Inertial Microfluidics Enabling Clinical Research
Abstract
:1. Introduction
2. Endogenous Targets
2.1. Circulating Tumor Cells
- Introduction
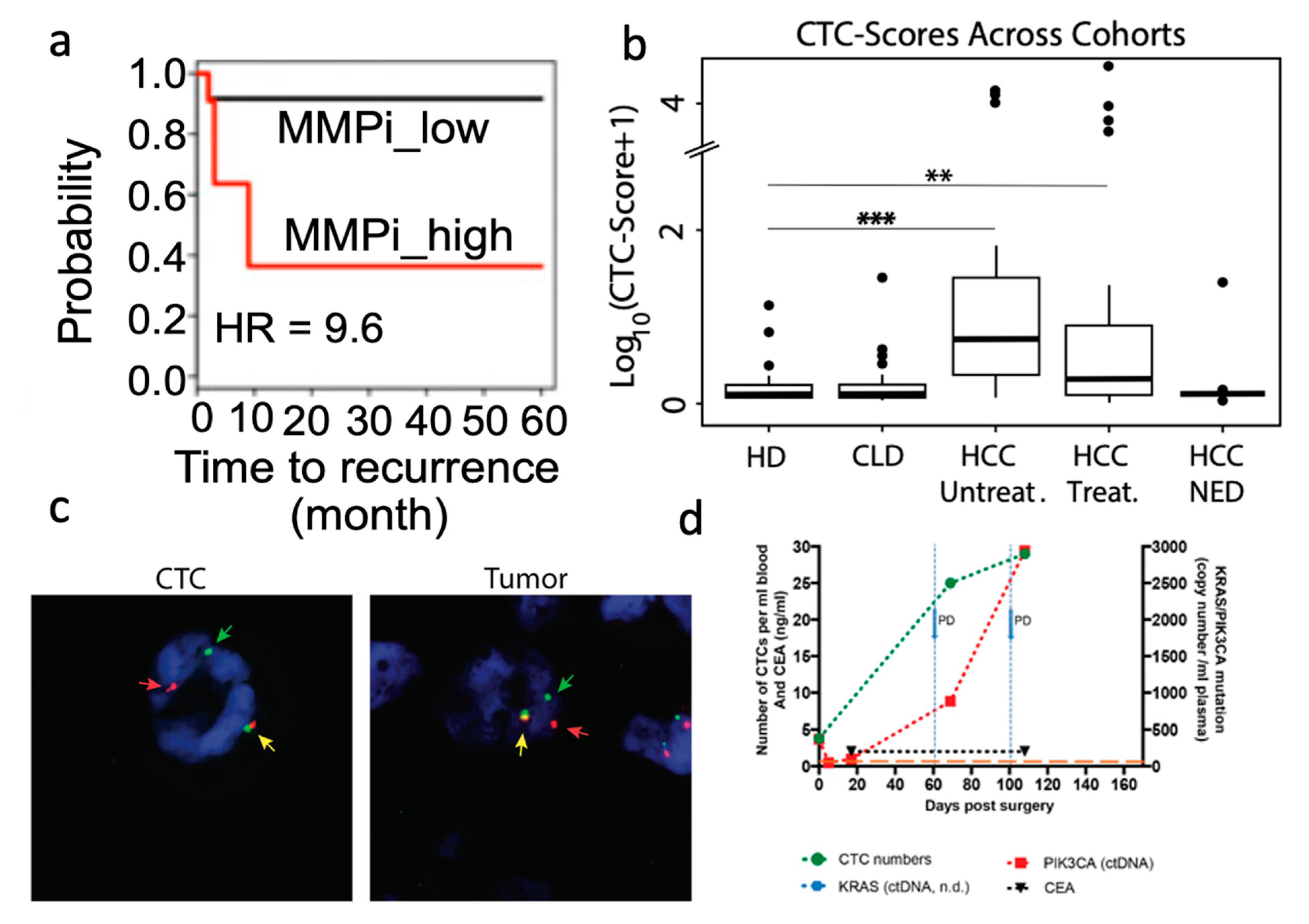
- Developing Predictors of Patient Outcome
- Guiding Therapeutic Selection and Monitoring Patient Response
- CTC Biomarker Exploration
- Investigations into CTC Biology
- Summary and Outlook
2.2. White Blood Cells
- Introduction
- Respiratory Illness
- Diabetes
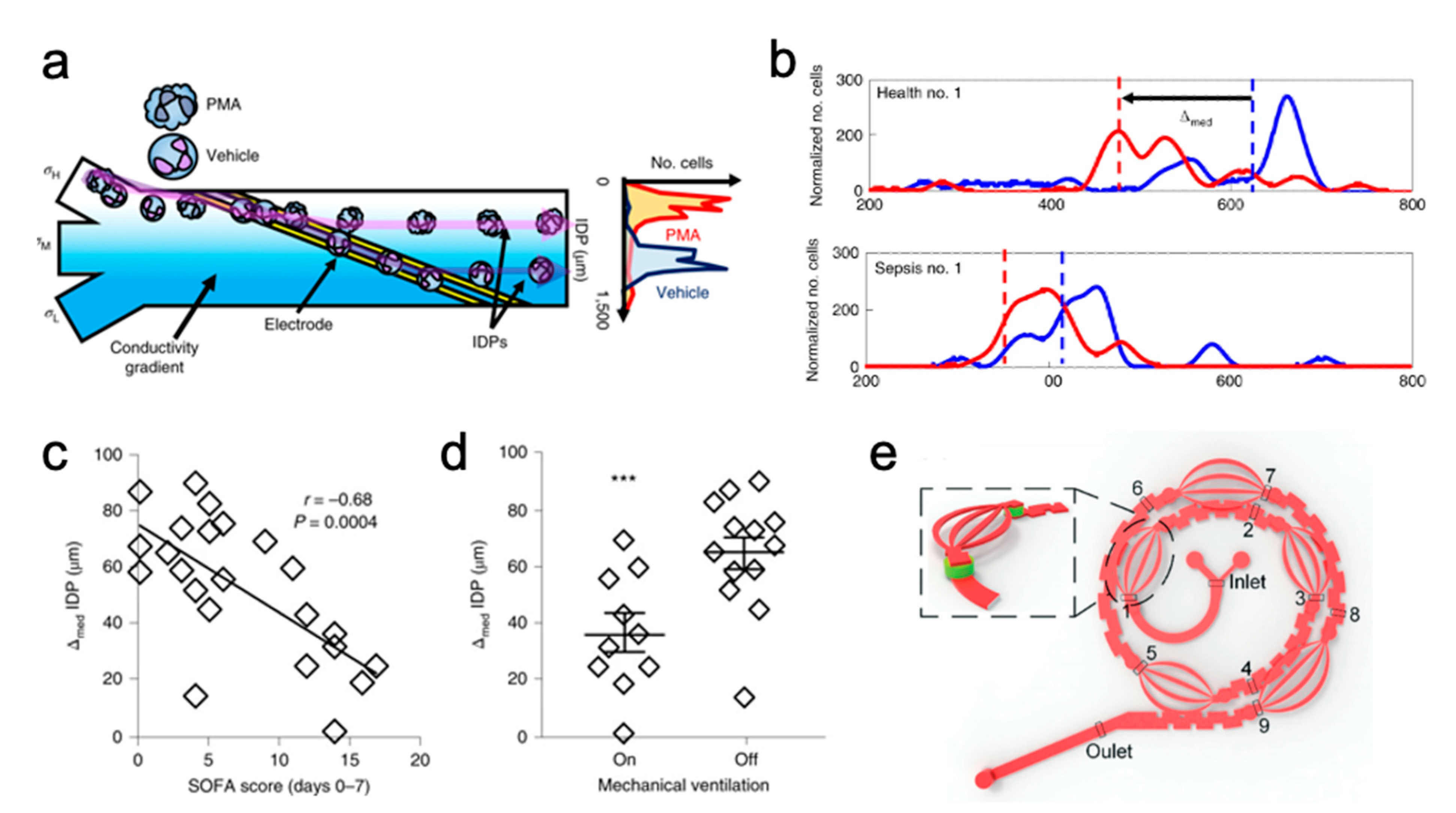
- Sepsis
- WBC Isolation with Hemolysis
- Summary and Outlook
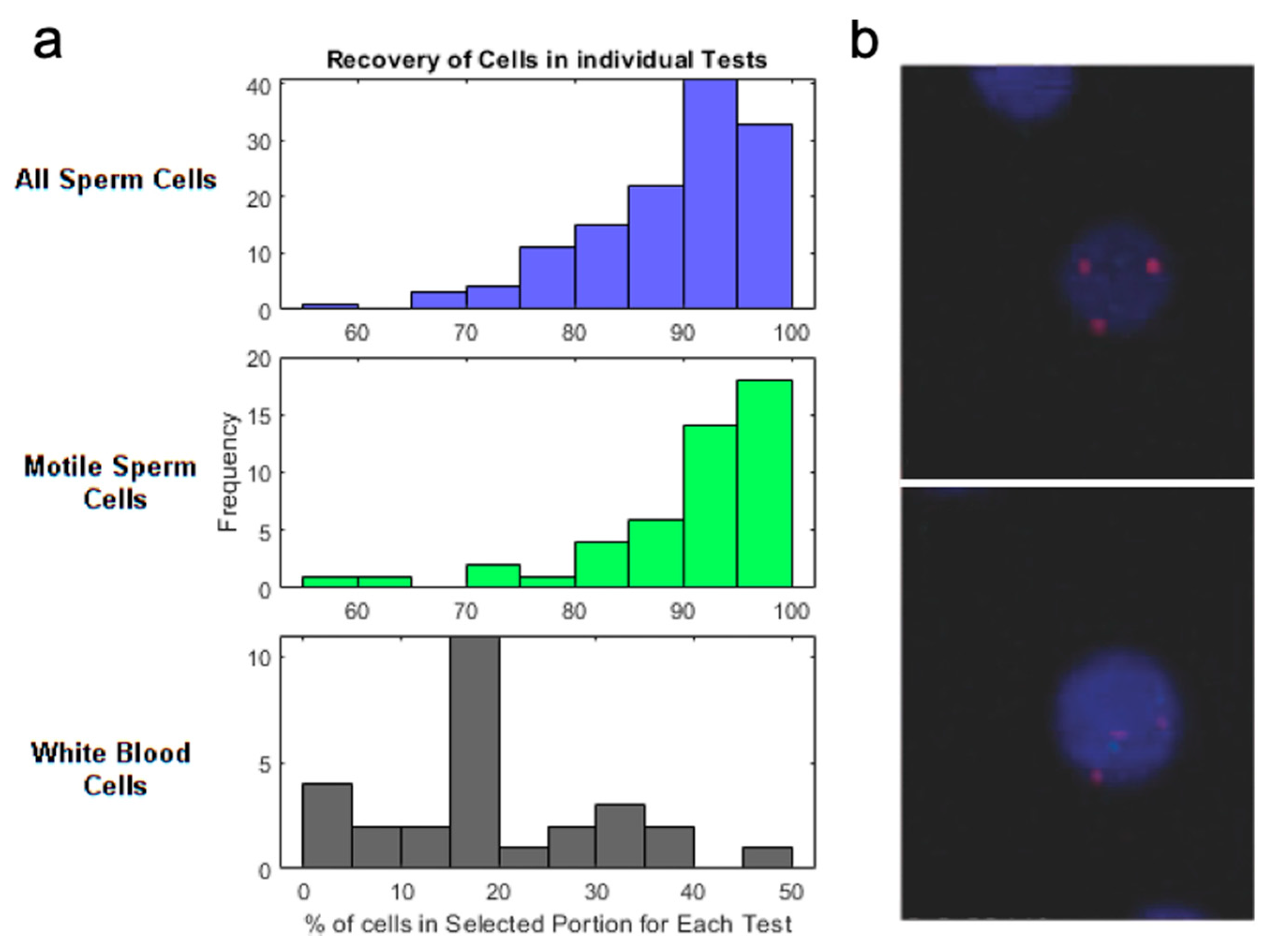
2.3. Reproductive Health Related Targets
- Introduction
- Purification of Reproductive Health Related Cells
- Summary and Outlook
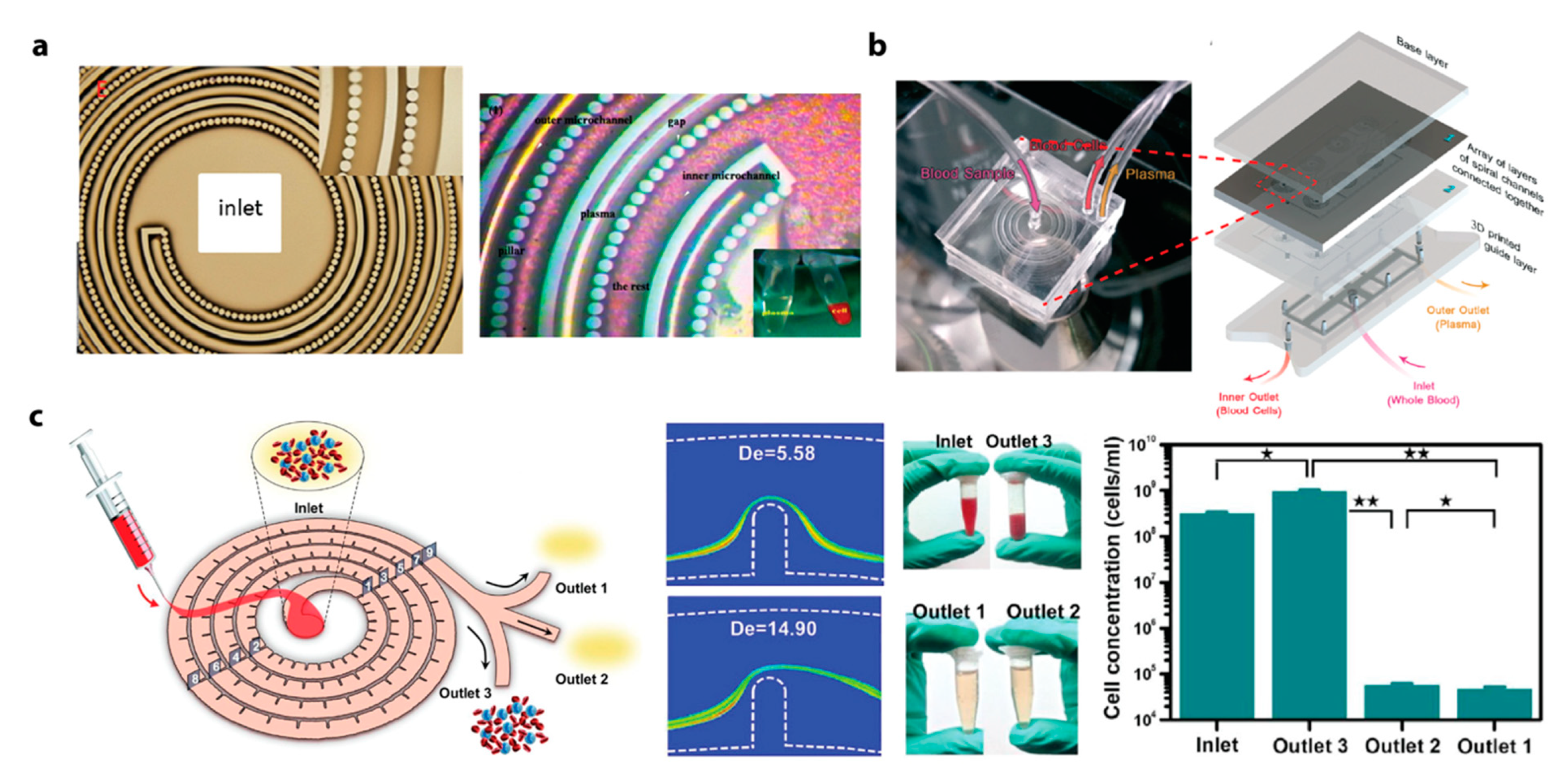
2.4. Plasma
- Introduction
- Recent Advances in Inertial Microfluidic Plasma Extraction
- Key Design Consideration for Inertial Microfluidic Plasma Extraction
- Summary and Outlook
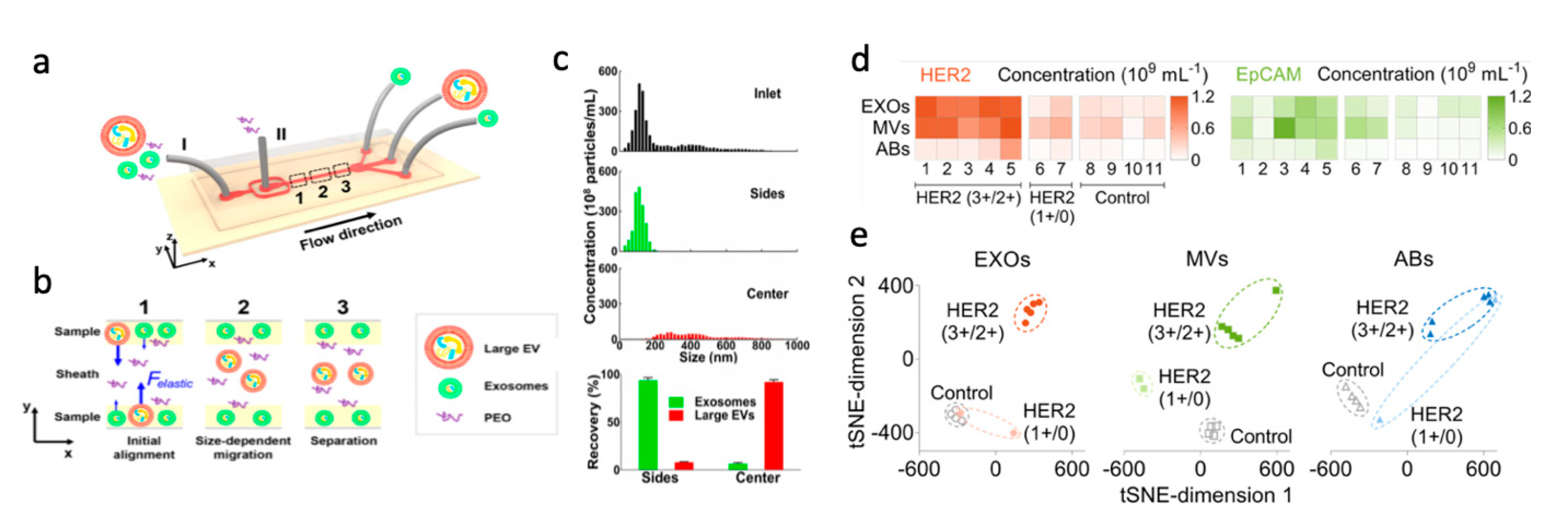
2.5. Extracellular Vesicles
- Introduction
- Purification of Extracellular Vesicles
- Summary and Outlook
3. Exogenous Targets
3.1. Bacteria and Fungus
- Introduction
- Enrichment and Analysis of Bacteria
- Enrichment and Analysis of Fungus
- Blood Cleansing
- Summary and Outlook
3.2. Viruses
- Introduction
- Virus Detection
- Viral Biomarker Identification
- Summary and Outlook
3.3. Parasites
- Introduction
- Enrichment and Detection of Parasites
- Biomarker Identification
- Summary and Outlook
3.4. Pathogens in Environmental Samples and Foodstuff
- Introduction
- Aerosol Sampling
- Food Sampling
- Water Sampling
- Summary and Outlook
4. Hybrid Systems for New Capabilities
5. Conclusions
- Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Martel, J.M.; Toner, M. Inertial Focusing in Microfluidics. Annu. Rev. Biomed. Eng. 2014, 16, 371–396. [Google Scholar] [CrossRef] [Green Version]
- Di Carlo, D. Inertial Microfluidics. Lab Chip 2009, 9, 3038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yan, S.; Yuan, D.; Alici, G.; Nguyen, N.-T.; Ebrahimi Warkiani, M.; Li, W. Fundamentals and Applications of Inertial Microfluidics: A Review. Lab Chip 2016, 16, 10–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volpe, A.; Gaudiuso, C.; Ancona, A. Sorting of Particles Using Inertial Focusing and Laminar Vortex Technology: A Review. Micromachines 2019, 10, 594. [Google Scholar] [CrossRef] [Green Version]
- Di Carlo, D.; Irimia, D.; Tompkins, R.G.; Toner, M. Continuous Inertial Focusing, Ordering, and Separation of Particles in Microchannels. Proc. Natl. Acad. Sci. USA 2007, 104, 18892–18897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segré, G.; Silberberg, A. Radial Particle Displacements in Poiseuille Flow of Suspensions. Nature 1961, 189, 209–210. [Google Scholar] [CrossRef]
- Amini, H.; Lee, W.; Di Carlo, D. Inertial Microfluidic Physics. Lab Chip 2014, 14, 2739. [Google Scholar] [CrossRef]
- Lee, M.G.; Choi, S.; Park, J.-K. Inertial Separation in a Contraction–Expansion Array Microchannel. J. Chromatogr. A 2011, 1218, 4138–4143. [Google Scholar] [CrossRef]
- Hur, S.C.; Mach, A.J.; Di Carlo, D. High-Throughput Size-Based Rare Cell Enrichment Using Microscale Vortices. Biomicrofluidics 2011, 5, 022206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martel, J.M.; Toner, M. Particle Focusing in Curved Microfluidic Channels. Sci. Rep. 2013, 3, 3340. [Google Scholar] [CrossRef] [Green Version]
- Gossett, D.R.; Carlo, D.D. Particle Focusing Mechanisms in Curving Confined Flows. Anal. Chem. 2009, 81, 8459–8465. [Google Scholar] [CrossRef]
- Kuntaegowdanahalli, S.S.; Bhagat, A.A.S.; Kumar, G.; Papautsky, I. Inertial Microfluidics for Continuous Particle Separation in Spiral Microchannels. Lab Chip 2009, 9, 2973–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, H.W.; Warkiani, M.E.; Khoo, B.L.; Li, Z.R.; Soo, R.A.; Tan, D.S.-W.; Lim, W.-T.; Han, J.; Bhagat, A.A.S.; Lim, C.T. Isolation and Retrieval of Circulating Tumor Cells Using Centrifugal Forces. Sci. Rep. 2013, 3, 1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Marusyk, A.; Polyak, K. Tumor Heterogeneity: Causes and Consequences. Biochim. Biophys. Acta Rev. Cancer 2010, 1805, 105–117. [Google Scholar] [CrossRef] [Green Version]
- Alix-Panabières, C.; Pantel, K. Circulating Tumor Cells: Liquid Biopsy of Cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Plaks, V.; Koopman, C.D.; Werb, Z. Circulating Tumor Cells. Science 2013, 341, 1186–1188. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabières, C. Liquid Biopsy and Minimal Residual Disease—Latest Advances and Implications for Cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Haber, D.A.; Velculescu, V.E. Blood-Based Analyses of Cancer: Circulating Tumor Cells and Circulating Tumor DNA. Cancer Discov. 2014, 4, 650–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but Not in Healthy Subjects or Patients with Nonmalignant Diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [Green Version]
- Shaw Bagnall, J.; Byun, S.; Begum, S.; Miyamoto, D.T.; Hecht, V.C.; Maheswaran, S.; Stott, S.L.; Toner, M.; Hynes, R.O.; Manalis, S.R. Deformability of Tumor Cells versus Blood Cells. Sci. Rep. 2015, 5, 18542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diez-Silva, M.; Dao, M.; Han, J.; Lim, C.-T.; Suresh, S. Shape and Biomechanical Characteristics of Human Red Blood Cells in Health and Disease. MRS Bull. 2010, 35, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Shevkoplyas, S.S.; Yoshida, T.; Munn, L.L.; Bitensky, M.W. Biomimetic Autoseparation of Leukocytes from Whole Blood in a Microfluidic Device. Anal. Chem. 2005, 77, 933–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabriel, M.T.; Calleja, L.R.; Chalopin, A.; Ory, B.; Heymann, D. Circulating Tumor Cells: A Review of Non–EpCAM-Based Approaches for Cell Enrichment and Isolation. Clin. Chem. 2016, 62, 571–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.; Kim, J.; Song, H.; Yong Sohn, K.; Jeon, M.; Han, K.-H. Microfluidic Technologies for Circulating Tumor Cell Isolation. Analyst 2018, 143, 2936–2970. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating Tumor Cell Technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [Green Version]
- Sollier, E.; Go, D.E.; Che, J.; Gossett, D.R.; O’Byrne, S.; Weaver, W.M.; Kummer, N.; Rettig, M.; Goldman, J.; Nickols, N.; et al. Size-Selective Collection of Circulating Tumor Cells Using Vortex Technology. Lab Chip 2013, 14, 63–77. [Google Scholar] [CrossRef]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.; Morgan, B.; Trautwein, J.; et al. Inertial Focusing for Tumor Antigen–Dependent and –Independent Sorting of Rare Circulating Tumor Cells. Sci. Transl. Med. 2013, 5, 179ra47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, S.; Kim, T.H.; Smith, K.J.; Delaney, R.; Park, G.-S.; Guo, H.; Lin, E.; Plegue, T.; Kuo, N.; Steffes, J.; et al. New Labyrinth Microfluidic Device Detects Circulating Tumor Cells Expressing Cancer Stem Cell Marker and Circulating Tumor Microemboli in Hepatocellular Carcinoma. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.B.; Yeo, T.; Lee, W.D.; Bhagat, A.A.S.; Tan, S.J.; Tan, D.S.W.; Lim, W.-T.; Lim, C.T. Addressing Cellular Heterogeneity in Tumor and Circulation for Refined Prognostication. Proc. Natl. Acad. Sci. USA 2019, 116, 17957–17962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, D.T.; Lee, R.J.; Kalinich, M.; LiCausi, J.A.; Zheng, Y.; Chen, T.; Milner, J.D.; Emmons, E.; Ho, U.; Broderick, K.; et al. An RNA-Based Digital Circulating Tumor Cell Signature Is Predictive of Drug Response and Early Dissemination in Prostate Cancer. Cancer Discov. 2018, 8, 288–303. [Google Scholar] [CrossRef] [Green Version]
