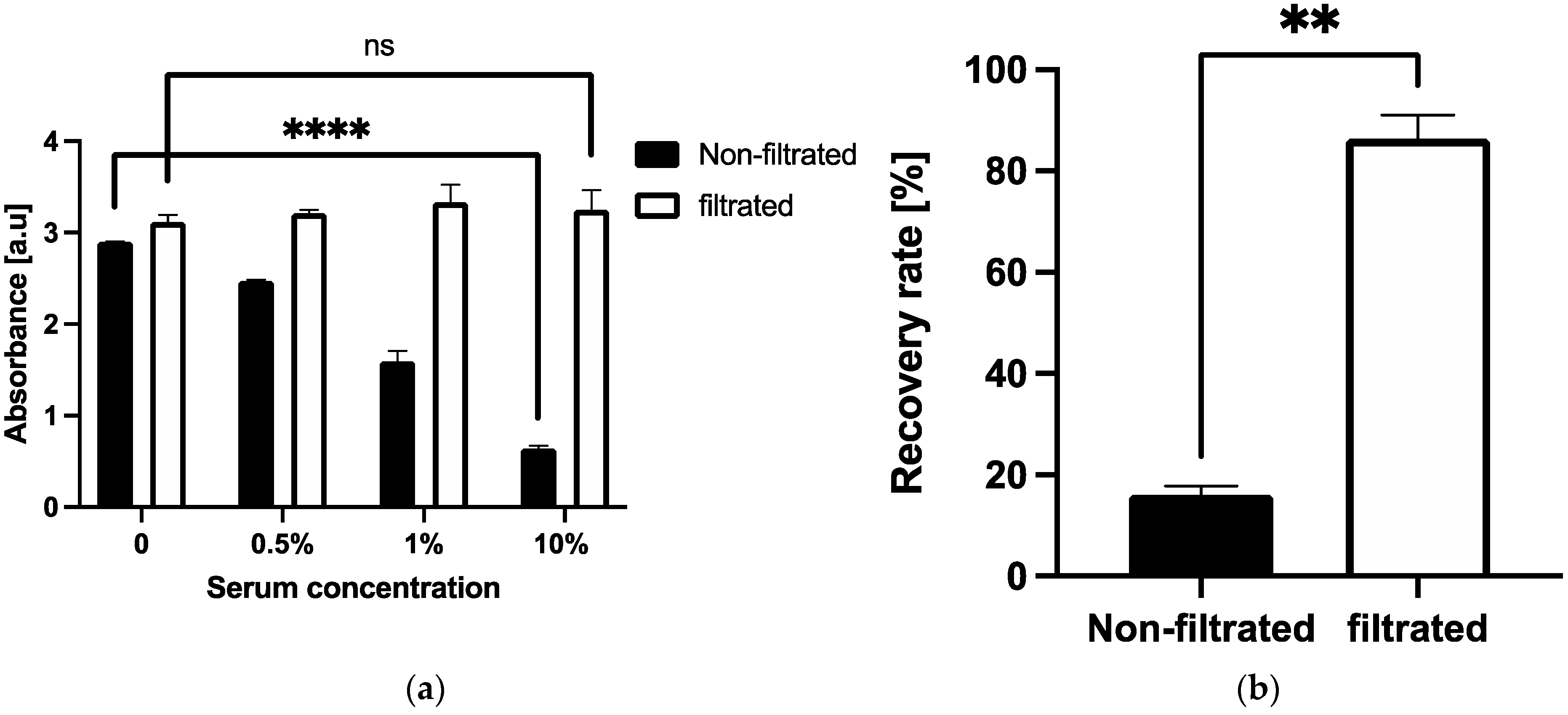

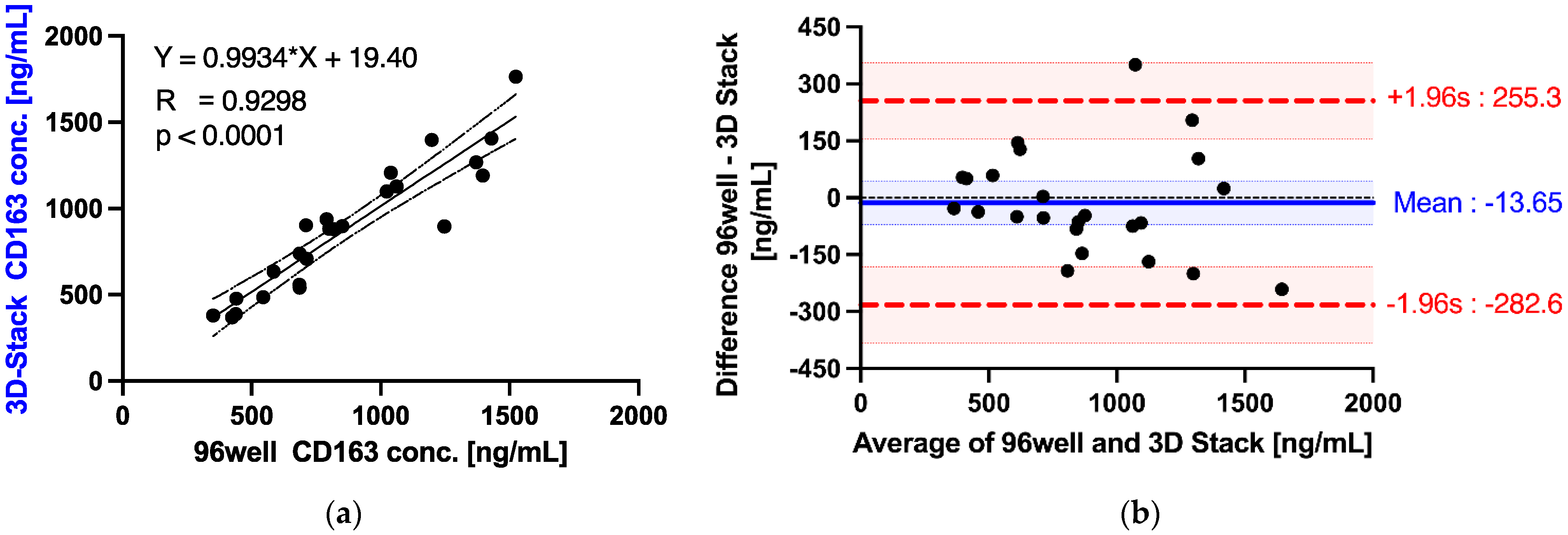
| Serum Dilution [%] | Recovery Rate (% ±SD) | |||
|---|---|---|---|---|
| Sample1 (96-Well) | Sample2 (96-Well) | Sample1 (3D Stack) | Sample2 (3D Stack) | |
| 0.5 | 81.7 ± 17.3 | 90.0 ± 0.81 | 92.6 ± 6.73 | 74.2 ± 6.24 |
| 1 | 83.6 ± 3.00 | 88.8 ± 7.47 | 50.0 ± 2.14 | 45.3 ± 12.5 |
| 10 | 122 ± 13.0 | 101 ± 19.9 | 5.00 ± 2.00 | 12.8 ± 3.79 |
| 96-Well Elisa | 3D Stack | Difference | |
|---|---|---|---|
| w | 0.9398 | 0.9536 | 0.9621 |
| p value | 0.1616 | 0.3244 | 0.483 |
| alpha | 0.05 | 0.05 | 0.05 |
| normality | yes | yes | yes |
| Parameter | Unit | Standard Error Formula | Standard Error (se) | t Value for 24 Degrees of Freedom | Confidence (se * t) | Confidence Intervals | |
|---|---|---|---|---|---|---|---|
| from | to | ||||||
| number (n) | 24 | ||||||
| degrees of freedom (n − 1) | 23 | ||||||
| ) | −13.65 | 28.01 | 2.06 | 57.69 | −71.34 | 44.05 | |
| standard deviation (s) | 137.2 | ||||||
| − 1.96s | −282.6 | 48.51 | 2.06 | 99.92 | −382.5 | −182.6 | |
| + 1.96s | 255.3 | 48.51 | 2.06 | 99.92 | 155.3 | 355.2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maeno, H.; Wong, P.-F.; AbuBakar, S.; Yang, M.; Sam, S.-S.; Jamil-Abd, J.; Shunmugarajoo, A.; Mustafa, M.; Said, R.M.; Mageswaren, E.; et al. A 3D Microfluidic ELISA for the Detection of Severe Dengue: Sensitivity Improvement and Vroman Effect Amelioration by EDC–NHS Surface Modification. Micromachines 2021, 12, 1503. https://doi.org/10.3390/mi12121503
