Raman Mapping of Biological Systems Interacting with a Disordered Nanostructured Surface: A Simple and Powerful Approach to the Label-Free Analysis of Single DNA Bases
Abstract
1. Introduction
2. Materials and Methods
2.1. Ag/SiNWs Fabrication
2.2. Morphological Characterisation of Ag/SiNWs
2.3. Single-Base Sample Preparation
2.4. Raman Mapping
2.5. Principal Component Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jain, K.K. A Handbook of Nanomedicina; Humana/Springer: Totowa, NJ, USA, 2008. [Google Scholar]
- Wong, X.Y.; Sena-Torralba, A.; Álvarez-Diduk, R.; Muthoosamy, K.; Merkoçi, A. Nanomaterials for Nanotheranostics: Tuning Their Properties According to Disease Needs. ACS Nano 2020, 14, 2585–2627. [Google Scholar] [CrossRef]
- Bathh, I.S.; Mitra, A.; Manier, S. Circulating tumor markers: Harmonizing the yin and yang of CTCs and ctDNA for precision medicine. Ann. Oncol. 2017, 28, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 1–10. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Schuster, S.C. Next-generation sequencing transforms today’s biology. Nat. Methods 2008, 5, 16–18. [Google Scholar] [CrossRef]
- Hoorfar, J.; Wolffs, P.; Rådstro, P. Diagnostic PCR: Validation and sample preparation are two sides of the same coin. APMIS 2004, 112, 808–814. [Google Scholar] [CrossRef]
- Mullis, K.B.; Faloona, F.A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987, 155, 335–350. [Google Scholar]
- Patolsky, F.; Lieber, C.M. Nanowire nanosensors. Mater. Today 2005, 8, 20–28. [Google Scholar] [CrossRef]
- Bunimovich, Y.L.; Shin, Y.S.; Yeo, W.-S.; Amori, M.; Kwong, G.; Heath, J.R. Quantitative Real-Time Measurements of DNA Hybridization with Alkylated Nonoxidized Silicon Nanowires in Electrolyte Solution. J. Am. Chem. Soc. 2006, 128, 16323–16331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-J.; Zhang, G.; Chua, J.H.; Chee, R.-E.; Wong, E.H.; Agarwal, A.; Buddharaju, K.D.; Singh, N.; Gao, Z.; Balasubramanian, N. DNA Sensing by Silicon Nanowire: Charge Layer Distance Dependence. Nanoletters 2008, 8, 1066–1070. [Google Scholar] [CrossRef]
- Gao, Z.; Agarwal, A.; Trigg, A.D.; Singh, N.; Fang, C.-H.; Tung, C.-H.; Fan, Y.; Buddharaju, K.D.; Kong, J.N. Silicon Nanowire Arrays for Label-Free Detection of DNA. Anal. Chem. 2007, 79, 3291–3297. [Google Scholar] [CrossRef]
- Leonardi, A.A.; Lo Faro, M.J.; Petralia, S.; Fazio, B.; Musumeci, P.; Conoci, S.; Irreara, A.; Priolo, F. Ultrasensitive Label- and PCR-Free Genome Detection Based on Cooperative Hybridization of Silicon Nanowires Optical Biosensors. ACS Sens. 2018, 3, 1690–1697. [Google Scholar] [CrossRef]
- Convertino, A.; Mussi, V.; Maiolo, L. Disordered array of Au covered silicon nanowires for SERS biosensing combined with electrochemical detection. Sci. Rep. 2016, 6, 25099. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.H.; Wang, H.S.; Lu, L.H.; Ai, K.L.; Zhang, G.; Cheng, X.L. Large-area silver-coated silicon nanowire arrays for molecular sensing using surface-enhanced Raman spectroscopy. Adv. Funct. Mater. 2008, 18, 2348–2355. [Google Scholar] [CrossRef]