- Rzhevskiy, A.S.; Razavi Bazaz, S.; Ding, L.; Kapitannikova, A.; Sayyadi, N.; Campbell, D.; Walsh, B.; Gillatt, D.; Ebrahimi Warkiani, M.; Zvyagin, A.V. Rapid and Label-Free Isolation of Tumour Cells from the Urine of Patients with Localised Prostate Cancer Using Inertial Microfluidics. Cancers 2020, 12, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalinich, M.; Bhan, I.; Kwan, T.T.; Miyamoto, D.T.; Javaid, S.; LiCausi, J.A.; Milner, J.D.; Hong, X.; Goyal, L.; Sil, S.; et al. An RNA-Based Signature Enables High Specificity Detection of Circulating Tumor Cells in Hepatocellular Carcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, 1123–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, X.; Sullivan, R.J.; Kalinich, M.; Kwan, T.T.; Giobbie-Hurder, A.; Pan, S.; LiCausi, J.A.; Milner, J.D.; Nieman, L.T.; Wittner, B.S.; et al. Molecular Signatures of Circulating Melanoma Cells for Monitoring Early Response to Immune Checkpoint Therapy. Proc. Natl. Acad. Sci. USA 2018, 115, 2467–2472. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.L.; Lim, T.H.; Lim, T.K.; Tan, D.S.-W.; Chua, Y.W.; Ang, M.K.; Pang, B.; Lim, C.T.; Takano, A.; Lim, A.S.-T.; et al. Concordance of Anaplastic Lymphoma Kinase (ALK) Gene Rearrangements between Circulating Tumor Cells and Tumor in Non-Small Cell Lung Cancer. Oncotarget 2016, 7, 23251–23262. [Google Scholar] [CrossRef] [Green Version]
- Kidess-Sigal, E.; Liu, H.E.; Triboulet, M.M.; Che, J.; Ramani, V.C.; Visser, B.C.; Poultsides, G.A.; Longacre, T.A.; Marziali, A.; Vysotskaia, V.; et al. Enumeration and Targeted Analysis of KRAS, BRAF and PIK3CA Mutations in CTCs Captured by a Label-Free Platform: Comparison to CtDNA and Tissue in Metastatic Colorectal Cancer. Oncotarget 2016, 7, 85349–85364. [Google Scholar] [CrossRef] [Green Version]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1–Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, J.; Mirenska, A.; Tan, M.; Lee, Y.; Oh, J.; Hong, L.Z.; Wnek, R.; Yap, Y.-S.; Shih, S.-J.S.; Bhagat, A.A.; et al. A Preliminary Study for the Assessment of PD-L1 and PD-L2 on Circulating Tumor Cells by Microfluidic-Based Chipcytometry. Future Sci. OA 2017, 3, FSO244. [Google Scholar] [CrossRef] [PubMed]
- Dhar, M.; Wong, J.; Che, J.; Matsumoto, M.; Grogan, T.; Elashoff, D.; Garon, E.B.; Goldman, J.W.; Sollier Christen, E.; Di Carlo, D.; et al. Evaluation of PD-L1 Expression on Vortex-Isolated Circulating Tumor Cells in Metastatic Lung Cancer. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; Kapeleris, J.; Cooper, C.; Warkiani, M.E.; O’Byrne, K.; Punyadeera, C. Phenotypic Characterization of Circulating Lung Cancer Cells for Clinically Actionable Targets. Cancers 2019, 11, 380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulasinghe, A.; Kapeleris, J.; Kimberley, R.; Mattarollo, S.R.; Thompson, E.W.; Thiery, J.; Kenny, L.; O’Byrne, K.; Punyadeera, C. The Prognostic Significance of Circulating Tumor Cells in Head and Neck and Non-small-cell Lung Cancer. Cancer Med. 2018, 7, 5910–5919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuoka, M.; Wu, Y.-L.; Thongprasert, S.; Sunpaweravong, P.; Leong, S.-S.; Sriuranpong, V.; Chao, T.-Y.; Nakagawa, K.; Chu, D.-T.; Saijo, N.; et al. Biomarker Analyses and Final Overall Survival Results from a Phase III, Randomized, Open-Label, First-Line Study of Gefitinib Versus Carboplatin/Paclitaxel in Clinically Selected Patients With Advanced Non–Small-Cell Lung Cancer in Asia (IPASS). J. Clin. Oncol. 2011, 29, 2866–2874. [Google Scholar] [CrossRef]
- Zeinali, M.; Lee, M.; Nadhan, A.; Mathur, A.; Hedman, C.; Lin, E.; Harouaka, R.; Wicha, M.S.; Zhao, L.; Palanisamy, N.; et al. High-Throughput Label-Free Isolation of Heterogeneous Circulating Tumor Cells and CTC Clusters from Non-Small-Cell Lung Cancer Patients. Cancers 2020, 12, 127. [Google Scholar] [CrossRef] [Green Version]
- Kulasinghe, A.; Perry, C.; Kenny, L.; Warkiani, M.E.; Nelson, C.; Punyadeera, C. PD-L1 Expressing Circulating Tumour Cells in Head and Neck Cancers. BMC Cancer 2017, 17, 333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulasinghe, A.; Tran, T.H.P.; Blick, T.; O’Byrne, K.; Thompson, E.W.; Warkiani, M.E.; Nelson, C.; Kenny, L.; Punyadeera, C. Enrichment of Circulating Head and Neck Tumour Cells Using Spiral Microfluidic Technology. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, T.; Tan, S.J.; Lim, C.L.; Lau, D.P.X.; Chua, Y.W.; Krisna, S.S.; Iyer, G.; Tan, G.S.; Lim, T.K.H.; Tan, D.S.W.; et al. Microfluidic Enrichment for the Single Cell Analysis of Circulating Tumor Cells. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onidani, K.; Shoji, H.; Kakizaki, T.; Yoshimoto, S.; Okaya, S.; Miura, N.; Sekikawa, S.; Furuta, K.; Lim, C.T.; Shibahara, T.; et al. Monitoring of Cancer Patients via Next-generation Sequencing of Patient-derived Circulating Tumor Cells and Tumor DNA. Cancer Sci. 2019, 110, 2590–2599. [Google Scholar] [CrossRef] [Green Version]
- Shaw, A.T.; Kim, D.-W.; Mehra, R.; Tan, D.S.W.; Felip, E.; Chow, L.Q.M.; Camidge, D.R.; Vansteenkiste, J.; Sharma, S.; De Pas, T.; et al. Ceritinib in ALK-Rearranged Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2014, 370, 1189–1197. [Google Scholar] [CrossRef] [Green Version]
- Dhar, M.; Pao, E.; Renier, C.; Go, D.E.; Che, J.; Montoya, R.; Conrad, R.; Matsumoto, M.; Heirich, K.; Triboulet, M.; et al. Label-Free Enumeration, Collection and Downstream Cytological and Cytogenetic Analysis of Circulating Tumor Cells. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Houssami, N.; Macaskill, P.; Balleine, R.L.; Bilous, M.; Pegram, M.D. HER2 Discordance between Primary Breast Cancer and Its Paired Metastasis: Tumor Biology or Test Artefact? Insights through Meta-Analysis. Breast Cancer Res. Treat. 2011, 129, 659–674. [Google Scholar] [CrossRef]
- Fachin, F.; Spuhler, P.; Martel-Foley, J.M.; Edd, J.F.; Barber, T.A.; Walsh, J.; Karabacak, M.; Pai, V.; Yu, M.; Smith, K.; et al. Monolithic Chip for High-Throughput Blood Cell Depletion to Sort Rare Circulating Tumor Cells. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Warkiani, M.E.; Guan, G.; Luan, K.B.; Lee, W.C.; Bhagat, A.A.S.; Chaudhuri, P.K.; Tan, D.S.-W.; Lim, W.T.; Lee, S.C.; Chen, P.C.Y.; et al. Slanted Spiral Microfluidics for the Ultra-Fast, Label-Free Isolation of Circulating Tumor Cells. Lab Chip 2013, 14, 128–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warkiani, M.E.; Khoo, B.L.; Tan, D.S.-W.; Bhagat, A.A.S.; Lim, W.-T.; Yap, Y.S.; Lee, S.C.; Soo, R.A.; Han, J.; Lim, C.T. An Ultra-High-Throughput Spiral Microfluidic Biochip for the Enrichment of Circulating Tumor Cells. Analyst 2014, 139, 3245–3255. [Google Scholar] [CrossRef] [PubMed]
