Collective Polarization of Cancer Cells at the Monolayer Boundary
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Generation and Sorting
2.2. Cell Culture
2.3. In Vitro Wound Healing Assay
2.4. Chemicals and Reagents
2.5. Golgi Apparatus Staining
2.6. Immunofluorescence Staining
2.7. Time-Lapse Imaging and Image Analysis
2.8. Time-Lapse Cell Motility Assay and Cell Trajectory Analysis
2.9. Alignment Characterization of F-Actin Filaments
3. Results
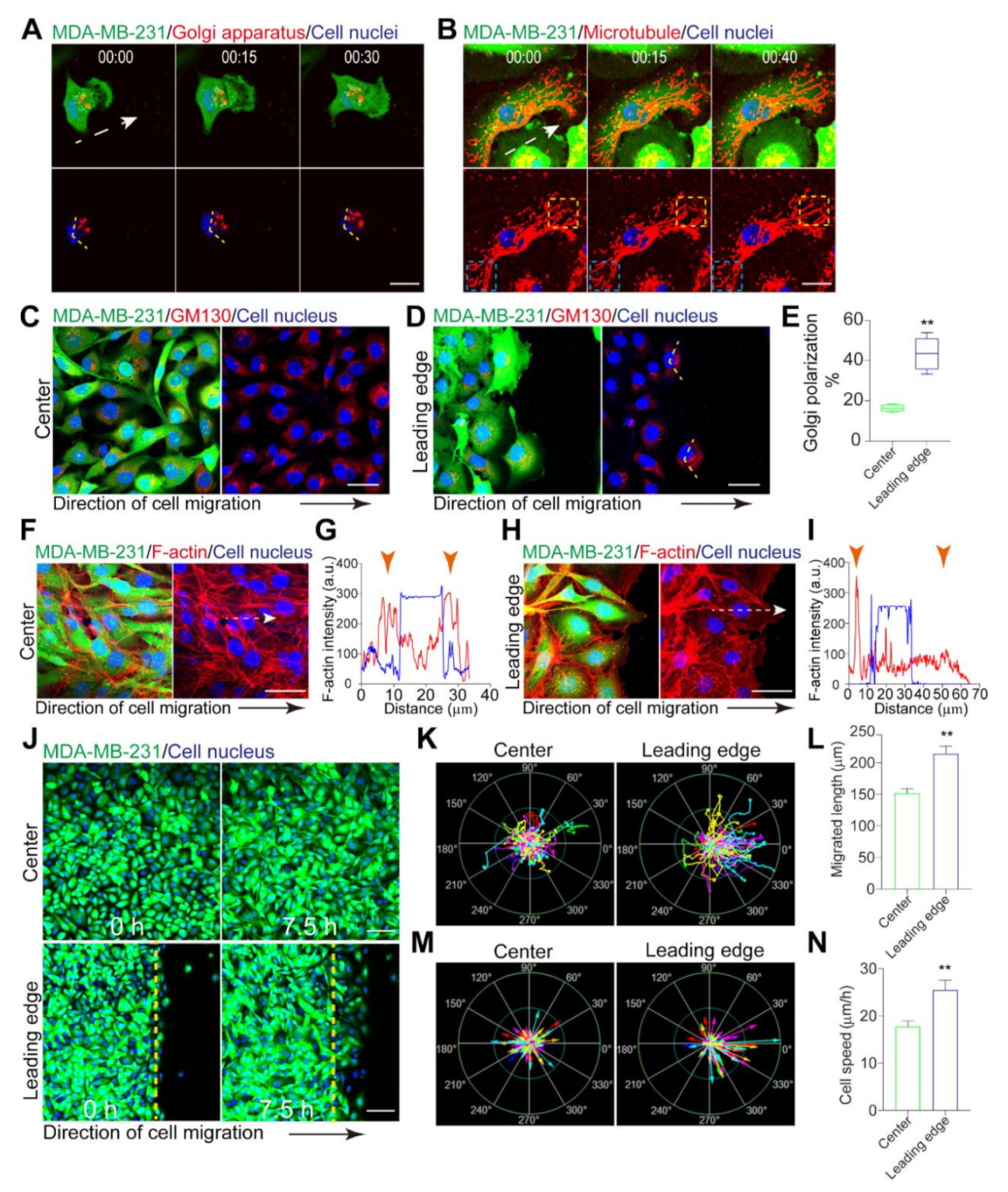
3.1. Leader Cells Exhibit a Stronger Polarization than Follower Cells
3.2. Polarity Transmits by Principal Membrane Protrusions
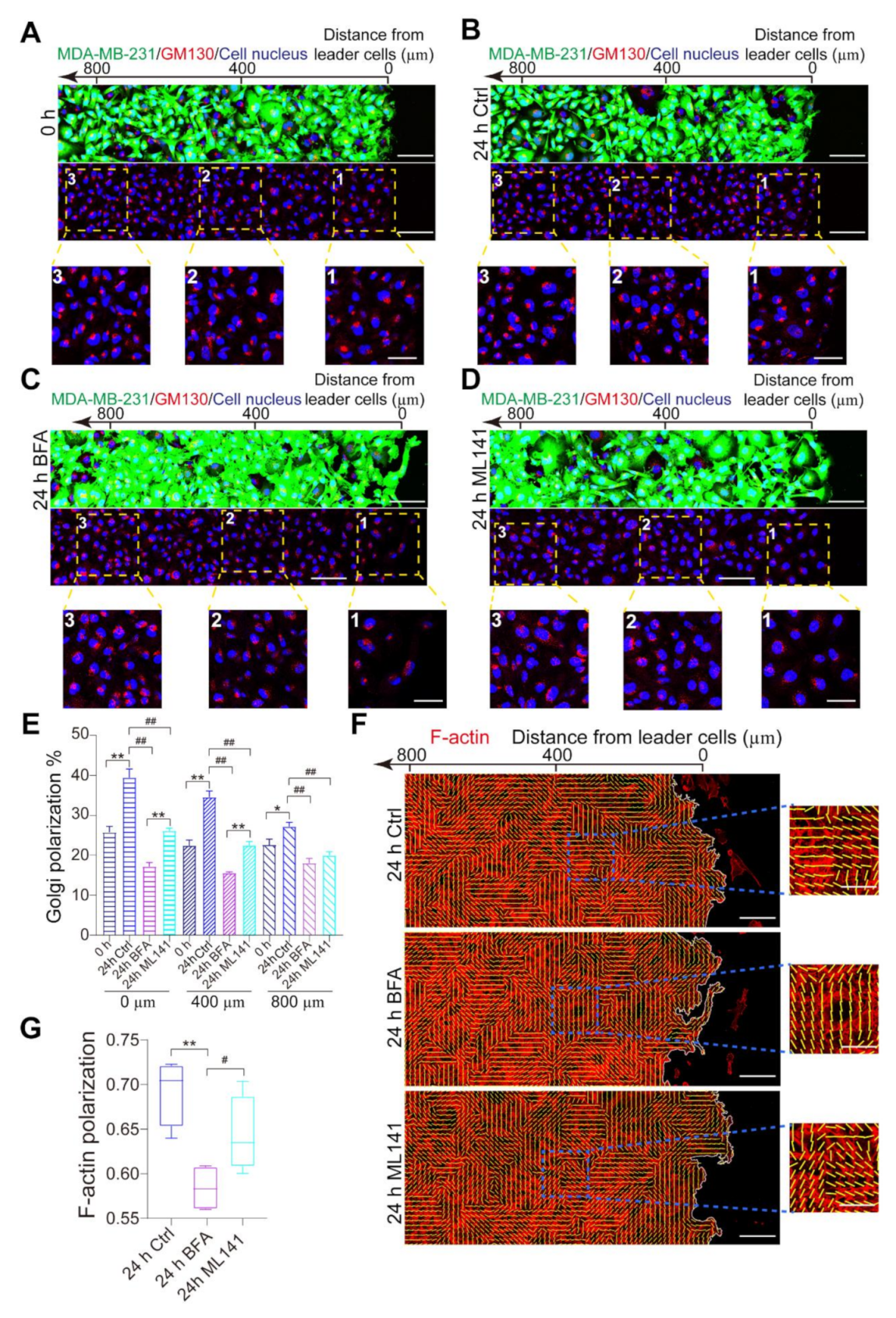
3.3. Collective Cell Polarization is Destructed by BFA and ML141
3.4. Polarized Golgi Apparatus Regulates Collective Polarization of MDA-MB-231 Cells
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Friedl, P.; Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009, 10, 445–457. [Google Scholar]
- Mayor, R.; Etienne-Manneville, S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 2016, 17, 97–109. [Google Scholar] [PubMed] [Green Version]
- Zhang, J.; Goliwas, K.F.; Wang, W.; Taufalele, P.V.; Bordeleau, F.; Reinhart-King, C.A. Energetic regulation of coordinated leader–follower dynamics during collective invasion of breast cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7867–7872. [Google Scholar]
- Mishra, A.K.; Campanale, J.P.; Mondo, J.A.; Montell, D.J. Cell interactions in collective cell migration. Development 2019, 146, dev172056. [Google Scholar]
- Lin, S.Z.; Chen, P.C.; Guan, L.Y.; Shao, Y.; Hao, Y.K.; Li, Q.; Li, B.; Weitz, D.A.; Feng, X.Q. Universal statistical laws for the velocities of collective migrating cells. Adv. Biosyst. 2020, 4, 2000065. [Google Scholar]
- Li, X.Z.; Das, A.; Bi, D. Mechanical heterogeneity in tissues promotes rigidity and controls cellular invasion. Phys. Rev. Lett. 2019, 123, 058101. [Google Scholar] [PubMed] [Green Version]
- Han, Y.L.; Pegoraro, A.F.; Li, H.; Li, K.F.; Yuan, Y.; Xu, G.Q.; Gu, Z.C.; Sun, J.W.; Hao, Y.K.; Gupta, S.K.; et al. Cell swelling, softening and invasion in a three-dimensional breast cancer model. Nat. Phys. 2020, 16, 101–108. [Google Scholar]
- Ladoux, B.; Mège, R.-M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 2017, 18, 743–757. [Google Scholar]