- Galopin, E.; Barbillat, J.; Coffinier, Y.; Szunerits, S.; Patriarche, G.; Boukherroub, R. Silicon nanowires coated with silver nanostructures as ultrasensitive interfaces for surface-enhanced Raman spectroscopy. ACS Appl. Mater. Interfaces 2009, 7, 1396–1403. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.L.; Fan, X.; Zhou, H.W.; Shao, M.W.; Zapien, J.A.; Wong, N.B.; Lee, S.T. A High efficiency surface-enhanced Raman scattering substrate based silicon nanowires array decorated with silver nanoparticles. J. Phys. Chem. C 2010, 114, 1969–1975. [Google Scholar] [CrossRef]
- Paria, D.; Convertino, A.; Mussi, V.; Maiolo, L.; Barman, I. A Silver-coated disordered silicon nanowires provide highly sensitive label-free glycated albumin detection through molecular trapping and plasmonic hotspot formation. Adv. Healthc. Mater. 2021, 10, 2001110. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.S.; Hübner, J.; Boisen, A. Large area fabrication of leaning silicon nanopillars for surface enhanced Raman Spectroscopy. Adv. Mater. 2012, 24, OP11. [Google Scholar] [CrossRef]
- Mussi, V.; Ledda, M.; Polese, D.; Maiolo, L.; Paria, D.; Barman, I.; Lolli, M.G.; Lisi, A.; Convertino, A. Silver-coated silicon nanowire platform discriminates genomic DNA from normal and malignant human epithelial cells using label-free Raman spectroscopy. Mater. Sci. Eng. C 2021, 122, 111951. [Google Scholar] [CrossRef] [PubMed]
- Convertino, A.; Mussi, V.; Maiolo, L.; Ledda, M.; Lolli, M.G.; Bovino, F.; Fortunato, G.; Rocchia, M.; Lisi, A. Array of disordered silicon nanowires coated by a gold film for combined NIR photothermal treatment of cancer cells and Raman monitoring of the process evolution. Nanotechnology 2018, 29, 415102. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Ng, J.K.; Kunitake, M.E.; Conklin, B.R.; Yang, P. A Interfacing Silicon Nanowires with Mammalian Cells. J. Am. Chem. Soc. 2007, 129, 7228. [Google Scholar] [CrossRef] [PubMed]
- Maiolo, L.; Polese, D.; Pecora, A.; Fortunato, G.; Shacham-Diamand, Y.; Convertino, A. Highly disordered array of silicon nanowires: An effective and scalable approach for performing and flexible electrochemical biosensors. Adv. Healthc. Mater. 2016, 5, 575–583. [Google Scholar] [CrossRef]
- Convertino, A.; Cuscunà, M.; Nicotra, G.; Spinella, C.; Felisari, L.; Fortunato, G.; Martell, F. Low-temperature growth of In-assisted silicon nanowires. J. Cryst. Growth 2011, 335, 10–16. [Google Scholar] [CrossRef]
- Otto, C.; van den Tweel, T.J.J.; de Mu, F.F.M.; Greve, J. Surface-enhanced Raman spectroscopy of DNA bases. J. Raman Spectrosc. 1986, 17, 289–298. [Google Scholar] [CrossRef]
- Madzharova, F.; Heiner, Z.; Guhlke, M.; Kneipp, J. Surface-Enhanced Hyper-Raman Spectra of Adenine, Guanine, Cytosine, Thymine, and Uracil. J. Phys. Chem. C 2016, 120, 15415–15423. [Google Scholar] [CrossRef]
- Suh, J.S.; Moskovits, M. Surface-Enhanced Raman Spectroscopy of Amino Acids and Nucleotide Bases Adsorbed on Silver. J. Am. Chem. Soc. 1986, 108, 4711–4718. [Google Scholar] [CrossRef]