- Jordan, N.V.; Bardia, A.; Wittner, B.S.; Benes, C.; Ligorio, M.; Zheng, Y.; Yu, M.; Sundaresan, T.K.; Licausi, J.A.; Desai, R.; et al. HER2 Expression Identifies Dynamic Functional States within Circulating Breast Cancer Cells. Nature 2016, 537, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Medford, A.J.; Dubash, T.D.; Juric, D.; Spring, L.; Niemierko, A.; Vidula, N.; Peppercorn, J.; Isakoff, S.; Reeves, B.A.; LiCausi, J.A.; et al. Blood-Based Monitoring Identifies Acquired and Targetable Driver HER2 Mutations in Endocrine-Resistant Metastatic Breast Cancer. NPJ Precis. Oncol. 2019, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, D.T.; Zheng, Y.; Wittner, B.S.; Lee, R.J.; Zhu, H.; Broderick, K.T.; Desai, R.; Fox, D.B.; Brannigan, B.W.; Trautwein, J.; et al. RNA-Seq of Single Prostate CTCs Implicates Noncanonical Wnt Signaling in Antiandrogen Resistance. Science 2015, 349, 1351–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, K.H.K.; Tessier, S.N.; Miyamoto, D.T.; Miller, K.L.; Bookstaver, L.D.; Carey, T.R.; Stannard, C.J.; Thapar, V.; Tai, E.C.; Vo, K.D.; et al. Whole Blood Stabilization for the Microfluidic Isolation and Molecular Characterization of Circulating Tumor Cells. Nat. Commun. 2017, 8, 1733. [Google Scholar] [CrossRef]
- Wang, J.; Lu, W.; Tang, C.; Liu, Y.; Sun, J.; Mu, X.; Zhang, L.; Dai, B.; Li, X.; Zhuo, H.; et al. Label-Free Isolation and MRNA Detection of Circulating Tumor Cells from Patients with Metastatic Lung Cancer for Disease Diagnosis and Monitoring Therapeutic Efficacy. Anal. Chem. 2015, 87, 11893–11900. [Google Scholar] [CrossRef] [PubMed]
- Drapkin, B.J.; George, J.; Christensen, C.L.; Mino-Kenudson, M.; Dries, R.; Sundaresan, T.; Phat, S.; Myers, D.T.; Zhong, J.; Igo, P.; et al. Genomic and Functional Fidelity of Small Cell Lung Cancer Patient-Derived Xenografts. Cancer Discov. 2018, 8, 600–615. [Google Scholar] [CrossRef] [Green Version]
- Sinkala, E.; Sollier-Christen, E.; Renier, C.; Rosàs-Canyelles, E.; Che, J.; Heirich, K.; Duncombe, T.A.; Vlassakis, J.; Yamauchi, K.A.; Huang, H.; et al. Profiling Protein Expression in Circulating Tumour Cells Using Microfluidic Western Blotting. Nat. Commun. 2017, 8, 14622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abouleila, Y.; Onidani, K.; Ali, A.; Shoji, H.; Kawai, T.; Lim, C.T.; Kumar, V.; Okaya, S.; Kato, K.; Hiyama, E.; et al. Live Single Cell Mass Spectrometry Reveals Cancer-specific Metabolic Profiles of Circulating Tumor Cells. Cancer Sci. 2019, 110, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.E.; Triboulet, M.; Zia, A.; Vuppalapaty, M.; Kidess-Sigal, E.; Coller, J.; Natu, V.S.; Shokoohi, V.; Che, J.; Renier, C.; et al. Workflow Optimization of Whole Genome Amplification and Targeted Panel Sequencing for CTC Mutation Detection. NPJ Genom. Med. 2017, 2, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Wang, Z.; Li, G.; Lin, F.; Shao, K.; Cao, B.; Hou, Y. Characterization of Circulating Tumor Cells in Breast Cancer Patients by Spiral Microfluidics. Cell. Biol. Toxicol. 2019, 35, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.; Cai, Z.; Winkler, K.; Georgiou, K.; Inglis, D.; Lavranos, T.; Rezaei, M.; Warkiani, M.; Thierry, B. Circulating Tumour Cell RNA Characterisation from Colorectal Cancer Patient Blood after Inertial Microfluidic Enrichment. Methods X 2019, 6, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Chudasama, D.Y.; Freydina, D.V.; Freidin, M.B.; Leung, M.; Montero Fernandez, A.; Rice, A.; Nicholson, A.G.; Karteris, E.; Anikin, V.; Lim, E. Inertia Based Microfluidic Capture and Characterisation of Circulating Tumour Cells for the Diagnosis of Lung Cancer. Ann. Transl. Med. 2016, 4, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, J.; Yu, V.; Garon, E.B.; Goldman, J.W.; Carlo, D.D. Biophysical Isolation and Identification of Circulating Tumor Cells. Lab Chip 2017, 17, 1452–1461. [Google Scholar] [CrossRef] [Green Version]
- Che, J.; Yu, V.; Dhar, M.; Renier, C.; Matsumoto, M.; Heirich, K.; Garon, E.B.; Goldman, J.; Rao, J.; Sledge, G.W.; et al. Classification of Large Circulating Tumor Cells Isolated with Ultra-High Throughput Microfluidic Vortex Technology. Oncotarget 2016, 7, 12748–12760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramani, V.C.; Lemaire, C.A.; Triboulet, M.; Casey, K.M.; Heirich, K.; Renier, C.; Vilches-Moure, J.G.; Gupta, R.; Razmara, A.M.; Zhang, H.; et al. Investigating Circulating Tumor Cells and Distant Metastases in Patient-Derived Orthotopic Xenograft Models of Triple-Negative Breast Cancer. Breast Cancer Res. 2019, 21, 98. [Google Scholar] [CrossRef]
- Lin, E.; Rivera-Báez, L.; Fouladdel, S.; Yoon, H.J.; Guthrie, S.; Wieger, J.; Deol, Y.; Keller, E.; Sahai, V.; Simeone, D.M.; et al. High-Throughput Microfluidic Labyrinth for the Label-Free Isolation of Circulating Tumor Cells. Cell Syst. 2017, 5, 295–304. [Google Scholar] [CrossRef] [Green Version]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulasinghe, A.; Schmidt, H.; Perry, C.; Whitfield, B.; Kenny, L.; Nelson, C.; Warkiani, M.E.; Punyadeera, C. A Collective Route to Head and Neck Cancer Metastasis. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulasinghe, A.; Zhou, J.; Kenny, L.; Papautsky, I.; Punyadeera, C. Capture of Circulating Tumour Cell Clusters Using Straight Microfluidic Chips. Cancers 2019, 11, 89. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Miyamoto, D.T.; Wittner, B.S.; Sullivan, J.P.; Aceto, N.; Jordan, N.V.; Yu, M.; Karabacak, N.M.; Comaills, V.; Morris, R.; et al. Expression of β-Globin by Cancer Cells Promotes Cell Survival during Blood-Borne Dissemination. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dhar, M.; Lam, J.N.; Walser, T.; Dubinett, S.M.; Rettig, M.B.; Di Carlo, D. Functional Profiling of Circulating Tumor Cells with an Integrated Vortex Capture and Single-Cell Protease Activity Assay. Proc. Natl. Acad. Sci. USA 2018, 115, 9986–9991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebright, R.Y.; Lee, S.; Wittner, B.S.; Niederhoffer, K.L.; Nicholson, B.T.; Bardia, A.; Truesdell, S.; Wiley, D.F.; Wesley, B.; Li, S.; et al. Deregulation of Ribosomal Protein Expression and Translation Promotes Breast Cancer Metastasis. Science 2020, 367, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Aya-Bonilla, C.A.; Marsavela, G.; Freeman, J.B.; Lomma, C.; Frank, M.H.; Khattak, M.A.; Meniawy, T.M.; Millward, M.; Warkiani, M.E.; Gray, E.S.; et al. Isolation and Detection of Circulating Tumour Cells from Metastatic Melanoma Patients Using a Slanted Spiral Microfluidic Device. Oncotarget 2017, 8, 67355–67368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, A.; Dubash, T.D.; Edd, J.F.; Jewett, M.K.; Garre, S.G.; Karabacak, N.M.; Rabe, D.C.; Mutlu, B.R.; Walsh, J.R.; Kapur, R.; et al. Ultrahigh-Throughput Magnetic Sorting of Large Blood Volumes for Epitope-Agnostic Isolation of Circulating Tumor Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 16839–16847. [Google Scholar] [CrossRef] [PubMed]
