Breaking the Third Wall: Implementing 3D-Printing Techniques to Expand the Complexity and Abilities of Multi-Organ-on-a-Chip Devices
Abstract
:1. Introduction
2. Materials and Methods
2.1. 3D-Printing
2.1.1. 3D-Printer and Software
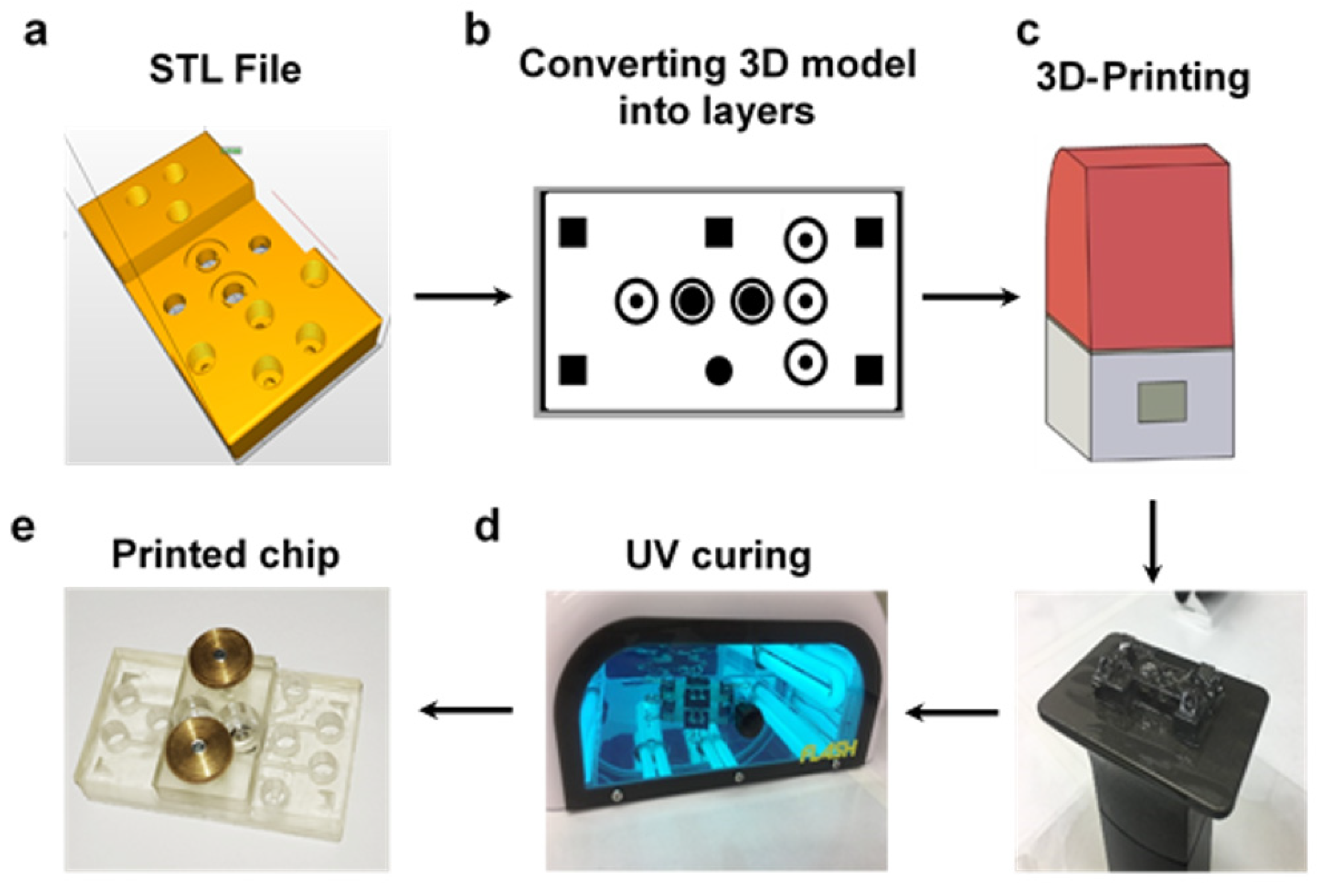
2.1.2. 3D-Printing Procedure
2.2. MOC Device Fabrication
2.2.1. PDMS-Based Microfluidic Unit Fabrication Using 3D-Templates
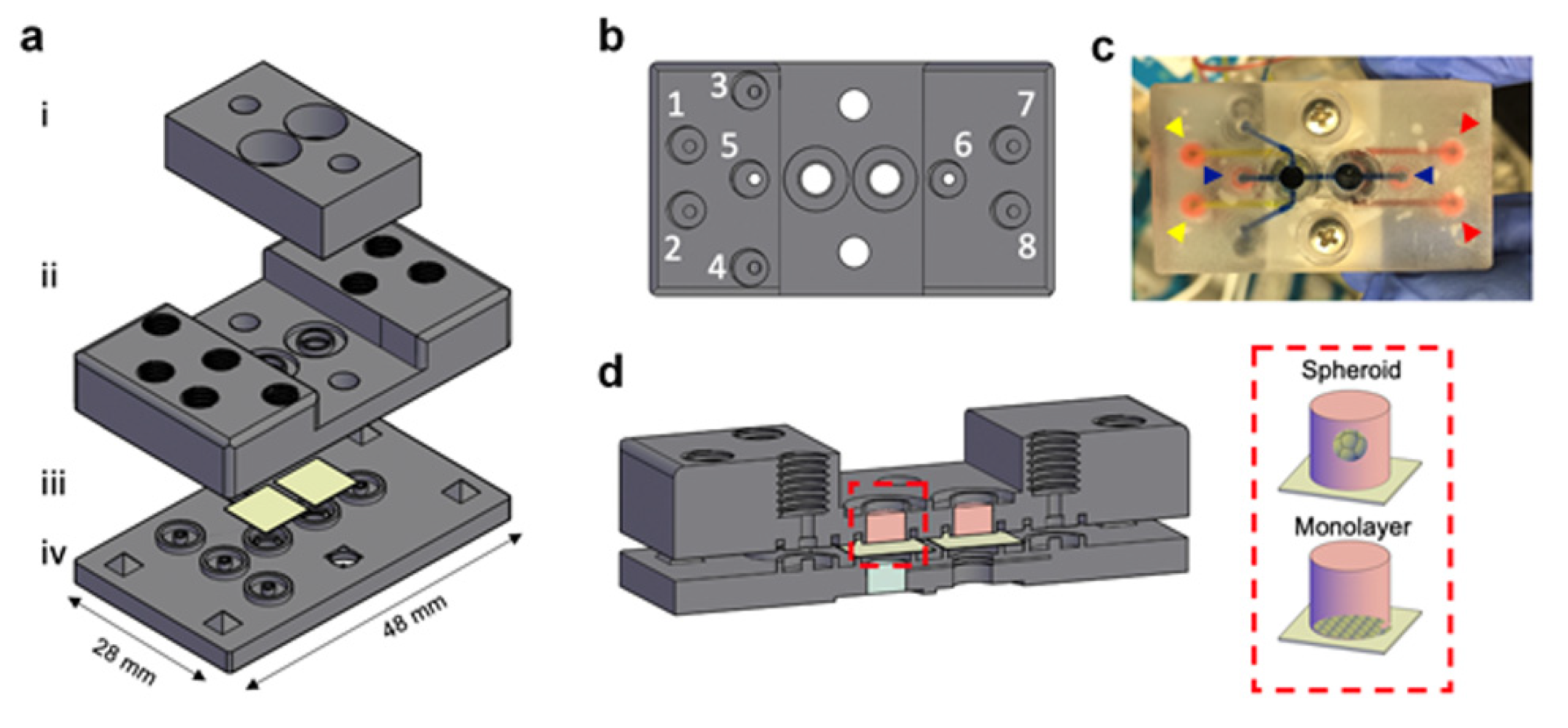
2.2.2. Whole 3D-Printed Device
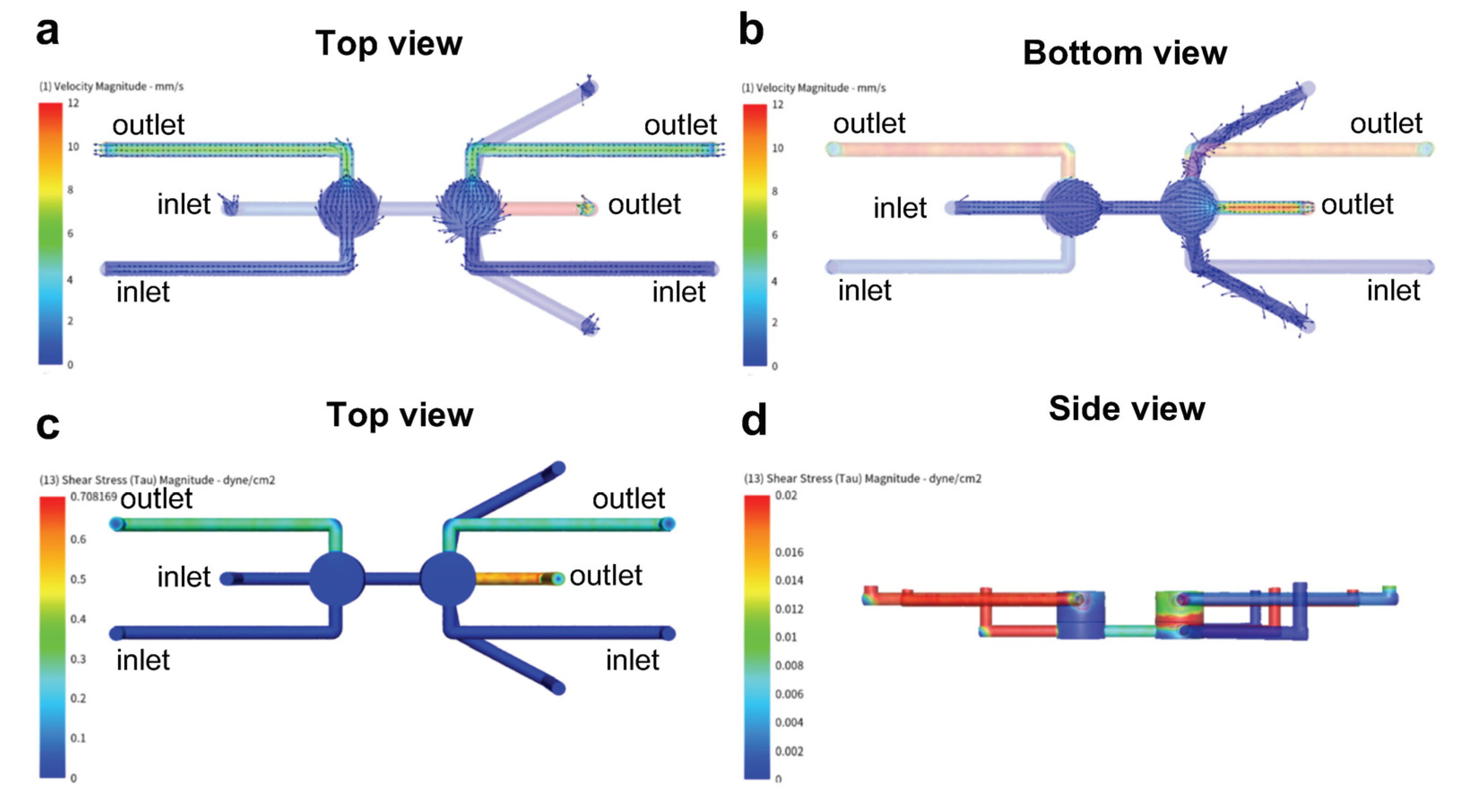
2.3. Computational Fluid Dynamics (CFD) Simulation
2.4. Cell Culture
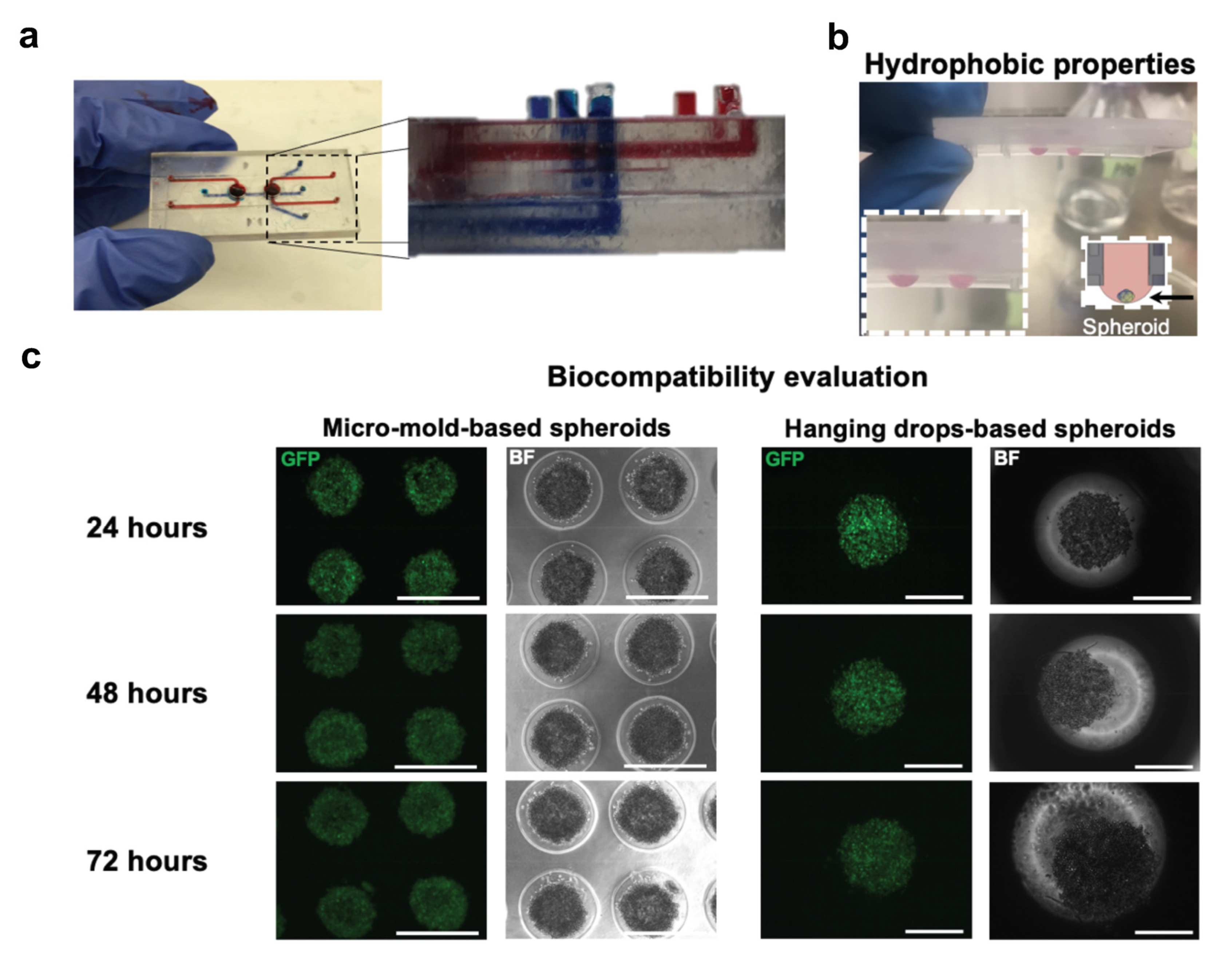
2.5. Cell Viability and Biocompatibility Evaluation
3. Results
3.1. MOCs Design Guidelines
3.2. PDMS Based MOCs Fabrication
3.3. 3D-Printing MOCs Fabrication
3.4. Shear Force Protection
3.5. 3D-Printed Based MOC Evaluation and Cytotoxic Effect of (Meth)Acrylate Resin
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Whitesides, G.M. The origins and the future of microfluidics. Nat. Cell Biol. 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Gravesen, P.; Branebjerg, J.A.; Jensen, O.S. Microfluidics—A review. J. Micromech. Microeng. 1993, 3, 168–182. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nat. Cell Biol. 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Stone, H.A.; Kim, S. Microfluidics: Basic issues, applications, and challenges. AIChE J. 2001, 47, 1250–1254. [Google Scholar] [CrossRef]
