Recent Advances in Microfluidic Devices for Contamination Detection and Quality Inspection of Milk
Abstract
:1. Introduction
2. Sample Pretreatment of Microfluidic Devices
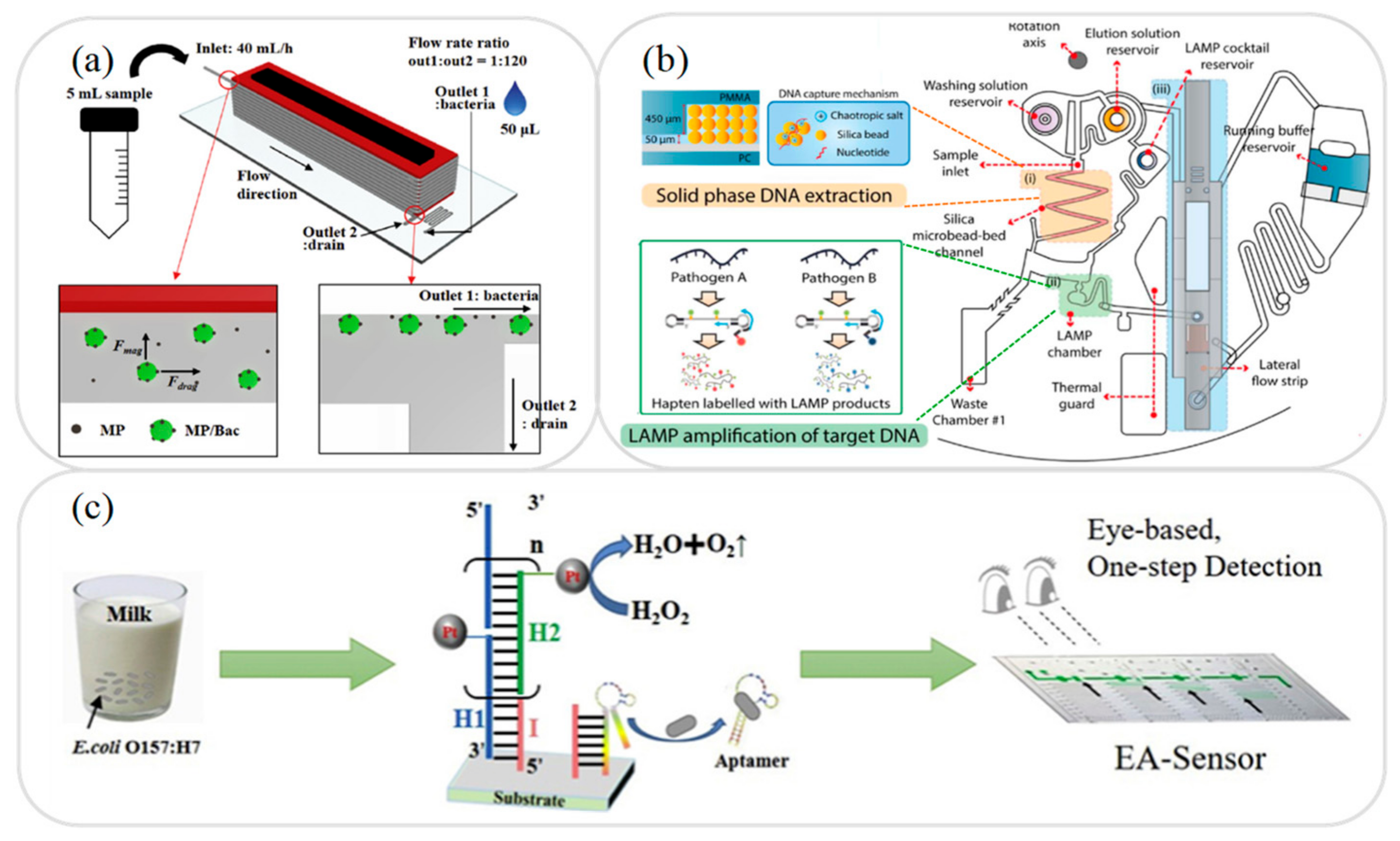
2.1. Sample Separation Microfluidic Devices
2.2. Sample Extraction Microfluidic Devices
2.3. Sample Amplification Microfluidic Devices
3. Microfluidic Platforms for Milk Sample Analysis
3.1. Aflatoxin Analysis
3.2. Melamine Analysis
3.3. Antibiotic and Drug Analysis
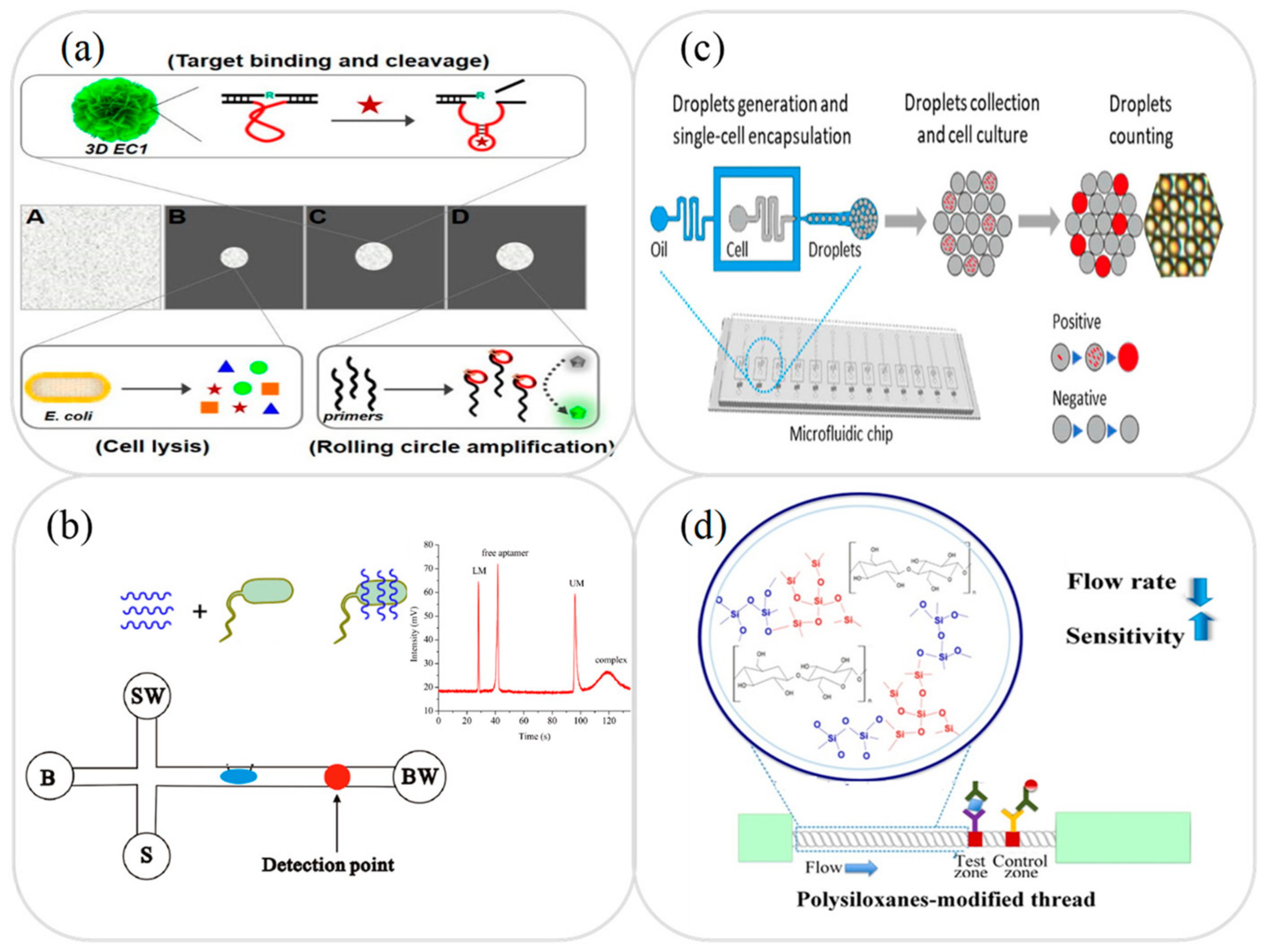
3.4. Foodborne Pathogens Analysis
3.5. Other Analysis and Applications
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wochner, K.F.; Becker-Algeri, T.A.; Colla, E.; Badiale-Furlong, E.; Drunkler, D.A. The action of probiotic microorganisms on chemical contaminants in milk. Crit. Rev. Microbiol. 2018, 44, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.Y.; Liu, C.C.; Yang, C.E.; Fu, L.M. Multifunctional microchip-based distillation apparatus I-Steam distillation for formaldehyde detection. Anal. Chim. Acta 2019, 1062, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Kung, C.T.; Gao, H.; Lee, C.Y.; Wang, Y.N.; Dong, W.; Ko, C.H.; Wang, G.; Fu, L.M. Microfluidic synthesis control technology and its application in drug delivery, bioimaging, biosensing, environmental analysis and cell analysis. Chem. Eng. J. 2020, 399, 125748. [Google Scholar] [CrossRef]
- Wu, Y.T.; Yang, C.E.; Ko, C.H.; Wang, Y.N.; Liu, C.C.; Fu, L.M. Microfluidic detection platform with integrated micro-spectrometer system. Chem. Eng. J. 2020, 393, 124700. [Google Scholar] [CrossRef]
- Khan, H.; Choi, J.H.; Ullah, A.; Kim, Y.H.; Kim, G.M. Continuous determination of glucose using a membraneless, microfluidic enzymatic biofuel cell. Micromachines 2020, 11, 1129. [Google Scholar]
- Zhang, Y.; Yang, N.; Xie, L.; Shu, F.; Shi, Q.; Shaheen, N. A New 3D cultured liver chip and real-time monitoringsystem based on microfluidic technology. Micromachines 2020, 11, 1118. [Google Scholar]
- Loessberg-Zahl, J.; Beumer, J.; van den Berg, A.; Eijkel, J.C.T.; van der Meer, A.D. Patterning biological gels for 3D cell culture inside microfluidic devices by local surface modification through Laminar Flow Patterning. Micromachines 2020, 11, 1112. [Google Scholar]
- Li, W.; Sun, X.; Ji, B.; Yang, X.; Zhou, B.; Lu, Z.; Gao, X. PLGA nanofiber/PDMS microporous composite membrane-sandwiched microchip for drug testing. Micromachines 2020, 11, 1054. [Google Scholar] [CrossRef]
- Fu, L.M.; Liu, C.C.; Yang, C.E.; Wang, Y.N.; Ko, C.H. A PET/paper chip platform for high resolution sulphur dioxide detection in foods. Food Chem. 2019, 286, 316–321. [Google Scholar] [CrossRef]
- Kung, C.T.; Hou, C.Y.; Wang, Y.N.; Fu, L.M. Microfluidic paper-based analytical devices for environmental analysis of soil, air, ecology and river water. Sens. Actuators B Chem. 2019, 301, 126855. [Google Scholar] [CrossRef]
- Soum, V.; Kim, Y.; Park, S.; Chuong, M.; Ryu, S.R.; Lee, S.H.; Tanev, G.; Madsen, J.; Kwon, O.-S.; Shin, K. Affordable fabrication of conductive electrodes and dielectric films for a paper-based digital microfluidic chip. Micromachines 2019, 10, 109. [Google Scholar] [CrossRef] [Green Version]
- Raj, N.; Breedveld, V.; Hess, D.W. Flow control in fully enclosed microfluidics paper based analytical devices using plasma processes. Sens. Actuators B Chem. 2020, 320, 128606. [Google Scholar] [CrossRef]
- Kalish, B.; Tan, M.K.; Tsutsui, H. Modifying wicking speeds in paper-based microfluidic devices by laser-etching. Micromachines 2020, 11, 773. [Google Scholar] [CrossRef]
- Amin, R.; Ghaderinezhad, F.; Bridge, C.; Temirel, M.; Jones, S.; Toloueinia, P.; Tasoglu, S. Pushing the limits of spatial assay resolution for paper-based microfluidics using low-cost and high-throughput pen plotter approach. Micromachines 2020, 11, 611. [Google Scholar] [CrossRef]
- Ko, C.H.; Liu, C.C.; Chen, K.H.; Sheu, F.; Fu, L.M.; Chen, S.J. Microfluidic colorimetric analysis system for sodium benzoate detection in foods. Food Chem. 2021, 345, 128773. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.C.; Ko, C.H.; Lu, S.Y.; Yang, C.E.; Fu, L.M.; Li, C.Y. Rapid electrochemical-biosensor microchip platform for determination of microalbuminuria in CKD patients. Anal. Chim. Acta 2021, 1146, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Fu, L.M. Recent advances and applications of micromixers. Sens. Actuators B Chem. 2018, 259, 677–702. [Google Scholar] [CrossRef]
- Li, Y.; Li, D.; Zhao, P.; Nandakumar, K.; Wang, L.; Song, Y. Microfluidics-based systems in diagnosis of Alzheimer’s disease and biomimetic modeling. Micromachines 2020, 11, 787. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Liu, Y.; Zhao, Y.; Zhang, L.; Zhang, L.; Mao, H.; Huang, C. Nanotechnology-assisted isolation and analysis of circulating tumor cells on microfluidic devices. Micromachines 2020, 11, 774. [Google Scholar] [CrossRef]
- Wu, S.; Wang, X.; Li, Z.; Zhang, S.; Xing, F. Recent advances in the fabrication and application of graphene microfluidic aensors. Micromachines 2020, 11, 1059. [Google Scholar] [CrossRef] [PubMed]