- Trepat, X.; Sahai, E. Mesoscale physical principles of collective cell organization. Nat. Phys. 2018, 14, 671–682. [Google Scholar]
- Li, B.; Sun, S.X. Coherent motions in confluent cell monolayer sheets. Biophys. J. 2014, 107, 1532–1541. [Google Scholar]
- De Pascalis, C.; Etienne-Manneville, S. Single and collective cell migration: The mechanics of adhesions. Mol. Biol. Cell 2017, 28, 1833–1846. [Google Scholar] [CrossRef] [PubMed]
- Shellard, A.; Mayor, R. Supracellular migration—beyond collective cell migration. J. Cell Sci. 2019, 132, jcs226142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amano, S.; Kaino, S.; Shinoda, S.; Harima, H.; Matsumoto, T.; Fujisawa, K.; Takami, T.; Yamamoto, N.; Yamasaki, T.; Sakaida, I. Invasion inhibition in pancreatic cancer using the oral iron chelating agent deferasirox. BMC Cancer 2020, 20, 681. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Zhang, D.Q.; Lin, S.Z.; Li, B. Pattern formation and defect ordering in active chiral nematics. Phys. Rev. Lett. 2020, 125, 098002. [Google Scholar] [CrossRef]
- Friedl, P.; Mayor, R. Tuning collective cell migration by cell–cell junction regulation. Cold Spring Harb. Perspect. Biol. 2017, 9, a029199. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.Z.; Ye, S.; Xu, G.K.; Li, B.; Feng, X.Q. Dynamic migration modes of collective cells. Biophys. J. 2018, 115, 1826–1835. [Google Scholar] [CrossRef] [Green Version]
- Capuana, L.; Boström, A.; Etienne-Manneville, S. Multicellular scale front-to-rear polarity in collective migration. Curr. Opin. Cell Biol. 2020, 62, 114–122. [Google Scholar] [CrossRef]
- He, S.J.; Li, X.M.; Ji, B.H. Mechanical force drives the polarization and orientation of cells. Acta Mech. Sin. 2019, 35, 275–288. [Google Scholar] [CrossRef]
- Trepat, X.; Wasserman, M.R.; Angelini, T.E.; Millet, E.; Weitz, D.A.; Butler, J.P.; Fredberg, J.J. Physical forces during collective cell migration. Nat. Phys. 2009, 5, 426–430. [Google Scholar] [CrossRef] [Green Version]
- Ishiyama, N.; Sarpal, R.; Wood, M.N.; Barrick, S.K.; Nishikawa, T.; Hayashi, H.; Kobb, A.B.; Flozak, A.S.; Yemelyanov, A.; Fernandez-Gonzalez, R.; et al. Force-dependent allostery of the α-catenin actin-binding domain controls adherens junction dynamics and functions. Nat. Commun. 2018, 9, 5121. [Google Scholar] [CrossRef]