- Verpoorte, E.; de Rooij, N.F. Microfluidics meets MEMS. Proc. IEEE 2003, 91, 930–953. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-chip: Recent breakthroughs and future prospects. Biomed. Eng. Online 2020, 19, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Samanipour, R.; Koo, K.-I.; Kim, K. Organ-on-a-Chip Platforms for Drug Delivery and Cell Characterization: A Review. Sens. Mater. 2015, 27, 487–506. [Google Scholar]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef]
- Global Agenda. 2016. Available online: https://www.weforum.org/agenda/2016/06/top-10-emerging-technologies-2016 (accessed on 21 April 2021).
- Haeberle, S.; Zengerle, R. Microfluidic platforms for lab-on-a-chip applications. Lab Chip 2007, 7, 1094–1110. [Google Scholar] [CrossRef]
- Sung, J.H.; Kam, C.; Shuler, M.L. A microfluidic device for a pharmacokinetic–pharmacodynamic (PK–PD) model on a chip. Lab Chip 2010, 10, 446–455. [Google Scholar] [CrossRef]
- Rogal, J.; Probst, C.; Loskill, P. Integration concepts for multi-organ chips: How to maintain flexibility?! Futur. Sci. OA 2017, 3, FSO180. [Google Scholar] [CrossRef] [Green Version]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2021, 20, 345–361. [Google Scholar] [CrossRef]
- Picollet-D’Hahan, N.; Zuchowska, A.; Lemeunier, I.; Le Gac, S. Multiorgan-on-a-Chip: A Systemic Approach To Model and Decipher Inter-Organ Communication. Trends Biotechnol. 2021, 2020. [Google Scholar] [CrossRef]
- Scott, S.; Ali, Z. Fabrication Methods for Microfluidic Devices: An Overview. Micromachines 2021, 12, 319. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Du, Z.; Xu, Y.; Sun, W. The crossing and integration between microfluidic technology and 3D printing for organ-on-chips. J. Mater. Chem. B 2018, 6, 6191–6206. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.K.; Whitesides, G.M. Basic microfluidic and soft lithographic techniques. In Optofluidics: Fundamantal, Devices, Applications; Yang, C., Ed.; The McGraw-Hill Companies: New York, NY, USA, 2010. [Google Scholar]