- Soum, V.; Park, S.; Brilian, A.I.; Kwon, O.-S.; Shin, K. Programmable paper-based microfluidic devices for biomarker detections. Micromachines 2019, 10, 516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaumburg, F.; Kler, P.A.; Berli, C.L.A. Comprehensive model of electromigrative transport in microfluidic paper based analytical devices. Electrophoresis 2020, 41, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Hahn, Y.K.; Kim, M.S. Recent advances of fluid manipulation technologies in microfluidic paper-based analytical devices (μPADs) toward multi-step assays. Micromachines 2020, 11, 269. [Google Scholar] [CrossRef] [Green Version]
- Tseng, C.C.; Kung, C.T.; Chen, R.F.; Tsai, M.H.; Chao, H.R.; Wang, Y.N.; Fu, L.M. Recent advances in microfluidic paper-based assay devices for diagnosis of human diseases using saliva, tears and sweat samples. Sens. Actuators B Chem. 2021, 342, 130078. [Google Scholar] [CrossRef]
- Fu, L.M.; Wang, Y.N. Detection methods and applications of microfluidic paper-based analytical devices. TrAC Trends Anal. Chem. 2018, 107, 196–211. [Google Scholar] [CrossRef]
- Guo, S.; Duan, X.; Xie, M.; Aw, K.C.; Xue, Q. Composites, fabrication and application of polyvinylidene fluoride for flexible electromechanical devices: A review. Micromachines 2020, 11, 1076. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, X.; Wang, J.; Wang, C.; Yan, Y.; Wu, A.; Ren, Y. Public-health-driven microfluidic technologies: From separation to detection. Micromachines 2021, 12, 391. [Google Scholar]
- Zhou, W.; Le, J.; Chen, Y.; Cai, Y.; Hong, Z.; Chai, Y. Recent advances in microfluidic devices for bacteria and fungus research. TrAC Trends Anal. Chem. 2019, 112, 175–195. [Google Scholar] [CrossRef]
- Khan, N.I.; Song, E. Lab-on-a-chip systems for aptamer-based biosensing. Micromachines 2020, 11, 220. [Google Scholar] [CrossRef] [Green Version]
- Tseng, C.C.; Yang, R.J.; Ju, W.J.; Fu, L.M. Microfluidic paper-based platform for whole blood creatinine detection. Chem. Eng. J. 2018, 348, 117–124. [Google Scholar] [CrossRef]
- Dayao, L.A.N.; Liu, C.C.; Hsu, S.Y.; Tayo, L.L.; Ju, W.J.; Fu, L.M. Multifunctional microchip-based distillation apparatus II-Aerated distillation for sulfur dioxide detection. Anal. Chim. Acta 2019, 1071, 44–52. [Google Scholar] [CrossRef]
- Hou, C.Y.; Fu, L.M.; Ju, W.J.; Wu, P.Y. Microfluidic colorimetric system for nitrite detection in foods. Chem. Eng. J. 2021, 398, 125573. [Google Scholar] [CrossRef]
- Singh, R.; Brockgreitens, J.; Saiapina, O.; Wu, Y.; Abbas, A. Microbial separation from a complex matrix by a hand-held microfluidic device. Chem. Commun. 2017, 53, 10788–10791. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Shen, Z.; Wang, J.; Zeng, J.; Wang, W.; Wu, H.; Wang, Q.; Gan, N. Simultaneously responsive microfluidic chip aptasensor for determination of kanamycin, aflatoxin M1, and 17β-estradiol based on magnetic tripartite DNA assembly nanostructure probes. Microchim. Acta 2020, 187, 176. [Google Scholar] [CrossRef]
- Zhao, Y.; Tang, M.; Liu, F.; Li, H.; Wang, H.; Xu, D. Highly integrated microfluidic chip coupled to mass spectrometry for online analysis of residual quinolones in milk. Anal. Chem. 2019, 91, 13418–13426. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; Jung, Y.; Ahn, J.; Yang, S. Continuous, rapid concentration of foodborne bacteria (Staphylococcus aureus, Salmonella typhimurium, and Listeria monocytogenes) using magnetophoresis-based microfluidic device. Food Control 2020, 114, 107229. [Google Scholar] [CrossRef]
- He, L.; Shen, Z.; Cao, Y.; Li, T.; Wu, D.; Dong, Y.; Gan, N. A microfluidic chip based ratiometric aptasensor for antibiotic detection in foods using stir bar assisted sorptive extraction and rolling circle amplification. Analyst 2019, 144, 2755–2764. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, Y.T.; Lee, S.J.; Lee, E.; Lee, K.G.; Im, S.G. Direct solvent-free modification of the inner wall of the microchip for rapid DNA extraction with enhanced capturing efficiency. Macromol. Res. 2020, 28, 249–256. [Google Scholar] [CrossRef]
- Park, B.H.; Oh, S.J.; Jung, J.H.; Choi, G.; Seo, J.H.; Lee, E.Y.; Seo, T.S. An integrated rotary microfluidic system with DNA extraction, loop-mediated isothermal amplification, and lateral flow strip based detection for point-of-care pathogen diagnostics. Biosensors Bioelectron. 2017, 91, 334–340. [Google Scholar] [CrossRef]
- Tang, R.; Yang, H.; Gong, Y.; You, M.; Liu, Z.; Choi, J.R.; Wen, T.; Qu, Z.; Mei, Q.; Xu, F. A fully disposable and integrated paper-based device for nucleic acid extraction, amplification and detection. Lab Chip 2017, 17, 1270–1279. [Google Scholar] [CrossRef]
- Yin, J.; Zou, Z.; Hu, Z.; Zhang, S.; Zhang, F.; Wang, B.; Lv, S.; Mu, Y. A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens. Lab Chip 2020, 20, 979–986. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, F.; Cai, G.; Li, Y.; Lin, J. Rapid and sensitive detection of Escherichia coli O157: H7 using coaxial channel-based DNA extraction and microfluidic PCR. J. Dairy Sci. 2018, 101, 9736–9746. [Google Scholar] [CrossRef]