- Edd, J.F.; Mishra, A.; Dubash, T.D.; Herrera, S.; Mohammad, R.; Williams, E.K.; Hong, X.; Mutlu, B.R.; Walsh, J.R.; de Carvalho, F.M.; et al. Microfluidic Concentration and Separation of Circulating Tumor Cell Clusters from Large Blood Volumes. Lab Chip 2020, 20, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Z. An Inertia-Deformability Hybrid Circulating Tumor Cell Chip: Design, Clinical Test, and Numerical Analysis. J. Med. Devices 2018, 12, 041004. [Google Scholar] [CrossRef]
- Xiang, N.; Wang, J.; Li, Q.; Han, Y.; Huang, D.; Ni, Z. Precise Size-Based Cell Separation via the Coupling of Inertial Microfluidics and Deterministic Lateral Displacement. Anal. Chem. 2019, 91, 10328–10334. [Google Scholar] [CrossRef] [PubMed]
- Xiang, N.; Li, Q.; Ni, Z. Combining Inertial Microfluidics with Cross-Flow Filtration for High-Fold and High-Throughput Passive Volume Reduction. Anal. Chem. 2020, 92, 6770–6776. [Google Scholar] [CrossRef]
- Gao, R.; Cheng, L.; Wang, S.; Bi, X.; Wang, X.; Wang, R.; Chen, X.; Zha, Z.; Wang, F.; Xu, X.; et al. Efficient Separation of Tumor Cells from Untreated Whole Blood Using a Novel Multistage Hydrodynamic Focusing Microfluidics. Talanta 2020, 207, 120261. [Google Scholar] [CrossRef]
- Wang, X.; Yang, X.; Papautsky, I. An Integrated Inertial Microfluidic Vortex Sorter for Tunable Sorting and Purification of Cells. Technology 2016, 4, 88–97. [Google Scholar] [CrossRef]
- Volpe, A.; Paiè, P.; Ancona, A.; Osellame, R. Polymeric Fully Inertial Lab-on-a-Chip with Enhanced-Throughput Sorting Capabilities. Microfluid Nanofluid 2019, 23, 37. [Google Scholar] [CrossRef]
- Lee, T.Y.; Hyun, K.-A.; Kim, S.-I.; Jung, H.-I. An Integrated Microfluidic Chip for One-Step Isolation of Circulating Tumor Cells. Sens. Actuators B Chem. 2017, 238, 1144–1150. [Google Scholar] [CrossRef]
- Zhou, J.; Kulasinghe, A.; Bogseth, A.; O’Byrne, K.; Punyadeera, C.; Papautsky, I. Isolation of Circulating Tumor Cells in Non-Small-Cell-Lung-Cancer Patients Using a Multi-Flow Microfluidic Channel. Microsyst. Nanoeng. 2019, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, F.; Cai, L.; Chang, J.; Li, S.; Liu, C.; Li, T.; Sun, J. Label-Free Isolation of Rare Tumor Cells from Untreated Whole Blood by Interfacial Viscoelastic Microfluidics. Lab Chip 2018, 18, 3436–3445. [Google Scholar] [CrossRef] [PubMed]
- Feher, J. White Blood Cells and Inflammation. In Quantitative Human Physiology, 2nd ed.; Feher, J., Ed.; Academic Press: Boston, MA, USA, 2012; pp. 507–515. ISBN 978-0-12-800883-6. [Google Scholar]
- Houwen, B. The Differential Count. Lab. Hematol. 2001, 7, 89–100. [Google Scholar]
- Farkas, J.D. The Complete Blood Count to Diagnose Septic Shock. J. Thorac. Dis. 2020, 12, S16–S21. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-Y.; Choi, C.H.; Sung, C.O.; Do, I.-G.; Huh, S.; Song, T.; Kim, M.K.; Kim, H.-J.; Kim, T.-J.; Lee, J.-W.; et al. Prognostic Value of Pre-Treatment Circulating Monocyte Count in Patients with Cervical Cancer: Comparison with SCC-Ag Level. Gynecol. Oncol. 2012, 124, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Shim, W.S.; Kim, H.J.; Kang, E.S.; Ahn, C.W.; Lim, S.K.; Lee, H.C.; Cha, B.S. The Association of Total and Differential White Blood Cell Count with Metabolic Syndrome in Type 2 Diabetic Patients. Diabetes Res. Clin. Pract. 2006, 73, 284–291. [Google Scholar] [CrossRef] [PubMed]
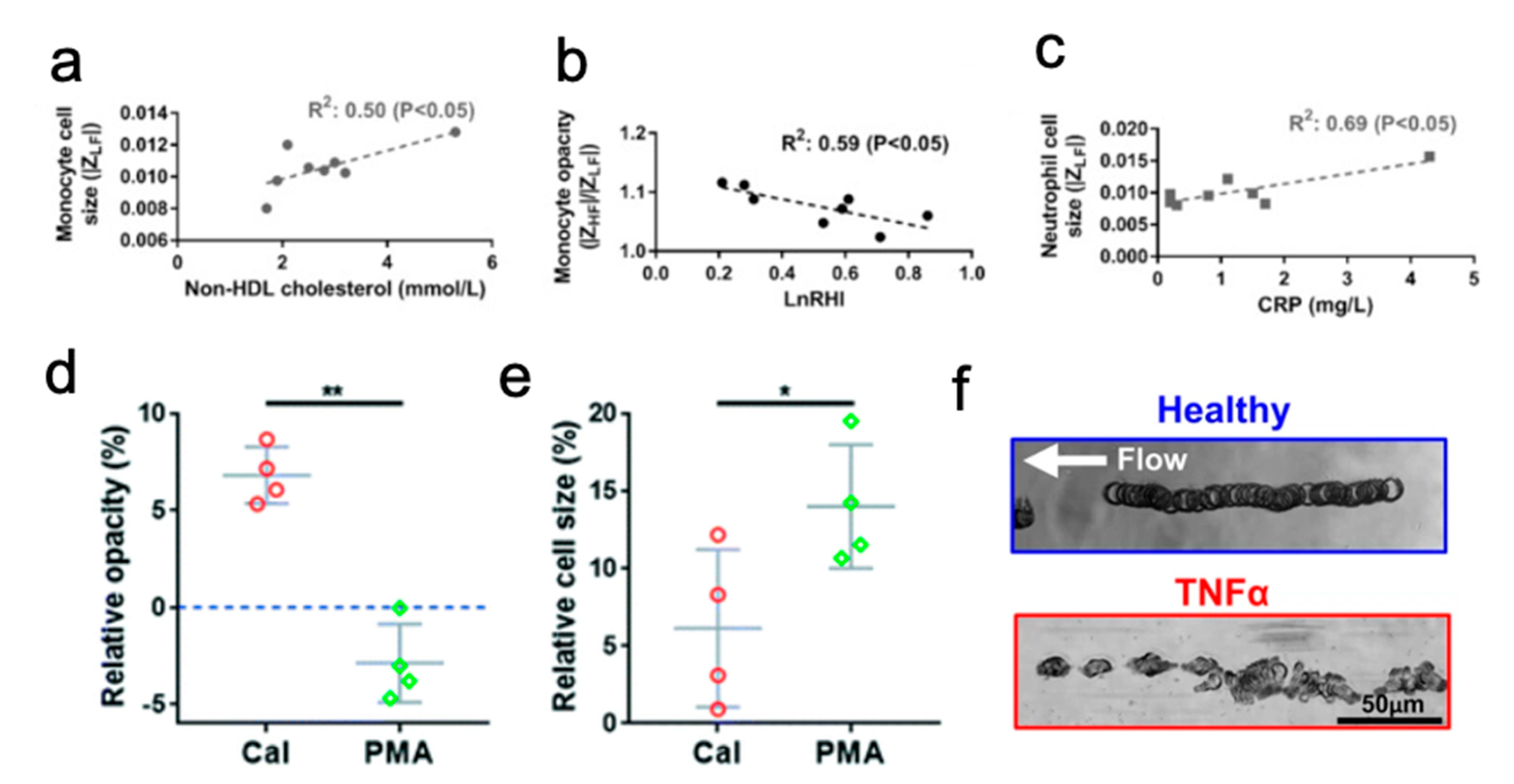
- Jundi, B.; Ryu, H.; Lee, D.-H.; Abdulnour, R.-E.E.; Engstrom, B.D.; Duvall, M.G.; Higuera, A.; Pinilla-Vera, M.; Benson, M.E.; Lee, J.; et al. Leukocyte Function Assessed via Serial Microlitre Sampling of Peripheral Blood from Sepsis Patients Correlates with Disease Severity. Nat. Biomed. Eng. 2019, 3, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Choi, K.; Qu, Y.; Kwon, T.; Lee, J.S.; Han, J. Patient-Derived Airway Secretion Dissociation Technique to Isolate and Concentrate Immune Cells Using Closed-Loop Inertial Microfluidics. Anal. Chem. 2017, 89, 5549–5556. [Google Scholar] [CrossRef] [Green Version]
- Petchakup, C.; Tay, H.M.; Yeap, W.H.; Dalan, R.; Wong, S.C.; Li, K.H.H.; Hou, H.W. Label-Free Leukocyte Sorting and Impedance-Based Profiling for Diabetes Testing. Biosens. Bioelectron. 2018, 118, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Petchakup, C.; Tay, H.M.; Li, K.H.H.; Hou, H.W. Integrated Inertial-Impedance Cytometry for Rapid Label-Free Leukocyte Isolation and Profiling of Neutrophil Extracellular Traps (NETs). Lab Chip 2019, 19, 1736–1746. [Google Scholar] [CrossRef]
- Tay, H.M.; Dalan, R.; Li, K.H.H.; Boehm, B.O.; Hou, H.W. A Novel Microdevice for Rapid Neutrophil Purification and Phenotyping in Type 2 Diabetes Mellitus. Small 2018, 14, 1702832. [Google Scholar] [CrossRef]
- Hou, H.W.; Petchakup, C.; Tay, H.M.; Tam, Z.Y.; Dalan, R.; Chew, D.E.K.; Li, K.H.H.; Boehm, B.O. Rapid and Label-Free Microfluidic Neutrophil Purification and Phenotyping in Diabetes Mellitus. Sci. Rep. 2016, 6, 29410. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wu, D.; Han, Y.; Wang, C.; Xiang, N.; Ni, Z. Inertial Microfluidic Cube for Automatic and Fast Extraction of White Blood Cells from Whole Blood. Lab Chip 2020, 20, 244–252. [Google Scholar] [CrossRef]
- Ramachandraiah, H.; Svahn, H.A.; Russom, A. Inertial Microfluidics Combined with Selective Cell Lysis for High Throughput Separation of Nucleated Cells from Whole Blood. RSC Adv. 2017, 7, 29505–29514. [Google Scholar] [CrossRef] [Green Version]
- Crawford, K.; DeWitt, A.; Brierre, S.; Caffery, T.; Jagneaux, T.; Thomas, C.; Macdonald, M.; Tse, H.; Shah, A.; Di Carlo, D.; et al. Rapid Biophysical Analysis of Host Immune Cell Variations Associated with Sepsis. Am. J. Respir. Crit. Care Med. 2018, 198, 280–282. [Google Scholar] [CrossRef] [PubMed]
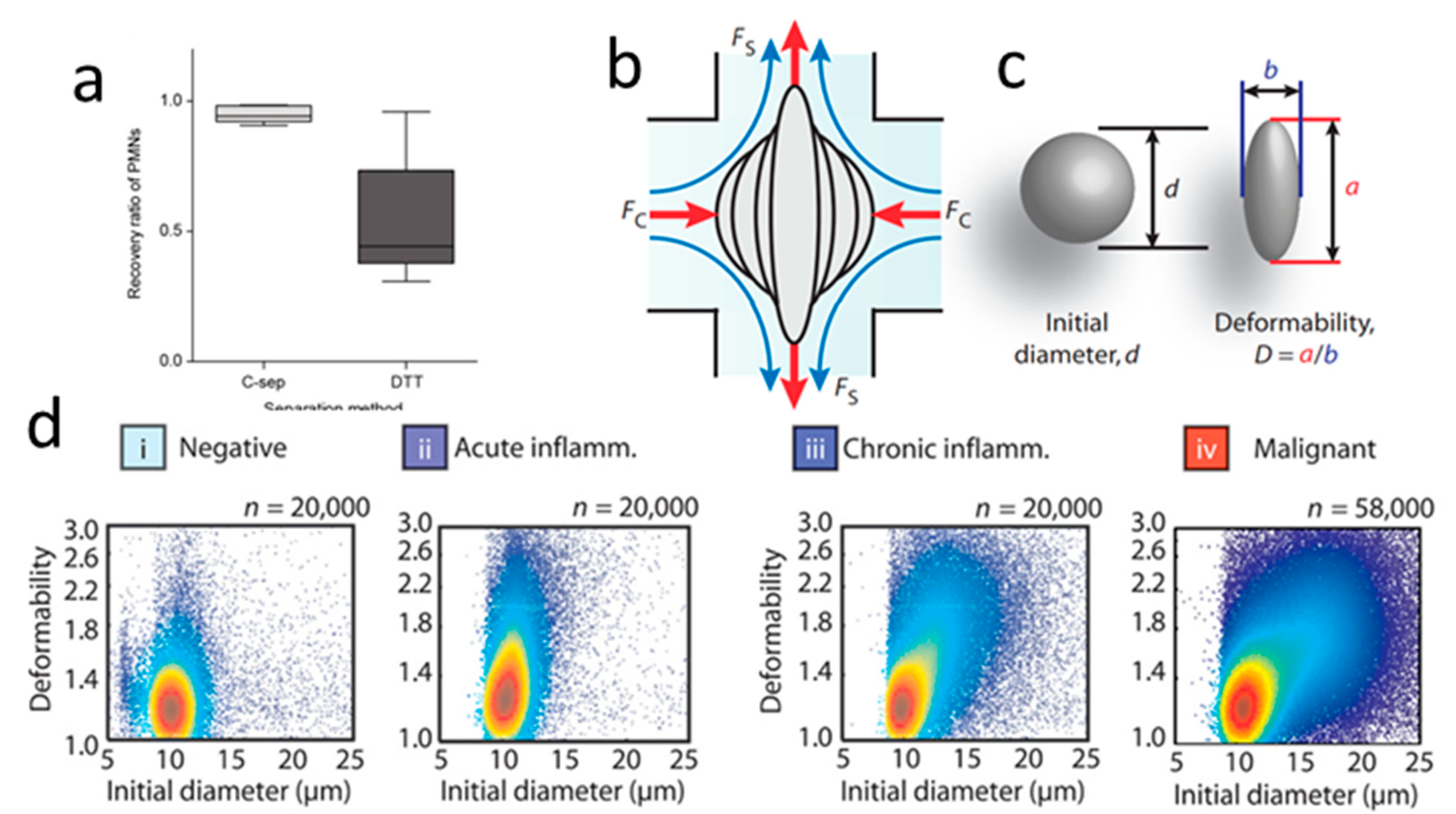
- Gossett, D.R.; Tse, H.T.K.; Lee, S.A.; Ying, Y.; Lindgren, A.G.; Yang, O.O.; Rao, J.; Clark, A.T.; Di Carlo, D. Hydrodynamic Stretching of Single Cells for Large Population Mechanical Phenotyping. Proc. Natl. Acad. Sci. USA 2012, 109, 7630–7635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houston, N.; Stewart, N.; Smith, D.S.; Bell, S.C.; Champion, A.C.; Reid, D.W. Sputum Neutrophils in Cystic Fibrosis Patients Display a Reduced Respiratory Burst. J. Cyst. Fibros. 2013, 12, 352–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suresh, S.; Spatz, J.; Mills, J.P.; Micoulet, A.; Dao, M.; Lim, C.T.; Beil, M.; Seufferlein, T. Connections between Single-Cell Biomechanics and Human Disease States: Gastrointestinal Cancer and Malaria. Acta Biomater. 2005, 1, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Guck, J.; Schinkinger, S.; Lincoln, B.; Wottawah, F.; Ebert, S.; Romeyke, M.; Lenz, D.; Erickson, H.M.; Ananthakrishnan, R.; Mitchell, D.; et al. Optical Deformability as an Inherent Cell Marker for Testing Malignant Transformation and Metastatic Competence. Biophys. J. 2005, 88, 3689–3698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gernez, Y.; Tirouvanziam, R.; Chanez, P. Neutrophils in Chronic Inflammatory Airway Diseases: Can We Target Them and How? Eur. Respir. J. 2010, 35, 467–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- 2017 Patient Registry Annual Data Report; Cystic Fibrosis Foundation: Bethesda, MD, USA, 2017; p. 96.