- Friend, J.; Yeo, L. Fabrication of microfluidic devices using polydimethylsiloxane. Biomicrofluidics 2010, 4, 026502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharjee, N.; Urrios, A.; Kang, S.; Folch, A. The upcoming 3D-printing revolution in microfluidics. Lab Chip 2016, 16, 1720–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waheed, S.; Cabot, J.M.; Macdonald, N.P.; Lewis, T.; Guijt, R.M.; Paull, B.; Breadmore, M.C. 3D printed microfluidic devices: Enablers and barriers. Lab Chip 2016, 16, 1993–2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macdonald, N.P.; Cabot, J.M.; Smejkal, P.; Guijt, R.M.; Paull, B.; Breadmore, M.C. Comparing Microfluidic Performance of Three-Dimensional (3D) Printing Platforms. Anal. Chem. 2017, 89, 3858–3866. [Google Scholar] [CrossRef] [PubMed]
- Van Der Linden, P.J.E.M.; Popov, A.M.; Pontoni, D. Accurate and rapid 3D printing of microfluidic devices using wavelength selection on a DLP printer. Lab Chip 2020, 20, 4128–4140. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, K.; Qureshi, A.J.; Montemagno, C. Tolerance Control Using Subvoxel Gray-Scale DLP 3D Printing. In Proceedings of the Micro- and Nano-Systems Engineering and Packaging; ASME International, Tampa, FL, USA, 3–9 November 2017; Volume 10. [Google Scholar] [CrossRef]