- Zhou, L.; Gan, N.; Hu, F.; Li, T.; Cao, Y.; Wu, D. Microchip electrophoresis array-based aptasensor for multiplex antibiotic detection using functionalized magnetic beads and polymerase chain reaction amplification. Sens. Actuators B Chem. 2018, 263, 568–574. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, Y.; He, P.; Zhang, Y.; Wang, Q. A multiplex PCR amplification strategy coupled with microchip electrophoresis for simultaneous and sensitive detection of three foodborne bacteria. J. Chromatogr. B 2018, 1093, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Nguyen, V.D.; Seo, T.S. Paper-based molecular diagnostics for the amplification and detection of pathogenic bacteria from human whole blood and milk without a sample preparation step. BioChip J. 2019, 13, 243–250. [Google Scholar] [CrossRef]
- Tsougeni, K.; Kastania, A.; Kaprou, G.; Eck, M.; Jobst, G.; Petrou, P.; Kakabakos, S.; Mastellos, D.; Gogolides, E.; Tserepi, A. A modular integrated lab-on-a-chip platform for fast and highly efficient sample preparation for foodborne pathogen screening. Sens. Actuators B Chem. 2019, 288, 171–179. [Google Scholar] [CrossRef]
- Li, T.; Ou, G.; Chen, X.; Li, Z.; Hu, R.; Li, Y.; Yang, Y.; Liu, M. Naked-eye based point-of-care detection of E. coli O157: H7 by a signal-amplified microfluidic aptasensor. Anal. Chim. Acta 2020, 1130, 20–28. [Google Scholar] [CrossRef]
- Luo, F.; Li, Z.; Dai, G.; Lu, Y.; He, P.; Wang, Q. Ultrasensitive biosensing pathogenic bacteria by combining aptamer-induced catalysed hairpin assembly circle amplification with microchip electrophoresis. Sens. Actuators B Chem. 2020, 306, 127577. [Google Scholar] [CrossRef]
- Feng, Y.; Lee, Y. Microfluidic assembly of food-grade delivery systems: Toward functional delivery structure design. Trends Food Sci. Technol. 2019, 86, 465–478. [Google Scholar] [CrossRef]
- Khoshbin, Z.; Housaindokht, M.R.; Izadyar, M.; Verdian, A.; Bozorgmehr, M.R. A simple paper-based aptasensor for ultrasensitive detection of lead (II) ion. Anal. Chim. Acta 2019, 1071, 70–77. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Kim, M.I. Nanomaterial-mediated paper-based biosensors for colorimetric pathogen detection. TrAC Trends Anal. Chem. 2020, 132, 116038. [Google Scholar] [CrossRef] [PubMed]
- Fang, B.; Xu, S.; Huang, Y.; Su, F.; Huang, Z.; Fang, H.; Peng, J.; Xiong, Y.; Lai, W. Gold nanorods etching-based plasmonic immunoassay for qualitative and quantitative detection of aflatoxin M1 in milk. Food Chem. 2020, 329, 127160. [Google Scholar] [CrossRef] [PubMed]
- Kasoju, A.; Shrikrishna, N.S.; Shahdeo, D.; Khan, A.A.; Alanazi, A.M.; Gandhi, S. Microfluidic paper device for rapid detection of aflatoxin B1 using an aptamer based colorimetric assay. RSC Adv. 2020, 10, 11843–11850. [Google Scholar] [CrossRef]
- Sheini, A. Colorimetric aggregation assay based on array of gold and silver nanoparticles for simultaneous analysis of aflatoxins, ochratoxin and zearalenone by using chemometric analysis and paper based analytical devices. Microchim. Acta 2020, 187, 167. [Google Scholar] [CrossRef] [PubMed]
- Soares, R.R.; Santos, D.R.; Pinto, I.F.; Azevedo, A.M.; Aires-Barros, M.R.; Chu, V.; Conde, J.P. Multiplexed microfluidic fluorescence immunoassay with photodiode array signal acquisition for sub-minute and point-of-need detection of mycotoxins. Lab Chip 2018, 18, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Aissa, S.B.; Mars, A.; Catanante, G.; Marty, J.-L.; Raouafi, N. Design of a redox-active surface for ultrasensitive redox capacitive aptasensing of aflatoxin M1 in milk. Talanta 2019, 195, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Gong, L.; Wang, J.; Zhang, X.; Jin, Y.; Zhao, R.; Yang, C.; He, L.; Feng, X.; Chen, Y. An octuplex lateral flow immunoassay for rapid detection of antibiotic residues, aflatoxin M1 and melamine in milk. Sens. Actuators B Chem. 2019, 292, 94–104. [Google Scholar] [CrossRef]
- Kasoju, A.; Shahdeo, D.; Khan, A.A.; Shrikrishna, N.S.; Mahari, S.; Alanazi, A.M.; Bhat, M.A.; Giri, J.; Gandhi, S. Fabrication of microfluidic device for Aflatoxin M1 detection in milk samples with specific aptamers. Sci. Rep. 2020, 10, 4627. [Google Scholar] [CrossRef] [Green Version]
- Tsounidi, D.; Koukouvinos, G.; Petrou, P.; Misiakos, K.; Zisis, G.; Goustouridis, D.; Raptis, I.; Kakabakos, S.E. Rapid and sensitive label-free determination of aflatoxin M1 levels in milk through a White Light Reflectance Spectroscopy immunosensor. Sens. Actuators B Chem. 2019, 282, 104–111. [Google Scholar] [CrossRef]
- Mu, W.Y.; Huang, P.Z.; Chen, Q.Y.; Wang, W. Determination of melamine and melamine–Cu (II) complexes in milk using a DNA-Ag hydrocolloid as the sensor. Food Chem. 2020, 311, 125889. [Google Scholar] [CrossRef]
- Ezhilan, M.; Gumpu, M.B.; Ramachandra, B.L.; Nesakumar, N.; Babu, K.J.; Krishnan, U.M.; Rayappan, J.B.B. Design and development of electrochemical biosensor for the simultaneous detection of melamine and urea in adulterated milk samples. Sens. Actuators B Chem. 2017, 238, 1283–1292. [Google Scholar] [CrossRef]