- Rubin, B.K. Secretion Properties, Clearance, and Therapy in Airway Disease. Transl. Respir. Med. 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lötvall, J.; Akdis, C.A.; Bacharier, L.B.; Bjermer, L.; Casale, T.B.; Custovic, A.; Lemanske, R.F.; Wardlaw, A.J.; Wenzel, S.E.; Greenberger, P.A. Asthma Endotypes: A New Approach to Classification of Disease Entities within the Asthma Syndrome. J. Allergy Clin. Immunol. 2011, 127, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Van Overveld, F.J.; Demkow, U.; Górecka, D.; Skopinska-Rozewska, E.; de Backer, W.A.; Zielinski, J. Effects of Homogenization of Induced Sputum by Dithiothreitol on Polymorphonuclear Cells. J. Physiol. Pharmacol. 2005, 56, 143–154. [Google Scholar] [PubMed]
- Qiu, D.; Tan, W.C. Dithiothreitol Has a Dose-Response Effect on Cell Surface Antigen Expression. J. Allergy Clin. Immunol. 1999, 103, 873–876. [Google Scholar] [CrossRef]
- Bloomfield, V.A. Hydrodynamic Properties of Mucous Glycoproteins. Biopolymers 1983, 22, 2141–2154. [Google Scholar] [CrossRef] [PubMed]
- Tse, H.T.K.; Gossett, D.R.; Moon, Y.S.; Masaeli, M.; Sohsman, M.; Ying, Y.; Mislick, K.; Adams, R.P.; Rao, J.; Carlo, D.D. Quantitative Diagnosis of Malignant Pleural Effusions by Single-Cell Mechanophenotyping. Sci. Transl. Med. 2013, 5, 212ra163. [Google Scholar] [CrossRef] [Green Version]
- Maher, G.G.; Berger, H.W. Massive Pleural Effusion: Malignant and Nonmalignant Causes in 46 Patients. Am. Rev. Respir. Dis. 1972, 105, 458–460. [Google Scholar] [PubMed]
- Noppen, M.; De Waele, M.; Li, R.; Gucht, K.V.; D’haese, J.; Gerlo, E.; Vincken, W. Volume and Cellular Content of Normal Pleural Fluid in Humans Examined by Pleural Lavage. Am. J. Respir. Crit. Care Med. 2000, 162, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Wild, S.; Roglic, G.; Green, A.; Sicree, R.; King, H. Global Prevalence of Diabetes: Estimates for the Year 2000 and Projections for 2030. Diabetes Care 2004, 27, 1047–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, H.M.; Hou, H.W.; Dalan, R.; Boehm, B.O. Advances in Neutrophil Testing In Type 2 Diabetes Mellitus. Curr. Res. Diabetes Obes. J. 2017, 1, 555572. [Google Scholar]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of Hypotension before Initiation of Effective Antimicrobial Therapy Is the Critical Determinant of Survival in Human Septic Shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, R.P.; Edmond, M.B. Managing Antibiotic Resistance. N. Engl. J. Med. 2000, 343, 1961–1963. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Salam, B.K.A.; Ebaid, H. Expression of CD11b and CD18 on Polymorphonuclear Neutrophils Stimulated with Interleukin-2. Cent. Eur. J. Immunol. 2014, 39, 209–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, B.S.; Chung, K.F. Blood Neutrophil Activation Markers in Severe Asthma: Lack of Inhibition by Prednisolone Therapy. Respir. Res. 2006, 7, 59. [Google Scholar] [CrossRef] [Green Version]
- Opasawatchai, A.; Amornsupawat, P.; Jiravejchakul, N.; Chan-in, W.; Spoerk, N.J.; Manopwisedjaroen, K.; Singhasivanon, P.; Yingtaweesak, T.; Suraamornkul, S.; Mongkolsapaya, J.; et al. Neutrophil Activation and Early Features of NET Formation Are Associated With Dengue Virus Infection in Human. Front. Immunol. 2019, 9, 3007. [Google Scholar] [CrossRef] [Green Version]
- Pentyala, S.; Lee, J.; Annam, S.; Alvarez, J.; Veerraju, A.; Yadlapalli, N.; Khan, S.A. Current Perspectives on Pyospermia: A Review. Asian J. Androl. 2007, 9, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Mujezinovic, F.; Alfirevic, Z. Procedure-Related Complications of Amniocentesis and Chorionic Villous Sampling: A Systematic Review. Obstet. Gynecol. 2007, 110, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Wallach, E.E.; Wolff, H. The Biologic Significance of White Blood Cells in Semen. Fertil. Steril. 1995, 63, 1143–1157. [Google Scholar] [CrossRef]
- Centola, G.M.; Ginsburg, K.A. Evaluation and Treatment of the Infertile Male; Cambridge University Press: Cambridge, UK, 2004; ISBN 978-0-521-60273-0. [Google Scholar]
- Schuster, T.G.; Cho, B.; Keller, L.M.; Takayama, S.; Smith, G.D. Isolation of Motile Spermatozoa from Semen Samples Using Microfluidics. Reprod. Biomed. Online 2003, 7, 75–81. [Google Scholar] [CrossRef]
- Son, J.; Samuel, R.; Murphy, K.; Sant, H.J.; Hockin, M.F.; Gale, B.K.; Hotaling, J.M.; Carrell, D.T. Sperm Cell Separation Using a Spiral Channel, Proceedings of the 18th International Conference on Miniaturized Systems for Chemistry and Life Sciences, San Antonio, TX, USA, 26–30 October 2014; Royal Society of Chemistry: London, UK, 2014. [Google Scholar]
- Son, J.; Murphy, K.; Samuel, R.; Gale, B.K.; Carrell, D.T.; Hotaling, J.M. Non-Motile Sperm Cell Separation Using a Spiral Channel. Anal. Methods 2015, 7, 8041–8047. [Google Scholar] [CrossRef]
- Son, J.; Samuel, R.; Gale, B.K.; Carrell, D.T.; Hotaling, J.M. Separation of Sperm Cells from Samples Containing High Concentrations of White Blood Cells Using a Spiral Channel. Biomicrofluidics 2017, 11. [Google Scholar] [CrossRef] [PubMed]
- Son, J. Active Sperm Separation Technique Using an Inertial Microfluidic Device. Ph.D. Thesis, University of Utah, Salt Lake City, UT, USA, 2017. [Google Scholar]
- Mielczarek, W.S.; Obaje, E.A.; Bachmann, T.T.; Kersaudy-Kerhoas, M. Microfluidic Blood Plasma Separation for Medical Diagnostics: Is It Worth It? Lab Chip 2016, 16, 3441–3448. [Google Scholar] [CrossRef] [Green Version]
- Lilja, H.; Ulmert, D.; Vickers, A.J. Prostate-Specific Antigen and Prostate Cancer: Prediction, Detection and Monitoring. Nat. Rev. Cancer 2008, 8, 268–278. [Google Scholar] [CrossRef]
- Rodbard, D. Continuous Glucose Monitoring: A Review of Successes, Challenges, and Opportunities. Diabetes Technol. Ther. 2016, 18, S2-3–S2-13. [Google Scholar] [CrossRef] [Green Version]
- Swarup, V.; Rajeswari, M.R. Circulating (Cell-Free) Nucleic Acids—A Promising, Non-Invasive Tool for Early Detection of Several Human Diseases. FEBS Lett. 2007, 581, 795–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Zhang, J.; Yan, S.; Yuan, D.; Du, H.; Alici, G.; Li, W. High-Throughput Sheathless and Three-Dimensional Microparticle Focusing Using a Microchannel with Arc-Shaped Groove Arrays. Sci. Rep. 2017, 7, 41153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jafek, A.; Feng, H.; Broberg, D.; Gale, B.; Samuel, R.; Aston, K.; Jenkins, T. Optimization of Dean Flow Microfluidic Chip for Sperm Preparation for Intrauterine Insemination. Microfluid Nanofluidics 2020, 24, 60. [Google Scholar] [CrossRef]