- Xing, J.-F.; Zheng, M.-L.; Duan, X.-M. Two-photon polymerization microfabrication of hydrogels: An advanced 3D printing technology for tissue engineering and drug delivery. Chem. Soc. Rev. 2015, 44, 5031–5039. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Gao, C.; Gritsenko, D.; Zhou, R.; Xu, J. Soft lithography based on photolithography and two-photon polymerization. Microfluid. Nanofluidics 2018, 22, 97. [Google Scholar] [CrossRef]
- Vanderpoorten, O.; Peter, Q.; Challa, P.K.; Keyser, U.F.; Baumberg, J.; Kaminski, C.F.; Knowles, T.P.J. Scalable integration of nano-, and microfluidics with hybrid two-photon lithography. Microsyst. Nanoeng. 2019, 5, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.Y.; Di Carlo, D.; Lee, L.P. Microfluidic self-assembly of tumor spheroids for anticancer drug discovery. Biomed. Microdevices 2008, 10, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.-Y.; Tseng, S.-Y.; Yang, S.-M.; Hsu, L.; Liu, C.-H.; Chang, H.-Y. A microfluidic chip with a U-shaped microstructure array for multicellular spheroid formation, culturing and analysis. Biofabrication 2014, 6, 015009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raj, M.K.; Chakraborty, S. PDMS microfluidics: A mini review. J. Appl. Polym. Sci. 2020, 137, 48958. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef]
- Walter, F.R.; Valkai, S.; Kincses, A.; Petneházi, A.; Czeller, T.; Veszelka, S.; Ormos, P.; Deli, M.A.; Dér, A. A versatile lab-on-a-chip tool for modeling biological barriers. Sens. Actuators B Chem. 2016, 222, 1209–1219. [Google Scholar] [CrossRef] [Green Version]