- Li, H.; Somerson, J.; Xia, F.; Plaxco, K.W. Electrochemical DNA-based sensors for molecular quality control: Continuous, real-time melamine detection in flowing whole milk. Anal. Chem. 2018, 90, 10641–10645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, N.; Huang, P.; Wu, F. Colorimetric detection of melamine in milk based on Triton X-100 modified gold nanoparticles and its paper-based application. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 192, 174–180. [Google Scholar] [CrossRef]
- Xie, L.; Zi, X.; Zeng, H.; Sun, J.; Xu, L.; Chen, S. Low-cost fabrication of a paper-based microfluidic using a folded pattern paper. Anal. Chim. Acta 2019, 1053, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; You, T.; Yang, N.; Gao, Y.; Jiang, L.; Yin, P. Hydrophobic paper-based SERS platform for direct-droplet quantitative determination of melamine. Food Chem. 2019, 287, 363–368. [Google Scholar] [CrossRef]
- Krafft, B.; Panneerselvam, R.; Geissler, D.; Belder, D. A microfluidic device enabling surface-enhanced Raman spectroscopy at chip-integrated multifunctional nanoporous membranes. Anal. Bioanal. Chem. 2020, 412, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hong, F.; Zhang, W.; Wu, D.; Li, T.; Hu, F.; Gan, N.; Lin, J.; Wang, Q. Microchip electrophoresis based multiplexed assay for silver and mercury ions simultaneous detection in complex samples using a stirring bar modified with encoded hairpin probes for specific extraction. J. Chromatogr. A 2019, 1589, 173–181. [Google Scholar] [CrossRef]
- Li, G.; Xu, L.; Wu, W.; Wang, D.; Jiang, J.; Chen, X.; Zhang, W.; Poapolathep, S.; Poapolathep, A.; Zhang, Z. On-Site ultrasensitive detection paper for multiclass chemical contaminants via universal bridge-antibody labeling: Mycotoxin and illegal additives in milk as an example. Anal. Chem. 2018, 91, 1968–1973. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, Y.; Shen, M.; Lu, Y.; Cheng, J.; Xu, Y. Rapid and automated detection of six contaminants in milk using a centrifugal microfluidic platform with two rotation axes. Anal. Chem. 2019, 91, 7958–7964. [Google Scholar] [CrossRef]
- Raza, N.; Kim, K.H. Quantification techniques for important environmental contaminants in milk and dairy products. TrAC Trends Anal. Chem. 2018, 98, 79–94. [Google Scholar] [CrossRef]
- Rosati, G.; Ravarotto, M.; Scaramuzza, M.; De Toni, A.; Paccagnella, A. Silver nanoparticles inkjet-printed flexible biosensor for rapid label-free antibiotic detection in milk. Sens. Actuators B Chem. 2019, 280, 280–289. [Google Scholar] [CrossRef]
- Hassan, S.u.; Zhang, X. Design and fabrication of optical flow cell for multiplex detection of β-lactamase in microchannels. Micromachines 2020, 11, 385. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, F.; Sun, Y.; Mi, T.; Wang, L.; Li, Q.; Li, J.; Ma, W.; Liu, W.; Zuo, J. Development of a highly sensitive lateral flow immunoassay based on receptor-antibody-amorphous carbon nanoparticles to detect 22 β-lactams in milk. Sens. Actuators B Chem. 2020, 321, 128458. [Google Scholar] [CrossRef]
- Xiao, L.; Zhang, Z.; Wu, C.; Han, L.; Zhang, H. Molecularly imprinted polymer grafted paper-based method for the detection of 17β-estradiol. Food Chem. 2017, 221, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Na, G.; Hu, X.; Yang, J.; Sun, Y.; Kwee, S.; Tang, L.; Xing, G.; Xing, Y.; Zhang, G. Colloidal gold-based immunochromatographic strip assay for the rapid detection of bacitracin zinc in milk. Food Chem. 2020, 327, 126879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Gan, N.; Shen, Z.; Cao, J.; Hu, F.; Li, T. Microchip electrophoresis based aptasensor for multiplexed detection of antibiotics in foods via a stir-bar assisted multi-arm junctions recycling for signal amplification. Biosens. Bioelectron. 2019, 130, 139–146. [Google Scholar] [CrossRef]
- Zhao, M.; Li, X.; Zhang, Y.; Wang, Y.; Wang, B.; Zheng, L.; Zhang, D.; Zhuang, S. Rapid quantitative detection of chloramphenicol in milk by microfluidic immunoassay. Food Chem. 2021, 339, 127857. [Google Scholar] [CrossRef]
- Zeng, L.; Liu, L.; Kuang, H.; Cui, G.; Xu, C. A paper-based colorimetric assay for rapid detection of four macrolides in milk. Mater. Chem. Front. 2019, 3, 2175–2183. [Google Scholar] [CrossRef]
- Ma, L.; Nilghaz, A.; Choi, J.R.; Liu, X.; Lu, X. Rapid detection of clenbuterol in milk using microfluidic paper-based ELISA. Food Chem. 2018, 246, 437–441. [Google Scholar] [CrossRef]
- Bosma, R.; Devasagayam, J.; Eswar, R.; Albuquerque, I.d.F.; Collier, C.M. Voltage control for microchip capillary electrophoresis analyses. Electrophoresis 2020, 41, 1961–1968. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, W.; Zheng, W.; Cao, F.; Jiang, X. Inkjet-printed barcodes for a rapid and multiplexed paper-based assay compatible with mobile devices. Lab Chip 2017, 17, 3874–3882. [Google Scholar] [CrossRef]