- Harris, T.J.C.; Tepass, U. Adherens junctions: From molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 2010, 11, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Rusu, A.D.; Georgiou, M. The multifarious regulation of the apical junctional complex. Open Biol. 2020, 10, 190278. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Pinto, C.S.; Sonawane, M. Polarized organization of the cytoskeleton: Regulation by cell polarity proteins. J. Mol. Biol. 2018, 430, 3565–3584. [Google Scholar] [CrossRef] [PubMed]
- Pichaud, F.; Walther, R.F.; Nunes de Almeida, F. Regulation of Cdc42 and its effectors in epithelial morphogenesis. J. Cell Sci. 2019, 132, jcs217869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Y.; Li, Y.H.; Zhang, L.Y.; Xu, G.K. Stochastic fluctuation-induced cell polarization on elastic substrates: A cytoskeleton-based mechanical model. J. Mech. Phys. Solids 2020, 137, 103872. [Google Scholar] [CrossRef]
- Butler, M.T.; Wallingford, J.B. Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 375–388. [Google Scholar] [CrossRef]
- Magi, S.; Tashiro, E.; Imoto, M. A chemical genomic study identifying diversity in cell migration signaling in cancer cells. Sci. Rep. 2012, 2, 823. [Google Scholar] [CrossRef] [Green Version]
- Haga, R.B.; Ridley, A.J. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016, 7, 207–221. [Google Scholar] [CrossRef] [Green Version]
- Bui, J.; Conway, D.E.; Heise, R.L.; Weinberg, S.H. Mechanochemical coupling and junctional forces during collective cell migration. Biophys. J. 2019, 117, 170–183. [Google Scholar] [CrossRef]
- Lawson, C.D.; Ridley, A.J. Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 2017, 217, 447–457. [Google Scholar] [CrossRef]
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal crosstalk in cell migration. Trends Cell Biol. 2020, 30, 720–735. [Google Scholar] [CrossRef] [PubMed]
- Vaidžiulytė, K.; Coppey, M.; Schauer, K. Intracellular organization in cell polarity—placing organelles into the polarity loop. J. Cell Sci. 2019, 132, jcs230995. [Google Scholar] [CrossRef] [PubMed]