- Taneja, P.A.; Snyder, H.L.; de Feo, E.; Kruglyak, K.M.; Halks-Miller, M.; Curnow, K.J.; Bhatt, S. Noninvasive Prenatal Testing in the General Obstetric Population: Clinical Performance and Counseling Considerations in over 85,000 Cases. Prenat. Diagn. 2016, 36, 237–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vestergaard, E.M.; Singh, R.; Schelde, P.; Hatt, L.; Ravn, K.; Christensen, R.; Lildballe, D.L.; Petersen, O.B.; Uldbjerg, N.; Vogel, I. On the Road to Replacing Invasive Testing with Cell-Based NIPT: Five Clinical Cases with Aneuploidies, Microduplication, Unbalanced Structural Rearrangement, or Mosaicism. Prenat. Diagn. 2017, 37, 1120–1124. [Google Scholar] [CrossRef]
- Kølvraa, S.; Singh, R.; Normand, E.A.; Qdaisat, S.; van den Veyver, I.B.; Jackson, L.; Hatt, L.; Schelde, P.; Uldbjerg, N.; Vestergaard, E.M.; et al. Genome-Wide Copy Number Analysis on DNA from Fetal Cells Isolated from the Blood of Pregnant Women. Prenat. Diagn. 2016, 36, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Mouawia, H.; Saker, A.; Jais, J.-P.; Benachi, A.; Bussières, L.; Lacour, B.; Bonnefont, J.-P.; Frydman, R.; Simpson, J.L.; Paterlini-Brechot, P. Circulating Trophoblastic Cells Provide Genetic Diagnosis in 63 Fetuses at Risk for Cystic Fibrosis or Spinal Muscular Atrophy. Reprod. Biomed. Online 2012, 25, 508–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, M.; Hardy, T.; Rezaei, M.; Nguyen, V.; Zander-Fox, D.; Warkiani, M.E.; Thierry, B. Isolation of Circulating Fetal Trophoblasts Using Inertial Microfluidics for Noninvasive Prenatal Testing. Adv. Mater. Technol. 2018, 3, 1800066. [Google Scholar] [CrossRef]
- Kersaudy-Kerhoas, M.; Sollier, E. Micro-Scale Blood Plasma Separation: From Acoustophoresis to Egg-Beaters. Lab Chip 2013, 13, 3323–3346. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.-J.; Frank, J.F. Biological Aerosols: A Review of Airborne Contamination and Its Measurement in Dairy Processing Plants. J. Food Prot. 1989, 52, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Urbina, A.; Godoy-Silva, R.; Hoyos, M.; Camacho, M. Acute Hydrodynamic Damage Induced by SPLITT Fractionation and Centrifugation in Red Blood Cells. J. Chromatogr. B 2016, 1020, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Sollier, E.; Rostaing, H.; Pouteau, P.; Fouillet, Y.; Achard, J.-L. Passive Microfluidic Devices for Plasma Extraction from Whole Human Blood. Sens. Actuators B Chem. 2009, 141, 617–624. [Google Scholar] [CrossRef]
- Lee, M.G.; Shin, J.H.; Choi, S.; Park, J.-K. Enhanced Blood Plasma Separation by Modulation of Inertial Lift Force. Sens Actuators B Chem. 2014, 190, 311–317. [Google Scholar] [CrossRef]
- Tripathi, S.; Varun Kumar, Y.V.B.; Prabhakar, A.; Joshi, S.S.; Agrawal, A. Passive Blood Plasma Separation at the Microscale: A Review of Design Principles and Microdevices. J. Micromech. Microeng. 2015, 25, 083001. [Google Scholar] [CrossRef]
- Tripathi, S.; Kumar, Y.V.B.; Agrawal, A.; Prabhakar, A.; Joshi, S.S. Microdevice for Plasma Separation from Whole Human Blood Using Bio-Physical and Geometrical Effects. Sci. Rep. 2016, 6, 26749. [Google Scholar] [CrossRef] [PubMed]
- Xiang, N.; Ni, Z. High-Throughput Blood Cell Focusing and Plasma Isolation Using Spiral Inertial Microfluidic Devices. Biomed. Microdevices 2015, 17, 110. [Google Scholar] [CrossRef]
- Rafeie, M.; Zhang, J.; Asadnia, M.; Li, W.; Warkiani, M.E. Multiplexing Slanted Spiral Microchannels for Ultra-Fast Blood Plasma Separation. Lab Chip 2016, 16, 2791–2802. [Google Scholar] [CrossRef]
- Shen, S.; Tian, C.; Li, T.; Xu, J.; Chen, S.-W.; Tu, Q.; Yuan, M.-S.; Liu, W.; Wang, J. Spiral Microchannel with Ordered Micro-Obstacles for Continuous and Highly-Efficient Particle Separation. Lab Chip 2017, 17, 3578–3591. [Google Scholar] [CrossRef]
- Zhao, Q.; Yuan, D.; Tang, S.-Y.; Yun, G.; Yan, S.; Zhang, J.; Li, W. Top Sheath Flow-Assisted Secondary Flow Particle Manipulation in Microchannels with the Slanted Groove Structure. Microfluid Nanofluidics 2019, 23, 6. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.G.; Choi, S.; Kim, H.-J.; Lim, H.K.; Kim, J.-H.; Huh, N.; Park, J.-K. Inertial Blood Plasma Separation in a Contraction–Expansion Array Microchannel. Appl. Phys. Lett. 2011, 98, 253702. [Google Scholar] [CrossRef] [Green Version]
- Geng, Z.; Ju, Y.; Wang, W.; Li, Z. Continuous Blood Separation Utilizing Spiral Filtration Microchannel with Gradually Varied Width and Micro-Pillar Array. Sens Actuators B Chem 2013, 180, 122–129. [Google Scholar] [CrossRef]
- Shen, S.; Zhang, F.; Wang, S.; Wang, J.; Long, D.; Wang, D.; Niu, Y. Ultra-Low Aspect Ratio Spiral Microchannel with Ordered Micro-Bars for Flow-Rate Insensitive Blood Plasma Extraction. Sens. Actuators B Chem. 2019, 287, 320–328. [Google Scholar] [CrossRef]
- Wu, L.; Guan, G.; Hou, H.W.; Bhagat, A.S.; Han, J. Separation of Leukocytes from Blood Using Spiral Channel with Trapezoid Cross-Section. Anal. Chem. 2012, 84, 9324–9331. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, S.; Yuan, D.; Alici, G.; Nguyen, N.-T.; Li, W. High Throughput Cell-Free Extraction of Plasma by an Integrated Microfluidic Device Combining Inertial Focusing and Membrane. J. Heat Transf. 2017, 139, 052404. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and Function of Extracellular Vesicles in Cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Raab-Traub, N.; Dittmer, D.P. Viral Effects on the Content and Function of Extracellular Vesicles. Nat. Rev. Microbiol. 2017, 15, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Tigges, J.; Toxavidis, V.; Ericsson, M.; Distel, R.J.; Ivanov, A.R.; Skog, J.; Kuo, W.P. Alternative Methods for Characterization of Extracellular Vesicles. Front. Physiol. 2012, 3, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 2018, 1–27. [Google Scholar] [CrossRef]
- Zhou, J.; Mukherjee, P.; Gao, H.; Luan, Q.; Papautsky, I. Label-Free Microfluidic Sorting of Microparticles. APL Bioeng. 2019, 3, 041504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudani, J.S.; Gossett, D.R.; Tse, H.T.K.; Lamm, R.J.; Kulkarni, R.P.; Carlo, D.D. Rapid Inertial Solution Exchange for Enrichment and Flow Cytometric Detection of Microvesicles. Biomicrofluidics 2015, 9, 014112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gossett, D.R.; Tse, H.T.K.; Dudani, J.S.; Goda, K.; Woods, T.A.; Graves, S.W.; Di Carlo, D. Inertial Manipulation and Transfer of Microparticles Across Laminar Fluid Streams. Small 2012, 8, 2757–2764. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, Z.; Tayebi, M.; Ai, Y. Submicron Particle Focusing and Exosome Sorting by Wavy Microchannel Structures within Viscoelastic Fluids. Anal. Chem. 2019, 91, 4577–4584. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Guo, J.; Tian, F.; Yang, N.; Yan, F.; Ding, Y.; Wei, J.; Hu, G.; Nie, G.; Sun, J. Field-Free Isolation of Exosomes from Extracellular Vesicles by Microfluidic Viscoelastic Flows. ACS Nano 2017, 11, 6968–6976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Zhao, J.; Tian, F.; Chang, J.; Zhang, W.; Sun, J. λ-DNA- and Aptamer-Mediated Sorting and Analysis of Extracellular Vesicles. J. Am. Chem. Soc. 2019, 141, 3817–3821. [Google Scholar] [CrossRef]