- Hu, M.; Hu, X.; Zhang, Y.; Teng, M.; Deng, R.; Xing, G.; Tao, J.; Xu, G.; Chen, J.; Zhang, Y. Label-free electrochemical immunosensor based on AuNPs/Zn/Ni-ZIF-8-800@ graphene composites for sensitive detection of monensin in milk. Sens. Actuators B Chem. 2019, 288, 571–578. [Google Scholar] [CrossRef]
- Zong, L.; Jiao, Y.; Guo, X.; Zhu, C.; Gao, L.; Han, Y.; Li, L.; Zhang, C.; Liu, Z.; Liu, J. based fluorescent immunoassay for highly sensitive and selective detection of norfloxacin in milk at picogram level. Talanta 2019, 195, 333–338. [Google Scholar] [CrossRef]
- Meng, X.; Gu, H.; Yi, H.; He, Y.; Chen, Y.; Sun, W. Sensitive detection of streptomycin in milk using a hybrid signal enhancement strategy of MOF-based bio-bar code and target recycling. Anal. Chim. Acta 2020, 1125, 1–7. [Google Scholar] [CrossRef]
- Li, X.; Wu, X.; Wang, J.; Hua, Q.; Wu, J.; Shen, X.; Sun, Y.; Lei, H. Three lateral flow immunochromatographic assays based on different nanoparticle probes for on-site detection of tylosin and tilmicosin in milk and pork. Sens. Actuators B Chem. 2019, 301, 127059. [Google Scholar] [CrossRef]
- Kim, H.J.; Kwon, C.; Lee, B.S.; Noh, H. One-step sensing of foodborne pathogenic bacteria using a 3D paper-based device. Analyst 2019, 144, 2248–2255. [Google Scholar] [CrossRef] [PubMed]
- Gorgannezhad, L.; Stratton, H.; Nguyen, N.-T. Microfluidic-based nucleic acid amplification systems in microbiology. Micromachines 2019, 10, 408. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Xu, Y.; Fohlerova, Z.; Chang, H.; Iliescu, C.; Neuzil, P. LAMP-on-a-chip: Revising microfluidic platforms for loop-mediated DNA amplification. Trends Anal. Chem. 2019, 113, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Suo, Y.; Zou, Z.; Sun, J.; Zhang, S.; Wang, B.; Xu, Y.; Darland, D.; Zhao, J.; Mu, Y. Integrated microfluidic systems with sample preparation and nucleic acid amplification. Lab Chip 2019, 19, 2769–2785. [Google Scholar] [CrossRef]
- Ilhan, H.; Guven, B.; Dogan, U.; Torul, H.; Evran, S.; Çetin, D.; Suludere, Z.; Saglam, N.; Boyaci, İ.H.; Tamer, U. The coupling of immunomagnetic enrichment of bacteria with paper-based platform. Talanta 2019, 201, 245–252. [Google Scholar] [CrossRef]
- Jiang, Y.; Qiu, Z.; Le, T.; Zou, S.; Cao, X. Developing a dual-RCA microfluidic platform for sensitive E. coli O157: H7 whole-cell detections. Anal. Chim. Acta 2020, 1127, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Fang, S.; Tian, Y.; Ma, J.; Wang, Z.; Xu, D.; Li, Y.; Hou, D.; Liu, Q. Rapid detection of Escherichia coli O157: H7 in milk, bread, and jelly by lac dye coloration-based bidirectional lateral flow immunoassay strip. J. Food Saf. 2021, 41, e12862. [Google Scholar] [CrossRef]
- Luo, K.; Ryu, J.; Seol, I.-H.; Jeong, K.-B.; You, S.-M.; Kim, Y.-R. Based Radial Chromatographic Immunoassay for the Detection of Pathogenic Bacteria in Milk. ACS Appl. Mater. Interfaces 2019, 11, 46472–46478. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.; Awan, F.R.; Khan, Q.M.; Ngamsom, B.; Pamme, N. based analytical devices for colorimetric detection of S. aureus and E. coli and their antibiotic resistant strains in milk. Analyst 2020, 145, 7320–7329. [Google Scholar] [CrossRef]
- Eissa, S.; Zourob, M. A dual electrochemical/colorimetric magnetic nanoparticle/peptide-based platform for the detection of Staphylococcus aureus. Analyst 2020, 145, 4606–4614. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.C.; Soares, J.C.; Rodrigues, V.C.; Oliveira, O.N.; Mattoso, L.H.C. Controlled molecular architectures in microfluidic immunosensors for detecting Staphylococcus aureus. Analyst 2020, 145, 6014–6023. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Su, Y.; Liang, Y.; Lai, W.; Jiang, J.; Wu, H.; Zhang, C. Ultrasensitive cloth-based microfluidic chemiluminescence detection of Listeria monocytogenes hlyA gene by hemin/G-quadruplex DNAzyme and hybridization chain reaction signal amplification. Anal. Bioanal. Chem. 2020, 412, 3787–3797. [Google Scholar] [CrossRef]
- Silva, N.F.; Neves, M.M.; Magalhães, J.M.; Freire, C.; Delerue-Matos, C. Electrochemical immunosensor towards invasion-associated protein p60: An alternative strategy for Listeria monocytogenes screening in food. Talanta 2020, 216, 120976. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, X.; Wang, Q. Sensitive and specific detection of E. coli, Listeria monocytogenes, and Salmonella enterica serovar Typhimurium in milk by microchip electrophoresis combined with multiplex PCR amplification. Microchem. J. 2020, 157, 104876. [Google Scholar] [CrossRef]
- Carrell, C.S.; Wydallis, R.M.; Bontha, M.; Boehle, K.E.; Beveridge, J.R.; Geiss, B.J.; Henry, C.S. Rotary manifold for automating a paper-based Salmonella immunoassay. RSC Adv. 2019, 9, 29078–29086. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, T.R.; Martucci, D.H.; Faria, R.C. Simple disposable microfluidic device for Salmonella typhimurium detection by magneto-immunoassay. Sens. Actuators B Chem. 2018, 255, 684–691. [Google Scholar] [CrossRef]