- Shafaq-Zadah, M.; Dransart, E.; Johannes, L. Clathrin-independent endocytosis, retrograde trafficking, and cell polarity. Curr. Opin. Cell Biol. 2020, 65, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Fourriere, L.; Jimenez, A.J.; Perez, F.; Boncompain, G. The role of microtubules in secretory protein transport. J. Cell Sci. 2020, 133, jcs237016. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Locker, J.; Sahai, E.; Segall, J.E. Classifying collective cancer cell invasion. Nat. Cell Biol. 2012, 14, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; He, L.; Liu, S.; Xie, J.; Huang, Z.; Jing, J.; Lee, Y.-T.; Wang, R.; Luo, H.; Han, W.; et al. Optogenetic engineering to probe the molecular choreography of STIM1-mediated cell signaling. Nat. Commun. 2020, 11, 1039. [Google Scholar] [CrossRef]
- Saw, T.B.; Doostmohammadi, A.; Nier, V.; Kocgozlu, L.; Thampi, S.; Toyama, Y.; Marcq, P.; Lim, C.T.; Yeomans, J.M.; Ladoux, B. Topological defects in epithelia govern cell death and extrusion. Nature 2017, 544, 212–216. [Google Scholar] [CrossRef]
- Nishiguchi, D.; Nagai, K.H.; Chaté, H.; Sano, M. Long-range nematic order and anomalous fluctuations in suspensions of swimming filamentous bacteria. Phys. Rev. E 2017, 95, 020601. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, S.; Augoff, K.; Rivera, L.; McCue, B.; Khoury, T.; Groman, A.; Zhang, L.; Tian, L.; Sossey-Alaoui, K. Increased expression levels of WAVE3 are associated with the progression and metastasis of triple negative breast cancer. PLoS ONE 2012, 7, e42895. [Google Scholar] [CrossRef]
- Huang, Z.; Yu, P.; Tang, J. Characterization of triple-negative breast cancer MDA-MB-231 cell spheroid model. OncoTargets Ther. 2020, 13, 5395–5405. [Google Scholar] [CrossRef]
- Ridley, A.J. Life at the leading edge. Cell 2011, 145, 1012–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauvin, T.J.; Young, L.E.; Higgs, H.N. The formin FMNL3 assembles plasma membrane protrusions that participate in cell–cell adhesion. Mol. Biol. Cell 2015, 26, 467–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.L.; Fucini, R.V.; Lacomis, L.; Erdjument-Bromage, H.; Tempst, P.; Stamnes, M. Coatomer-bound Cdc42 regulates dynein recruitment to COPI vesicles. J. Cell Biol. 2005, 169, 383–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, H.; Yu, J.; Zhang, L.; Carpenter, A.; Zhu, H.; Li, L.; Ma, D.; Yuan, J. A novel