Maeno H, Wong P-F, AbuBakar S, Yang M, Sam S-S, Jamil-Abd J, Shunmugarajoo A, Mustafa M, Said RM, Mageswaren E, et al. A 3D Microfluidic ELISA for the Detection of Severe Dengue: Sensitivity Improvement and Vroman Effect Amelioration by EDC–NHS Surface Modification. Micromachines. 2021; 12(12):1503. https://doi.org/10.3390/mi12121503
Chicago/Turabian StyleMaeno, Hinata, Pooi-Fong Wong, Sazaly AbuBakar, Ming Yang, Sing-Sin Sam, Juraina Jamil-Abd, Anusha Shunmugarajoo, Mahiran Mustafa, Rosaida Md Said, Eashwary Mageswaren, and et al. 2021. "A 3D Microfluidic ELISA for the Detection of Severe Dengue: Sensitivity Improvement and Vroman Effect Amelioration by EDC–NHS Surface Modification" Micromachines 12, no. 12: 1503. https://doi.org/10.3390/mi12121503
APA StyleMaeno, H., Wong, P.-F., AbuBakar, S., Yang, M., Sam, S.-S., Jamil-Abd, J., Shunmugarajoo, A., Mustafa, M., Said, R. M., Mageswaren, E., Azmel, A., & Mat Jelani, A. (2021). A 3D Microfluidic ELISA for the Detection of Severe Dengue: Sensitivity Improvement and Vroman Effect Amelioration by EDC–NHS Surface Modification. Micromachines, 12(12), 1503. https://doi.org/10.3390/mi12121503