- Luo, F.; Li, Z.; Dai, G.; Lu, Y.; He, P.; Wang, Q. Simultaneous detection of different bacteria by microchip electrophoresis combined with universal primer-duplex polymerase chain reaction. J. Chromatogr. A 2020, 1615, 460734. [Google Scholar] [CrossRef] [PubMed]
- Murasova, P.; Kovarova, A.; Kasparova, J.; Brozkova, I.; Hamiot, A.; Pekarkova, J.; Dupuy, B.; Drbohlavova, J.; Bilkova, Z.; Korecka, L. Direct culture-free electrochemical detection of Salmonella cells in milk based on quantum dots-modified nanostructured dendrons. J. Electroanal. Chem. 2020, 863, 114051. [Google Scholar] [CrossRef]
- Srbova, J.; Krulisova, P.; Holubova, L.; Pereiro, I.; Bendali, A.; Hamiot, A.; Podzemna, V.; Macak, J.; Dupuy, B.; Descroix, S. Advanced immunocapture of milk-borne Salmonella by microfluidic magnetically stabilized fluidized bed. Electrophoresis 2018, 39, 526–533. [Google Scholar] [CrossRef]
- Sun, Y.; Chang, Y.; Zhang, Q.; Liu, M. An origami paper-based device printed with DNAzyme-containing DNA superstructures for escherichia coli detection. Micromachines 2019, 10, 531. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Kwon, C.; Noh, H. Based diagnostic system facilitating escherichia coli assessments by duplex coloration. ACS Sens. 2019, 4, 2435–2441. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, L.; He, P.; Zi, F.; Hu, X.; Wang, Q. Sensitive assay of Escherichia coli in food samples by microchip capillary electrophoresis based on specific aptamer binding strategy. Talanta 2019, 197, 284–290. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Zuo, P.; Ye, B.-C. A single cell droplet microfluidic system for quantitative determination of food-borne pathogens. Talanta 2020, 209, 120571. [Google Scholar] [CrossRef]
- Choi, J.R.; Nilghaz, A.; Chen, L.; Chou, K.C.; Lu, X. Modification of thread-based microfluidic device with polysiloxanes for the development of a sensitive and selective immunoassay. Sens. Actuators B Chem. 2018, 260, 1043–1051. [Google Scholar] [CrossRef]
- Mahato, K.; Chandra, P. based miniaturized immunosensor for naked eye ALP detection based on digital image colorimetry integrated with smartphone. Biosensors Bioelectron. 2019, 128, 9–16. [Google Scholar] [CrossRef]
- Zhang, W.; Niu, X.; Li, X.; He, Y.; Song, H.; Peng, Y.; Pan, J.; Qiu, F.; Zhao, H.; Lan, M. A smartphone-integrated ready-to-use paper-based sensor with mesoporous carbon-dispersed Pd nanoparticles as a highly active peroxidase mimic for H2O2 detection. Sens. Actuators B Chem. 2018, 265, 412–420. [Google Scholar] [CrossRef]
- Khoshbin, Z.; Housaindokht, M.R.; Verdian, A. A low-cost paper-based aptasensor for simultaneous trace-level monitoring of mercury (II) and silver (I) ions. Anal. Biochem. 2020, 597, 113689. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, J.; Liu, H.; Gao, Y.; Zhao, Q.; Ling, S.; Wang, S. Sensitive immunoassays based on specific monoclonal IgG for determination of bovine lactoferrin in cow milk samples. Food Chem. 2021, 338, 127820. [Google Scholar] [CrossRef]
- Tůma, P.; Sommerová, B.; Daněček, V. On-line coupling of capillary electrophoresis with microdialysis for determining saccharides in dairy products and honey. Food Chem. 2020, 316, 126362. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Fu, H. Quantitative measurements of the somatic cell count of fat-free milk based on droplet microfluidics. J. Mater. Chem. C 2020, 8, 13770–13776. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, H.; Liu, S.; Zhang, B.; Zhang, Y. Milk carton with integrated paper-based microfluidics for milk quality rapid test. J. Food Saf. 2018, 38, e12548. [Google Scholar] [CrossRef]
- Younas, M.; Maryam, A.; Khan, M.; Nawaz, A.A.; Jaffery, S.H.I.; Anwar, M.N.; Ali, L. Parametric analysis of wax printing technique for fabricating microfluidic paper-based analytic devices (µPAD) for milk adulteration analysis. Microfluid. Nanofluid. 2019, 23, 38. [Google Scholar] [CrossRef]
- Puneeth, S.; Goel, S. Novel 3D printed microfluidic paper-based analytical device with integrated screen-printed electrodes for automated viscosity measurements. IEEE Trans. Electron. Devices 2019, 66, 3196–3201. [Google Scholar] [CrossRef]
- Bougadi, E.T.; Kalogianni, D.P. based DNA biosensor for food authenticity testing. Food Chem. 2020, 322, 126758. [Google Scholar] [CrossRef] [PubMed]
- Han, C.M.; Catoe, D.; Munro, S.A.; Khnouf, R.; Snyder, M.P.; Santiago, J.G.; Salit, M.L.; Cenik, C. Simultaneous RNA purification and size selection using on-chip isotachophoresis with an ionic spacer. Lab Chip 2019, 19, 2741–2749. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Schlecht, W.; Li, L.; Dong, W.J. Paper-based cascade cationic isotachophoresis: Multiplex detection of cardiac markers. Talanta 2019, 205, 120112. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.L.; Linz, T.H. Characterizing the impact of thermal gels on isotachophoresis in microfluidic devices. Electrophoresis 2020, 41, 691–696. [Google Scholar] [CrossRef]