- Jing, B.; Luo, Y.; Lin, B.; Li, J.; Wang, Z.A.; Du, Y. Establishment and application of a dynamic tumor-vessel microsystem for studying different stages of tumor metastasis and evaluating anti-tumor drugs. RSC Adv. 2019, 9, 17137–17147. [Google Scholar] [CrossRef] [Green Version]
- Tilles, A.W.; Baskaran, H.; Roy, P.; Yarmush, M.L.; Toner, M. Effects of oxygenation and flow on the viability and function of rat hepatocytes cocultured in a microchannel flat-plate bioreactor. Biotechnol. Bioeng. 2001, 73, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.D.; Ye, M.; Levy, A.F.; Rothstein, J.D.; Bergles, D.E.; Searson, P.C. The blood-brain barrier: An engineering perspective. Front. Neuroeng. 2013, 6, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, S.L.; Nelman-Gonzalez, M.; Nickerson, C.A.; Ott, C.M. Induction of Attachment-Independent Biofilm Formation and Repression ofhfqExpression by Low-Fluid-Shear Culture of Staphylococcus aureus. Appl. Environ. Microbiol. 2011, 77, 6368–6378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Song, W.; Li, P.; Huang, Y.; Gong, X.; Zhou, G.; Jia, X.; Zheng, L.; Fan, Y. Galanin Protects against Nerve Injury after Shear Stress in Primary Cultured Rat Cortical Neurons. PLoS ONE 2013, 8, e63473. [Google Scholar] [CrossRef]
- Konduri, G.G.; Afolayan, A.J.; Eis, A.; Pritchard, K.A.; Teng, R.-J. Interaction of endothelial nitric oxide synthase with mitochondria regulates oxidative stress and function in fetal pulmonary artery endothelial cells. Am. J. Physiol. Cell. Mol. Physiol. 2015, 309, L1009–L1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García, O.; Almeida, A.; Massieu, L.; Bolaños, J.P. Increased mitochondrial respiration maintains the mitochondrial membrane potential and promotes survival of cerebellar neurons in an endogenous model of glutamate receptor activation. J. Neurochem. 