- Tay, H.M.; Kharel, S.; Dalan, R.; Chen, Z.J.; Tan, K.K.; Boehm, B.O.; Loo, S.C.J.; Hou, H.W. Rapid Purification of Sub-Micrometer Particles for Enhanced Drug Release and Microvesicles Isolation. NPG Asia Mater. 2017, 9, e434. [Google Scholar] [CrossRef] [Green Version]
- Nozaki, T.; Sugiyama, S.; Koga, H.; Sugamura, K.; Ohba, K.; Matsuzawa, Y.; Sumida, H.; Matsui, K.; Jinnouchi, H.; Ogawa, H. Significance of a Multiple Biomarkers Strategy Including Endothelial Dysfunction to Improve Risk Stratification for Cardiovascular Events in Patients at High Risk for Coronary Heart Disease. J. Am. Coll. Cardiol. 2009, 54, 601–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtis, A.M.; Zhang, L.; Medenilla, E.; Gui, M.; Wilkinson, P.F.; Hu, E.; Giri, J.; Doraiswamy, V.; Gunda, S.; Burgert, M.E.; et al. Relationship of Microparticles to Progenitor Cells as a Measure of Vascular Health in a Diabetic Population. Cytom. Part B Clin. Cytom. 2010, 78B, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Pilecky, M.; Schildberger, A.; Orth-Höller, D.; Weber, V. Pathogen Enrichment from Human Whole Blood for the Diagnosis of Bloodstream Infection: Prospects and Limitations. Diagn. Microbiol. Infect. Dis. 2019, 94, 7–14. [Google Scholar] [CrossRef]
- Antfolk, M.; Laurell, T. Continuous Flow Microfluidic Separation and Processing of Rare Cells and Bioparticles Found in Blood—A Review. Anal. Chim. Acta 2017, 965, 9–35. [Google Scholar] [CrossRef]
- Ohlsson, P.; Evander, M.; Petersson, K.; Mellhammar, L.; Lehmusvuori, A.; Karhunen, U.; Soikkeli, M.; Seppä, T.; Tuunainen, E.; Spangar, A.; et al. Integrated Acoustic Separation, Enrichment, and Microchip Polymerase Chain Reaction Detection of Bacteria from Blood for Rapid Sepsis Diagnostics. Anal. Chem. 2016, 88, 9403–9411. [Google Scholar] [CrossRef] [Green Version]
- Faridi, M.A.; Ramachandraiah, H.; Banerjee, I.; Ardabili, S.; Zelenin, S.; Russom, A. Elasto-Inertial Microfluidics for Bacteria Separation from Whole Blood for Sepsis Diagnostics. J. Nanobiotechnol. 2017, 15, 3. [Google Scholar] [CrossRef]
- Wu, Z.; Willing, B.; Bjerketorp, J.; Jansson, J.K.; Hjort, K. Soft Inertial Microfluidics for High Throughput Separation of Bacteria from Human Blood Cells. Lab Chip 2009, 9, 1193–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, H.W.; Bhattacharyya, R.P.; Hung, D.T.; Han, J. Direct Detection and Drug-Resistance Profiling of Bacteremias Using Inertial Microfluidics. Lab Chip 2015, 15, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Mach, A.J.; Di Carlo, D. Continuous Scalable Blood Filtration Device Using Inertial Microfluidics. Biotechnol. Bioeng. 2010, 107, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.W.; Gan, H.Y.; Bhagat, A.A.S.; Li, L.D.; Lim, C.T.; Han, J. A Microfluidics Approach towards High-Throughput Pathogen Removal from Blood Using Margination. Biomicrofluidics 2012, 6, 024115. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, B.B.; Eatemadpour, S.; Martel-Foley, J.M.; Stott, S.; Toner, M.; Mylonakis, E. Rapid Isolation and Concentration of Pathogenic Fungi Using Inertial Focusing on a Chip-Based Platform. Front. Cell Infect. Microbiol. 2019, 9, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monard, C.; Rimmelé, T.; Ronco, C. Extracorporeal Blood Purification Therapies for Sepsis. Blood Purif. 2019, 47, 1–14. [Google Scholar] [CrossRef]
- Martel, J.M.; Smith, K.C.; Dlamini, M.; Pletcher, K.; Yang, J.; Karabacak, M.; Haber, D.A.; Kapur, R.; Toner, M. Continuous Flow Microfluidic Bioparticle Concentrator. Sci. Rep. 2015, 5, 11300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Le, J.; Chen, Y.; Cai, Y.; Hong, Z.; Chai, Y. Recent Advances in Microfluidic Devices for Bacteria and Fungus Research. Trends Analyt. Chem. 2019, 112, 175–195. [Google Scholar] [CrossRef]
- Xia, Y.; Tang, Y.; Yu, X.; Wan, Y.; Chen, Y.; Lu, H.; Zheng, S.-Y. Label-Free Virus Capture and Release by a Microfluidic Device Integrated with Porous Silicon Nanowire Forest. Small 2017, 13, 1603135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, K.; Ryu, H.; Siddle, K.J.; Piantadosi, A.; Freimark, L.; Park, D.J.; Sabeti, P.; Han, J. Negative Selection by Spiral Inertial Microfluidics Improves Viral Recovery and Sequencing from Blood. Anal. Chem. 2018, 90, 4657–4662. [Google Scholar] [CrossRef]
- Sarkar, A.; Hou, H.W.; Mahan, A.E.; Han, J.; Alter, G. Multiplexed Affinity-Based Separation of Proteins and Cells Using Inertial Microfluidics. Sci. Rep. 2016, 6, 23589. [Google Scholar] [CrossRef]
- Fook Kong, T.; Ye, W.; Peng, W.K.; Wei Hou, H.M.; Preiser, P.R.; Nguyen, N.-T.; Han, J. Enhancing Malaria Diagnosis through Microfluidic Cell Enrichment and Magnetic Resonance Relaxometry Detection. Sci. Rep. 2015, 5, 11425. [Google Scholar] [CrossRef] [Green Version]
- Geislinger, T.M.; Chan, S.; Moll, K.; Wixforth, A.; Wahlgren, M.; Franke, T. Label-Free Microfluidic Enrichment of Ring-Stage Plasmodium Falciparum-Infected Red Blood Cells Using Non-Inertial Hydrodynamic Lift. Malar. J. 2014, 13, 375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, H.W.; Bhagat, A.A.S.; Lin Chong, A.G.; Mao, P.; Wei Tan, K.S.; Han, J.; Lim, C.T. Deformability Based Cell Margination—A Simple Microfluidic Design for Malaria-Infected Erythrocyte Separation. Lab Chip 2010, 10, 2605–2613. [Google Scholar] [CrossRef] [PubMed]
- Warkiani, M.E.; Tay, A.K.P.; Khoo, B.L.; Xiaofeng, X.; Han, J.; Lim, C.T. Malaria Detection Using Inertial Microfluidics. Lab Chip 2015, 15, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.; Shin, Y.; Tan, J.K.S.; Lim, Y.B.; Lim, C.T.; Kim, S. High-Throughput Malaria Parasite Separation Using a Viscoelastic Fluid for Ultrasensitive PCR Detection. Lab Chip 2016, 16, 2086–2092. [Google Scholar] [CrossRef] [PubMed]
- Birch, C.M.; Hou, H.W.; Han, J.; Niles, J.C. Identification of Malaria Parasite-Infected Red Blood Cell Surface Aptamers by Inertial Microfluidic SELEX (I-SELEX). Sci. Rep. 2015, 5, 11347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.C.; Kang, J.S.; Lee, J.E.; Kim, S.S.; Jung, J.H. Continuous Aerosol Size Separator Using Inertial Microfluidics and Its Application to Airborne Bacteria and Viruses. Lab Chip 2015, 15, 1889–1897. [Google Scholar] [CrossRef]
- Lednicky, J.A.; Lauzardo, M.; Fan, Z.H.; Jutla, A.; Tilly, T.B.; Gangwar, M.; Usmani, M.; Shankar, S.N.; Mohamed, K.; Eiguren-Fernandez, A.; et al. Viable SARS-CoV-2 in the Air of a Hospital Room with COVID-19 Patients. Int. J. Infect. Dis. 2020, 100, 476–482. [Google Scholar] [CrossRef]
- Ganz, K.R.; Clime, L.; Farber, J.M.; Corneau, N.; Veres, T.; Dixon, B.R. Enhancing the Detection of Giardia Duodenalis Cysts in Foods by Inertial Microfluidic Separation. Appl. Environ. Microbiol. 2015, 81, 3925–3933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimenez, M.; Miller, B.; Bridle, H.L. Efficient Separation of Small Microparticles at High Flowrates Using Spiral Channels: Application to Waterborne Pathogens. Chem. Eng. Sci. 2017, 157, 247–254. [Google Scholar] [CrossRef] [Green Version]