- Schaumburg, F.; Kler, P.A.; Carrell, C.S.; Berli, C.L.A.; Henry, C.S. USB powered microfluidic paper-based analytical device. Electrophoresis 2020, 41, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Somaweera, H.; Estlack, Z.; Devadhasan, J.P.; Kim, J.; Kim, J. Characterization and optimization of isotachophoresis parameters for pacific blue succinimidyl ester dye on a PDMS microfluidic chip. Micromachines 2020, 11, 951. [Google Scholar]
- Guo, S.; Xu, J.; Estell, A.P.; Ivory, C.F.; Du, D.; Lin, Y.; Dong, W.J. Paper-based ITP technology: An application to specific cancer-derived exosome detection and analysis. Biosens. Bioelectron. 2020, 164, 112292. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, B.P.; Bender, A.T.; Ngyuen, D.N.; Zhang, J.Y.; Posner, J.D. Nucleic acid sample preparation from whole blood in a paper microfluidic device using isotachophoresis. J. Chromatogr. B 2021, 1163, 122494. [Google Scholar] [CrossRef]
- Li, J.; Chen, D.; Ye, J.; Zhang, L.; Zhou, T.; Zhou, Y. Direct numerical simulation of seawater desalination based on ion concentration polarization. Micromachines 2019, 10, 562. [Google Scholar]
- Papadimitriou, V.A.; Segerink, L.I.; Eijkel, J.C.T. Continuous focusing, fractionation and extraction of anionic analytes in a microfluidic chip. Lab Chip 2019, 19, 3238–3248. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Lee, K.H.; Han, S.I.; Lee, D.; Chung, S.; Lee, D.; Lee, J.H. Origami-paper-based device for microvesicle/exosome preconcentration and isolation. Lab Chip 2019, 19, 3917–3921. [Google Scholar] [CrossRef]
- Subramanian, V.; Lee, S.; Jena, S.; Jana, S.K.; Ray, D.; Kim, S.J.; Thalappil, P. Enhancing the sensitivity of point-of-use electrochemical microfluidic sensors by ion concentration polarization—A case study on arsenic. Sens. Actuators B Chem. 2020, 304, 127340. [Google Scholar] [CrossRef]
- Perera, A.T.K.; Pudasaini, S.; Ahmed, S.S.U.; Phan, D.T.; Liu, Y.; Yang, C. Rapid pre-concentration of Escherichia coli in a microfluidic paper-based device using ion concentration polarization. Electrophoresis 2020, 41, 867–874. [Google Scholar] [CrossRef]
- Lee, J.; Yoo, Y.K.; Lee, D.; Kim, C.; Kim, K.H.; Lee, S.; Kwak, S.; Kang, J.Y.; Kim, H.; Yoon, D.S.; et al. Origami paper-based sample preconcentration using sequentially driven ion concentration polarization. Lab Chip 2021, 21, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Hu, X.; Ouyang, W.; Chen, Y.; Liu, S.; Han, J.; Liu, L. Femtomolar detection of lipopolysaccharide in injectables and serum samples using aptamer-coupled reduced graphene oxide in a continuous injection-electrostacking biochip. Anal. Chem. 2019, 91, 2360–2367. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xie, M.R.; Chen, Y.Z.; Wu, Z.Y. Simultaneous electrokinetic stacking and separation of anionic and cationic species on a paper fluidic channel. Lab Chip 2019, 19, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; Han, J. One-step nucleic acid purification and noise-resistant polymerase chain reaction by electrokinetic concentration for ultralow abundance nucleic acid detection. Angew. Chem. Int. Ed. 2020, 59, 10981–10988. [Google Scholar] [CrossRef]
- Xi, L.; De, R.; Dutta, D. Electrokinetic stacking of particle zones in confined channels enabling their UV absorbance detection on microchips. Anal. Chim. Acta 2020, 1135, 83–90. [Google Scholar] [CrossRef]
- Zhai, H.M.; Zhou, T.; Fang, F.; Wu, Z.Y. Colorimetric speciation of Cr on paper-based analytical devices based on field amplified stacking. Talanta 2020, 210, 120635. [Google Scholar] [CrossRef]
- Cai, Y.; Niu, J.C.; Du, X.L.; Fang, F.; Wu, Z.Y. Novel field amplification for sensitive colorimetric detection of microalbuminuria on a paper-based analytical device. Anal. Chim. Acta 2019, 1080, 146–152. [Google Scholar] [CrossRef]
- Zhang, X.X.; Liu, J.J.; Cai, Y.; Zhao, S.; Wu, Z.Y. A field amplification enhanced paper-based analytical device with a robust chemiluminescence detection module. Analyst 2019, 144, 498–503. [Google Scholar] [CrossRef]
- Zhou, T.; Liu, J.J.; Xu, Y.; Wu, Z.Y. Fast and sensitive screening detection of tetracyclines with a paper-based analytical device. Microchem. J. 2019, 145, 703–707. [Google Scholar] [CrossRef]
- Cai, Y.; Niu, J.C.; Liu, Y.Q.; Du, X.L.; Wu, Z.Y. Online sample clean-up and enrichment of proteins from salty media with dynamic double gradients on a paper fluidic channel. Anal. Chim. Acta 2020, 1100, 149–155. [Google Scholar] [CrossRef]
- Costa-Rama, E.; Nouws, H.P.A.; Delerue-Matos, C.; Blanco-Lopez, M.C.; Fernandez-Abedul, M.T. Preconcentration and sensitive determination of the anti-inflammatory drug diclofenac on a paper-based electroanalytical platform. Anal. Chim. Acta 2019, 1074, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Ng, H.Y.; Hou, C.Y.; Lee, C.T.; Fu, L.M. Recent advances in lab-on-paper diagnostic devices using blood samples. Lab Chip 2021, 21, 1433–1453. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Abafogi, A.T.; Tran, B.M.; Kim, J.; Lee, J.; Chen, Z.; Bae, P.K.; Park, K.; Shin, Y.-B.; van Noort, D.; et al. Integrated microfluidic preconcentration and nucleic amplification system for detection of influenza a virus H1N1 in saliva. Micromachines 2020, 11, 203. [Google Scholar] [CrossRef] [Green Version]