- Huang, J.-A.; Mousavi, M.Z.; Zhao, Y.; Hubarevich, A.; Omeis, F.; Giovannini, G.; Schütte, M.; Garoli, D.; De Angelis, F. SERS discrimination of single DNA bases in single oligonucleotides by electro-plasmonic trapping. Nat. Commun. 2019, 10, 5321. [Google Scholar] [CrossRef]
- Venkatesan, S.; Erdheim, G.; Lombardi, J.R.; Birke, R.L. Voltage dependence of the surface molecule line in the enhanced Raman spectrum of several nitrogen containing compounds. Surf. Sci. 1980, 101, 387–398. [Google Scholar] [CrossRef]
- Creighton, J.A.; Albrecht, M.G.; Hester, R.E.; Matthew, J.A.D. The dependence of the intensity of Raman bands of pyridine at a silver electrode on the wavelength of excitation. Chem. Phys. Lett. 1978, 55, 55–58. [Google Scholar] [CrossRef]
- Giese, B.; McNaughton, D. Surface-enhanced Raman spectroscopic and density functional theory study of adenine adsorption to silver surfaces. J. Phys. Chem. B 2002, 106, 101–112. [Google Scholar] [CrossRef]
- Jang, N.H. The coordination chemistry of DNA nucleotides on gold nanoparticles as a probe by SERS. Bull. Korean Chem. Soc. 2002, 23, 1790–1800. [Google Scholar]
- Panikkanvalappil, S.R.; Mackey, M.A.; El-Sayed, M.A. Probing the Unique Dehydration-Induced Structural Modifications in Cancer Cell DNA Using Surface Enhanced Raman Spectroscopy. J. Am. Chem. Soc. 2013, 135, 4815–4821. [Google Scholar] [CrossRef] [PubMed]

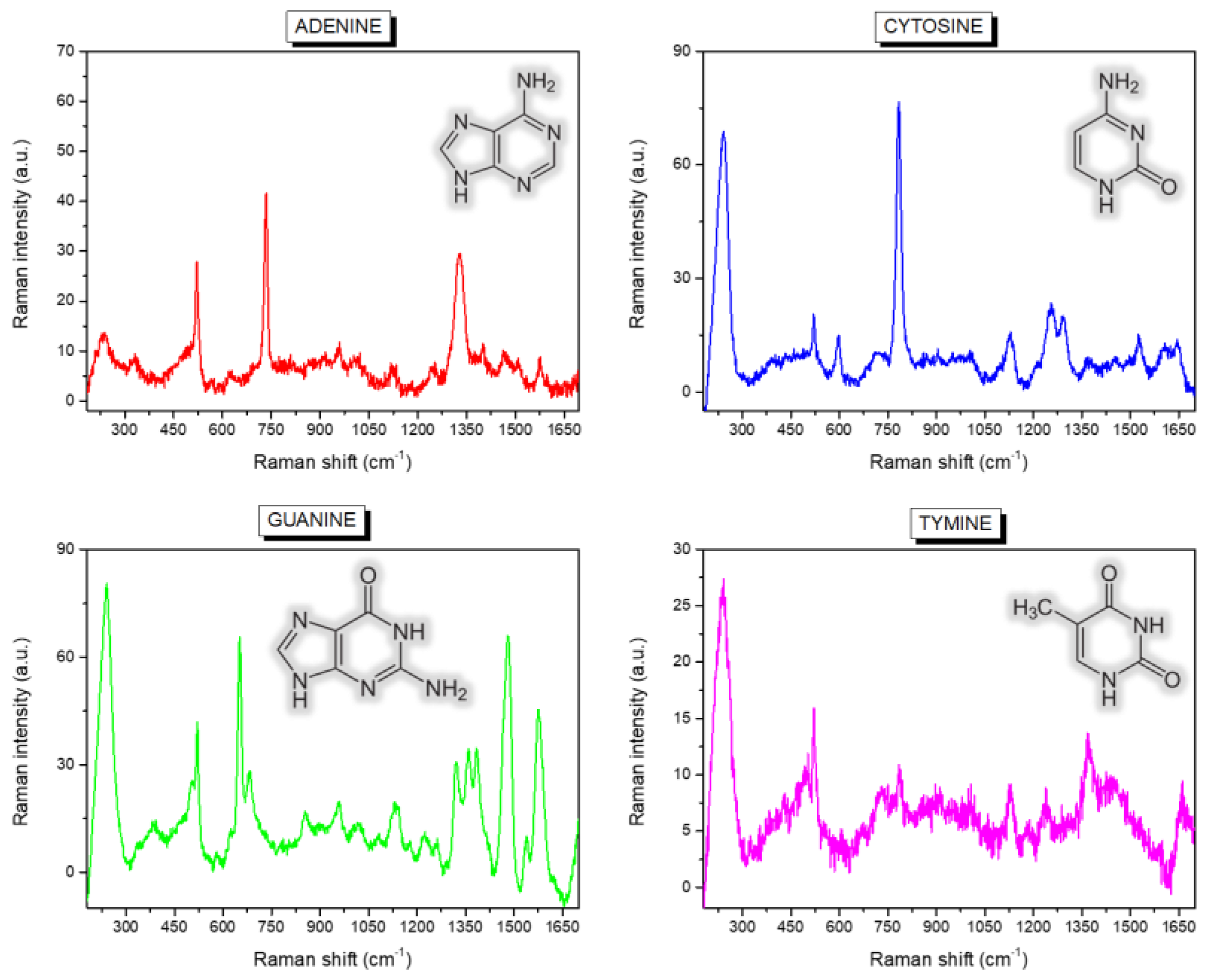



| Band Position (cm−1) | Assignment |
|---|---|
| 735 | A, Ring breath |
| 1329 | A, C-N str, C-H bend |
| 651 | G, Ring str |
| 959 | G, 5-ring def |
| 1322 | G, ring str C-N, C-C str, C-H bend, NH2 rock |
| 1360 | G, N-H bend, C-N str |
| 1384 | G, ring str C-N, C-C str, NH2 rock, N-H bend |
| 1460 | G, ring str C-N, C-H and N-H bend |
| 1577 | G, NH2 sciss, C-N str |
| 787 | C, ring breath |
| 1248 | C, ring str C-N |
| 1291 | C, ring str C-N |
| 1524 | C, N-C str |
| 776 | T, ring breath |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mussi, V.; Ledda, M.; Convertino, A.; Lisi, A. Raman Mapping of Biological Systems Interacting with a Disordered Nanostructured Surface: A Simple and Powerful Approach to the Label-Free Analysis of Single DNA Bases. Micromachines 2021, 12, 264. https://doi.org/10.3390/mi12030264
Mussi V, Ledda M, Convertino A, Lisi A. Raman Mapping of Biological Systems Interacting with a Disordered Nanostructured Surface: A Simple and Powerful Approach to the Label-Free Analysis of Single DNA Bases. Micromachines. 2021; 12(3):264. https://doi.org/10.3390/mi12030264
Chicago/Turabian StyleMussi, Valentina, Mario Ledda, Annalisa Convertino, and Antonella Lisi. 2021. "Raman Mapping of Biological Systems Interacting with a Disordered Nanostructured Surface: A Simple and Powerful Approach to the Label-Free Analysis of Single DNA Bases" Micromachines 12, no. 3: 264. https://doi.org/10.3390/mi12030264
APA StyleMussi, V., Ledda, M., Convertino, A., & Lisi, A. (2021). Raman Mapping of Biological Systems Interacting with a Disordered Nanostructured Surface: A Simple and Powerful Approach to the Label-Free Analysis of Single DNA Bases. Micromachines, 12(3), 264. https://doi.org/10.3390/mi12030264