2005, 92, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.U.; Delude, R.L.; Han, Y.Y.; Sappington, P.L.; Han, X.; Carcillo, J.A.; Fink, M.P. Liposomal NAD+ prevents diminished O2consumption by immunostimulated Caco-2 cells. Am. J. Physiol. Cell. Mol. Physiol. 2002, 282, L1082–L1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Zhao, Z.; Rahim, N.A.A.; Van Noort, D.; Yu, H. Towards a human-on-chip: Culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip 2009, 9, 3185–3192. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Murphy, S.V.; Devarasetty, M.; Mead, I.; Kang, H.W.; Seol, Y.J.; Zhang, Y.S.; Shin, S.R.; Zhao, L.; Aleman, J.; et al. Multi-Tissue Interactions in an Integrated Three-Tissue Organ-on-a-Chip Platform. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Esch, M.B.; Smith, A.S.; Prot, J.-M.; Oleaga, C.; Hickman, J.J.; Shuler, M.L. How multi-organ microdevices can help foster drug development. Adv. Drug Deliv. Rev. 2014, 69-70, 158–169. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Rennert, K.; Steinborn, S.; Gröger, M.; Ungerböck, B.; Jank, A.M.; Ehgartner, J.; Nietzsche, S.; Dinger, J.; Kiehntopf, M.; Funke, H.; et al. A Microfluidically Perfused Three Dimensional Human Liver Model. Biomaterials 2015, 71, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Gao, D.; Liu, W.; Wei, H.; Lin, J.-M. Imitation of drug metabolism in human liver and cytotoxicity assay using a microfluidic device coupled to mass spectrometric detection. Lab Chip 2011, 12, 219–226. [Google Scholar] [CrossRef]