- Kalsi, S.; Valiadi, M.; Turner, C.; Sutton, M.; Morgan, H. Sample pre-concentration on a digital microfluidic platform for rapid AMR detection in urine. Lab Chip 2019, 19, 168–177. [Google Scholar] [CrossRef] [Green Version]
- Kopp, M.R.G.; Linsenmeier, M.; Hettich, B.; Prantl, S.; Stavrakis, S.; Leroux, J.C.; Arosio, P. Microfluidic shrinking droplet concentrator for analyte detection and phase separation of protein solutions. Anal. Chem. 2020, 92, 5803–5812. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Jafry, A.T.; Lee, J. Fabrication, flow control, and applications of microfluidic paper-based analytical devices. Molecules 2019, 24, 2869. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Device Type, Materials, and Structures | Fabrication Methods | Detection Methods | Target | LOD | Ref. |
|---|---|---|---|---|---|
| Lab-on-a-chip, 3-D Plastic | Injection Molding | CM | Aflatoxin M1 | 0.11 ng/mL | [52] |
| Lab-on-paper, 2-D Filter paper | Soft Lithography | CM | Aflatoxin B1 | 10 nM | [53] |
| Lab-on-paper, 2-D Nitrocellulose MB | Spotting | CM | Aflatoxin M1 Melamine Β-Lactams | 0.016 ng/mL 2.5 ng/mL 0.13 ng/mL | [57] |
| Lab-on-paper, 2-D Filter paper | Soft Lithography | CM | Aflatoxin M1 | 3 pM @Water 10 nM @milk | [58] |
| Lab-on-a-chip, 3-D Si/SiO2 chip | Deposition | WLRS | Aflatoxin M1 | 6 pg/mL | [59] |
| Electrode, 3-D Pt/ZnO/AChE | Coating | EC | Melamine Urea | 3 pM, 1 pM | [61] |
| Lab-on-paper, 3-D Filter paper/PDMS | Cutting and Coating | CM | Melamine | 0.1 ppm | [64] |
| Lab-on-paper, 2-D Filter paper | Dip-coating | SERS | Melamine | 1 ppm | [65] |
| Lab-on-a-Chip, 3-D gold/Quartz | Soft Lithography | Flu | Ag+ Hg2+ | 0.038 nM, 0.054 nM | [67] |
| Lab-on-paper, 2-D Nitrocellulose MB | Spotting | Flu | Aflatoxin M1 Melamine | 0.009 ng/mL 0.024 ng/mL | [68] |
| Device Type, Materials, and Structures | Fabrication Methods | Detection Methods | Target | LOD | Ref. |
|---|---|---|---|---|---|
| Lab-on-paper, 2-D Paper/AgNPs | Printing | EC | Antibiotic | 10 μg/mL | [71] |
| Lab-on-a-chip, 3-D PMMA | Cutting and Adhesive | CM | Β-Lactamase | 0.05 mg/mL | [72] |
| Lab-on-paper, 2-D Millipore MB | Dip-coating | CM | 17 Β-Estradiol | 0.25 μg/L | [74] |
| Lab-on-paper, 2-D Nitrocellulose MB | Spotting | CM | Bacitracin Zinc | 0.82 ng/mL | [75] |
| Lab-on-a-Chip, 3-D Gold/Quartz | Soft Lithography and Modify | Flu | CP Kanamycin | 0.52 pg/mL 0.41 pg/mL | [76] |
| Lab-on-paper, 3-D CG paper | Waxing and Coating | CM | Clenbuterol | 0.2 ppb | [79] |
| Electrode, 3-D AuNPs | Coating | EC | Monensin | 0.11 ng/mL | [82] |
| Lab-on-paper, 2-D Filter paper | Waxing and spotting | Flu | Norfloxacin | 10 pg/mL | [83] |
| Lab-on-paper, 2-D Nitrocellulose MB | Spotting | Flu | Tylosin | 2 ng/mL | [85] |
| Lab-on-a-chip, 3-D PDMS | Soft Lithography | Flu | E. Coli | 80 cells/m | [91] |
| Lab-on-a-Chip, 3-D Quartz | Soft Lithography | Flu | S. typhimurium P. aeruginosa | 15 CFU/mL 5 CFU/mL | [93] |
| Lab-on-paper, 3-D chitosan/chondroitin sulfate | Deposited layer-by-layer | EC | S. aureus | 2.8 CFU /mL | [96] |
| Lab-on-a-Chip, 3-D cloth-based | Cutting and screen-printing | CL | L. monocytogenes | 1.1 fM. | [97] |
| Lab-on-a-Chip, 3-D Quartz | Soft Lithography | Flu | E. coli L. monocytogenes S. typhimurium | 2.1 ng/μL 1.8 ng/μL 2.4 ng/μL | [99] |
| Lab-on-a-Chip, 3-D Ag/AgCl polystyrene | Screen printing and Cutting | EC | S. typhimurium | 7.7 cells/mL | [101] |
| Lab-on-a-Chip, 3-D Electrodes | Deposition modified | EC | Salmonella Cells | 4 CFU/mL | [103] |
| Lab-on-paper, 3-D Filter paper/PL | Waxing | CM | E. coli | 10 CFU/mL | [106] |
| Lab-on-a-chip, 3-D PLA/PDMS | 3D-printing and Soft Lithography | CM | E. coli O157:H7 | 50 CFU/mL | [107] |
| Lab-on-paper, 2-D Filter paper | Cutting and Dip-coating | CM | H2O2 | 1 μM | [111] |
| Lab-on-a-Chip, 3-D Quartz | Fused | CE analysis | Lactose | 2.2 mg/L | [114] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ng, H.-Y.; Lee, W.-C.; Kung, C.-T.; Li, L.-C.; Lee, C.-T.; Fu, L.-M. Recent Advances in Microfluidic Devices for Contamination Detection and Quality Inspection of Milk. Micromachines 2021, 12, 558. https://doi.org/10.3390/mi12050558
Ng H-Y, Lee W-C, Kung C-T, Li L-C, Lee C-T, Fu L-M. Recent Advances in Microfluidic Devices for Contamination Detection and Quality Inspection of Milk. Micromachines. 2021; 12(5):558. https://doi.org/10.3390/mi12050558
Chicago/Turabian StyleNg, Hwee-Yeong, Wen-Chin Lee, Chia-Te Kung, Lung-Chih Li, Chien-Te Lee, and Lung-Ming Fu. 2021. "Recent Advances in Microfluidic Devices for Contamination Detection and Quality Inspection of Milk" Micromachines 12, no. 5: 558. https://doi.org/10.3390/mi12050558
APA StyleNg, H.-Y., Lee, W.-C., Kung, C.-T., Li, L.-C., Lee, C.-T., & Fu, L.-M. (2021). Recent Advances in Microfluidic Devices for Contamination Detection and Quality Inspection of Milk. Micromachines, 12(5), 558. https://doi.org/10.3390/mi12050558