small molecule regulator of guanine nucleotide exchange activity of the ADP-ribosylation factor and golgi membrane trafficking. J. Biol. Chem. 2008, 283, 31087–31096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisel, B.; Calamai, M.; Vanzi, F.; Pavone, F.S. Decoupling polarization of the Golgi apparatus and GM1 in the plasma membrane. PLoS ONE 2013, 8, e80446. [Google Scholar] [CrossRef] [Green Version]
- Zeeh, J.C.; Zeghouf, M.; Grauffel, C.; Guibert, B.; Martin, E.; Dejaegere, A.; Cherfils, J. Dual specificity of the interfacial inhibitor brefeldin A for Arf proteins and Sec7 domains. J. Biol. Chem. 2006, 281, 11805–11814. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Wang, Z.; Wang, K.; Zhang, T.; Ding, J. Structural basis for targeting BIG1 to Golgi apparatus through interaction of its DCB domain with Arl1. J. Mol. Cell Biol. 2016, 8, 459–461. [Google Scholar] [CrossRef]
- Hong, L.; Kenney, S.R.; Phillips, G.K.; Simpson, D.; Schroeder, C.E.; Nöth, J.; Romero, E.; Swanson, S.; Waller, A.; Strouse, J.J.; et al. Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J. Biol. Chem. 2013, 288, 8531–8543. [Google Scholar] [CrossRef] [Green Version]
- Xing, M.K.; Peterman, M.C.; Davis, R.L.; Oegema, K.; Shiau, A.K.; Field, S.J. GOLPH3 drives cell migration by promoting Golgi reorientation and directional trafficking to the leading edge. Mol. Biol. Cell 2016, 27, 3828–3840. [Google Scholar] [CrossRef]
- Perdomo, D.; Aït-Ammar, N.; Syan, S.; Sachse, M.; Jhingan, G.D.; Guillén, N. Cellular and proteomics analysis of the endomembrane system from the unicellular Entamoeba histolytica. J. Proteom. 2015, 112, 125–140. [Google Scholar] [CrossRef] [Green Version]
- Jackson, C.L. Mechanisms of transport through the Golgi complex. J. Cell Sci. 2009, 122, 443–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stalder, D.; Gershlick, D.C. Direct trafficking pathways from the Golgi apparatus to the plasma membrane. Semin. Cell Dev. Biol. 2020, 107, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Pakdel, M.; von Blume, J. Exploring new routes for secretory protein export from the trans-Golgi network. Mol. Biol. Cell 2018, 29, 235–240. [Google Scholar] [CrossRef]