- WHO Guidelines for Indoor Air Quality: Dampness and Mould; Heseltine, E.; Rosen, J.; World Health Organization (Eds.) WHO: Copenhagen, Denmark, 2009; ISBN 978-92-890-4168-3. [Google Scholar]
- Rao, C.Y.; Burge, H.A.; Chang, J.C.S. Review of Quantitative Standards and Guidelines for Fungi in Indoor Air. J. Air Waste Manag. Assoc. 1996, 46, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Lee, K.S.; Kim, S.S.; Bae, G.-N.; Jung, J.H. Real-Time Detection of an Airborne Microorganism Using Inertial Impaction and Mini-Fluorescent Microscopy. Lab Chip 2014, 14, 244–251. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Hong, S.C.; Kim, W.; Jung, J.H. Highly Enriched, Controllable, Continuous Aerosol Sampling Using Inertial Microfluidics and Its Application to Real-Time Detection of Airborne Bacteria. ACS Sens. 2017, 2, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kwon, D.; Choi, W.; Jung, G.Y.; Au, A.K.; Folch, A.; Jeon, S. 3D-Printed Microfluidic Device for the Detection of Pathogenic Bacteria Using Size-Based Separation in Helical Channel with Trapezoid Cross-Section. Sci. Rep. 2015, 5, 7717. [Google Scholar] [CrossRef] [PubMed]
- Clime, L.; Hoa, X.D.; Corneau, N.; Morton, K.J.; Luebbert, C.; Mounier, M.; Brassard, D.; Geissler, M.; Bidawid, S.; Farber, J.; et al. Microfluidic Filtration and Extraction of Pathogens from Food Samples by Hydrodynamic Focusing and Inertial Lateral Migration. Biomed. Microdevices 2015, 17, 17. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, M.; Bridle, H. Microfluidics for Effective Concentration and Sorting of Waterborne Protozoan Pathogens. J. Microbiol. Methods 2016, 126, 8–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raillon, C.; Che, J.; Thill, S.; Duchamp, M.; Desbiolles, B.X.E.; Millet, A.; Sollier, E.; Renaud, P. Toward Microfluidic Label-Free Isolation and Enumeration of Circulating Tumor Cells from Blood Samples. Cytom. A 2019, 95, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, M.; Hill, W.; Lee, J.H.; Hur, S.C. Microscale Symmetrical Electroporator Array as a Versatile Molecular Delivery System. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, H.-S.; Je, K.; Min, J.-W.; Park, D.; Han, K.-Y.; Shin, S.-H.; Park, W.-Y.; Eun Yoo, C.; Kim, S.-H. Inertial-Ordering-Assisted Droplet Microfluidics for High-Throughput Single-Cell RNA-Sequencing. Lab Chip 2018, 18, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Matuła, K.; Rivello, F.; Huck, W.T.S. Single-Cell Analysis Using Droplet Microfluidics. Adv. Biosyst. 2020, 4, 1900188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razavi Bazaz, S.; Rouhi, O.; Raoufi, M.A.; Ejeian, F.; Asadnia, M.; Jin, D.; Ebrahimi Warkiani, M. 3D Printing of Inertial Microfluidic Devices. Sci. Rep. 2020, 10, 5929. [Google Scholar] [CrossRef] [PubMed] [Green Version]

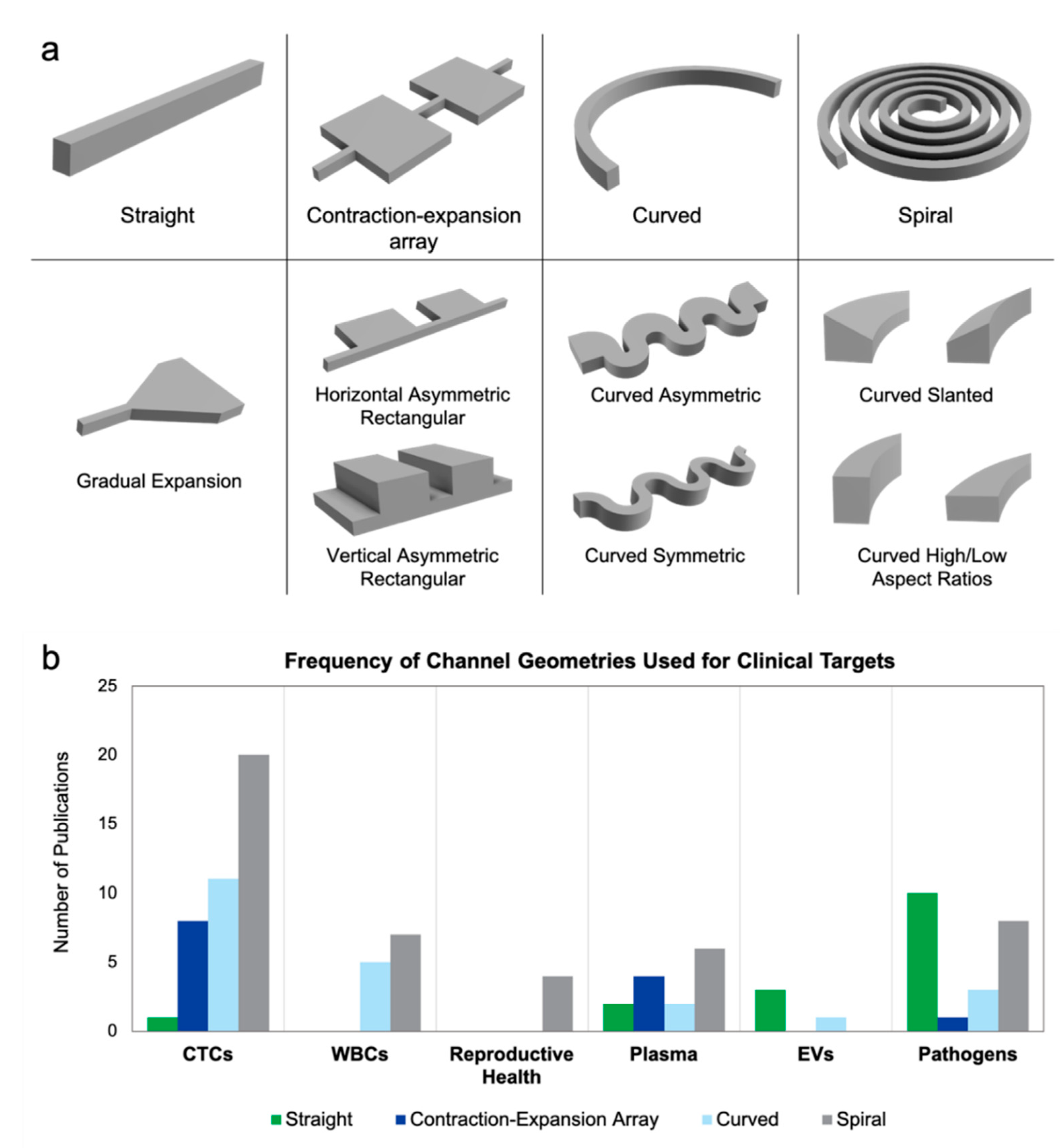

| Channel Geometry | Sample | Target | Dilution Factor | Throughput (min) * | Reference |
|---|---|---|---|---|---|
| Spiral | Tracheal secretions | Neutrophils | 1000× | 250 | [95] |
| Spiral | Blood | Granulocytes | 1000× | 250 | [94] |
| Spiral | Blood | Leukocytes | 10× | 76.9 | [96] |
| Spiral | Blood | Neutrophils and monocytes | 1000× | 6250 | [97] |
| Spiral | Blood | Neutrophils | 500× | 3125 | [98] |
| Spiral | Blood | Neutrophils | 5× | 50 | [99] |
| Spiral | Blood | Leukocytes | 14× | 5.83 | [100] |
| Spiral | Blood | All subpopulations | 10× | 10 | [101] |
| Asymmetrically curved | Blood | Neutrophils | N/A ** | N/A ** | [102] |
| Asymmetrically curved | Pleural fluid | Granulocytes | N/A ** | N/A ** | [103] |
| Channel Design | Plasma Purity | Dilution Factor * | Throughput ** | Extraction Efficiency | Reference |
|---|---|---|---|---|---|
| Contraction-expansion array in a straight channel | 92.60% | 1 a | 33.3 | 69.50% | [148] |
| Microchannel with the slanted groove structure | 99.90% | 1 a,b | 2 b | Not specified | [154] |
| Serpentine channel with a membrane filter | 100% | 20 | 7.1 | Not specified | [159] |
| Spiral microchannel | ~100% | 20 | 28.6 | 38.50% | [151] |
| Spiral microchannels with micropillar filters | Not clear c | 20 | 2000 | 53.90% | [156] |
| Spiral microchannel with a trapezoidal cross-section | 100% | 45 or 90 | 60 or 120 | Not specified | [152] |
| Spiral microchannel with microbar obstacles | 99.96% | 18 | 6 | 67.60% | [153] |
| Spiral channel with a low-aspect-ratio cross-section and microbar obstacles | 99.99% | 15 | 3-15 | 67.57% | [157] |
| Channel geometry | Target | Dilution Factor | Throughput * | Pathogen load (/mL) | Separation Efficiency | Purity | Reference |
|---|---|---|---|---|---|---|---|
| straight | E. coli | undiluted | 33 hr | 106 | 76% | 92% | [176] |
| straight | E. coli | 20× | 18 hr | 1.6x107 | 62% | 99.71% | [177] |
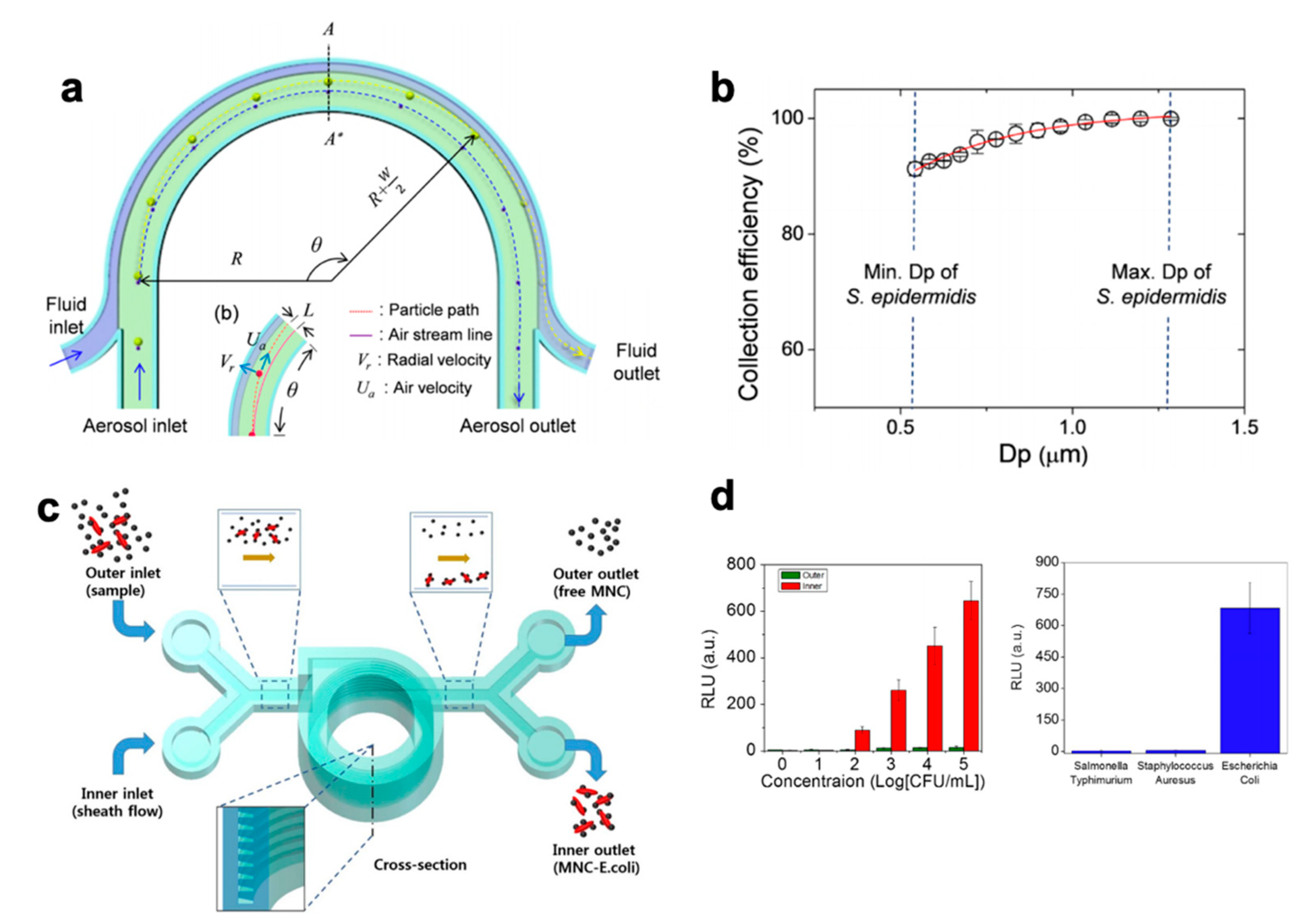
| straight | E. coli | 90× | 11.25 min | 108 | 80% | Not specified | [179] |
| straight | E. coli and S. cerevisiae | undiluted | 10 min | 106–107 | 80% (E. coli), 90% (S. cerevisiae) | Not specified | [180] |
| spiral | C. albicans/Candida | 10× | 25 min | 1600 | 44.60% | Not specified | [181] |
| spiral | E. coli, K. pneumoniae, P.aeruginosa, and S. aureus | 3× | 20 min | 102–104 | >65% | Not specified | [178] |
| Channel Shape | Sample | Hematocrit | Flow Rate (µL/min) | Pathogen Load | Parasite Enrichment Factor | Separation Efficiency | Reference |
|---|---|---|---|---|---|---|---|
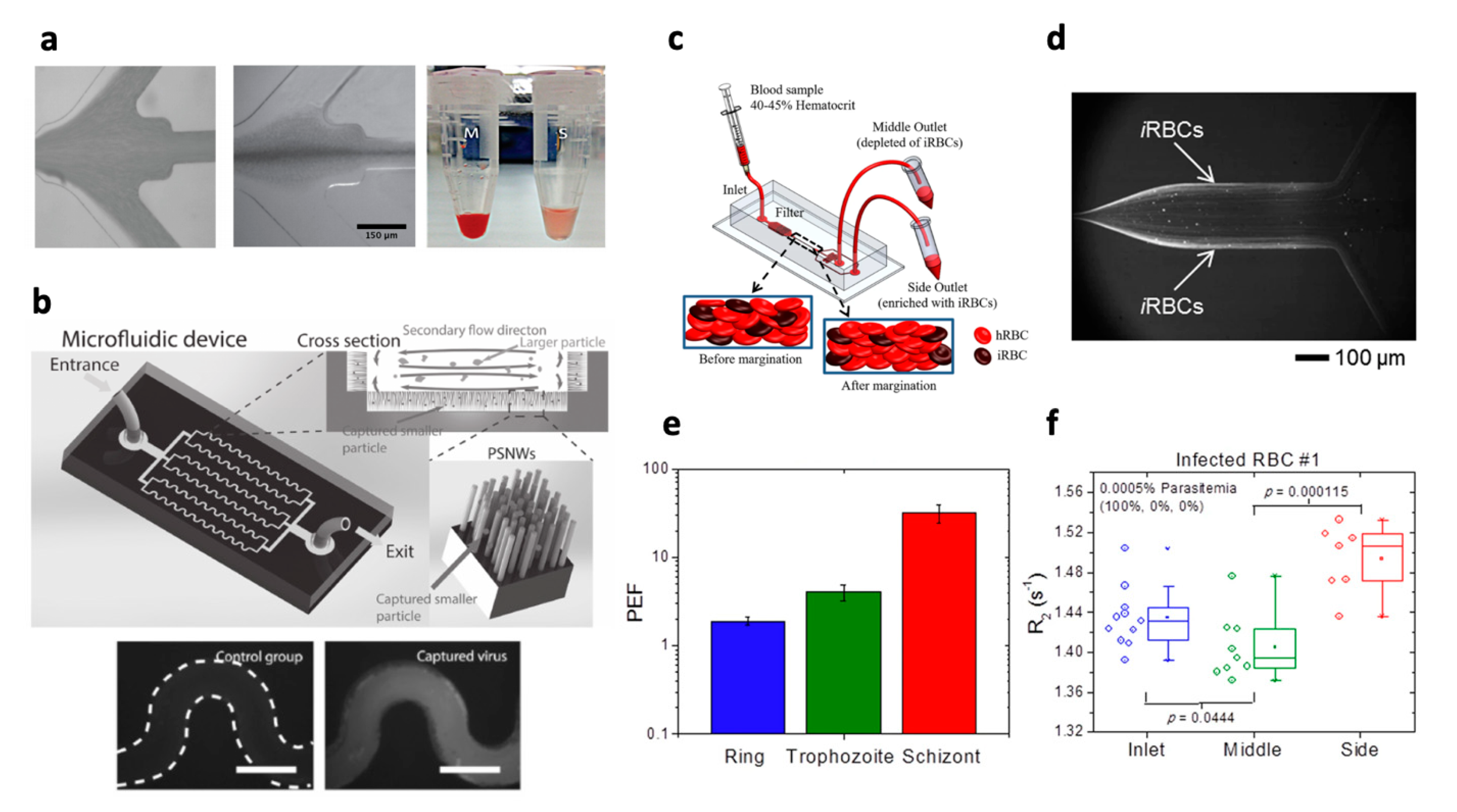
| straight | P. falciparum iRBCs (all stages) | 45% | 10 | 0.0005–5% parasitemia | 1.9 (ring) 4.1 (trophozoite) 32.1 (schizont) | Not specified | [188] |
| straight | P. falciparum | 45% | 400 | 500 /mL | 6.4 | 94% | [192] |
| straight | P. falciparum iRBCs (all stages) | 40% | 5 | 0.01–1% parasitemia | 2 | 75% (ring) >90% (trophozoite/schizont) | [190] |
| straight | P. falciparum iRBCs (ring stage) | 2% | 0.67 | 1.6–3.4% parasitemia | 4.3 | Not specified | [189] |
| CEA | P. falciparum (ring stage) | ~10% | 400 | 103–104 /mL | Not specified | 70.9% | [191] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalyan, S.; Torabi, C.; Khoo, H.; Sung, H.W.; Choi, S.-E.; Wang, W.; Treutler, B.; Kim, D.; Hur, S.C. Inertial Microfluidics Enabling Clinical Research. Micromachines 2021, 12, 257. https://doi.org/10.3390/mi12030257
Kalyan S, Torabi C, Khoo H, Sung HW, Choi S-E, Wang W, Treutler B, Kim D, Hur SC. Inertial Microfluidics Enabling Clinical Research. Micromachines. 2021; 12(3):257. https://doi.org/10.3390/mi12030257
Chicago/Turabian StyleKalyan, Srivathsan, Corinna Torabi, Harrison Khoo, Hyun Woo Sung, Sung-Eun Choi, Wenzhao Wang, Benjamin Treutler, Dohyun Kim, and Soojung Claire Hur. 2021. "Inertial Microfluidics Enabling Clinical Research" Micromachines 12, no. 3: 257. https://doi.org/10.3390/mi12030257
APA StyleKalyan, S., Torabi, C., Khoo, H., Sung, H. W., Choi, S.-E., Wang, W., Treutler, B., Kim, D., & Hur, S. C. (2021). Inertial Microfluidics Enabling Clinical Research. Micromachines, 12(3), 257. https://doi.org/10.3390/mi12030257