- Janiszewska, M.; Primi, M.C.; Izard, T. Cell adhesion in cancer: Beyond the migration of single cells. J. Biol. Chem. 2020, 295, 2495–2505. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Kang, Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 2019, 49, 361–374. [Google Scholar] [CrossRef]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Solis, G.P.; Bilousov, O.; Koval, A.; Lüchtenborg, A.-M.; Lin, C.; Katanaev, V.L. Golgi-resident Gαo promotes protrusive membrane dynamics. Cell 2017, 170, 939–955. [Google Scholar] [CrossRef] [Green Version]
- Sutkeviciute, I.; Vilardaga, J.-P. Structural insights into emergent signaling modes of G protein–coupled receptors. J. Biol. Chem. 2020, 295, 11626–11642. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J.; Yuan, L.C.; Bonifacino, J.S.; Klausner, R.D. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: Evidence for membrane cycling from Golgi to ER. Cell 1989, 56, 801–813. [Google Scholar] [CrossRef]
- Donaldson, J.G.; Finazzi, D.; Klausner, R.D. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature 1992, 360, 350–352. [Google Scholar] [CrossRef]
- Donaldson, J.G.; Honda, A.; Weigert, R. Multiple activities for Arf1 at the Golgi complex. Biochim. Biophys. Acta 2005, 1744, 364–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.; Buel, G.R.; Nagiec, M.J.; Han, M.J.; Roux, P.P.; Blenis, J.; Yoon, S.O. ERK2 regulates epithelial-to-mesenchymal plasticity through DOCK10-dependent Rac1/FoxO1 activation. Proc. Natl. Acad. Sci. USA 2019, 116, 2967–2976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Wedlich-Soldner, R.; Altschuler, S.; Wu, L.; Li, R. Spontaneous Cell Polarization Through Actomyosin-Based Delivery of the Cdc42 GTPase. Science 2003, 299, 1231–1235. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, L.-Y.; Lv, J.-Q.; Zhang, D.-Q.; Li, B. Collective Polarization of Cancer Cells at the Monolayer Boundary. Micromachines 2021, 12, 112. https://doi.org/10.3390/mi12020112
Guan L-Y, Lv J-Q, Zhang D-Q, Li B. Collective Polarization of Cancer Cells at the Monolayer Boundary. Micromachines. 2021; 12(2):112. https://doi.org/10.3390/mi12020112
Chicago/Turabian StyleGuan, Liu-Yuan, Jian-Qing Lv, De-Qing Zhang, and Bo Li. 2021. "Collective Polarization of Cancer Cells at the Monolayer Boundary" Micromachines 12, no. 2: 112. https://doi.org/10.3390/mi12020112