- Kitsara, M.; Kontziampasis, D.; Agbulut, O.; Chen, Y. Heart on a chip: Micro-nanofabrication and microfluidics steering the future of cardiac tissue engineering. Microelectron. Eng. 2019, 203–204, 44–62. [Google Scholar] [CrossRef]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef] [Green Version]
- Kilic, O.; Pamies, D.; Lavell, E.; Schiapparelli, P.; Feng, Y.; Hartung, T.; Bal-Price, A.; Hogberg, H.T.; Quinones-Hinojosa, A.; Guerrero-Cazares, H.; et al. Brain-on-a-Chip Model Enables Analysis of Human Neuronal Differentiation and Chemotaxis. Lab Chip 2016, 16, 4152–4162. [Google Scholar] [CrossRef] [Green Version]
- Hübner, J.; Raschke, M.; Rütschle, I.; Gräßle, S.; Hasenberg, T.; Schirrmann, K.; Lorenz, A.; Schnurre, S.; Lauster, R.; Maschmeyer, I.; et al. Simultaneous Evaluation of Anti-EGFR-Induced Tumour and Adverse Skin Effects in a Microfluidic Human 3D Co-Culture Model. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Trapecar, M.; Communal, C.; Velazquez, J.; Maass, C.A.; Huang, Y.J.; Schneider, K.; Wright, C.W.; Butty, V.; Eng, G.; Yilmaz, O.; et al. Gut-Liver Physiomimetics Reveal Paradoxical Modulation of IBD-Related Inflammation by Short-Chain Fatty Acids. Cell Syst. 2020, 10, 223–239. [Google Scholar] [CrossRef]
- Bauer, S.; Huldt, C.W.; Kanebratt, K.P.; Durieux, I.; Gunne, D.; Andersson, S.; Ewart, L.; Haynes, W.; Maschmeyer, I.; Winter, A.; et al. Functional coupling of human pancreatic islets and liver spheroids on-a-chip: Towards a novel human ex vivo type 2 diabetes model. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A Four-Organ-Chip for Interconnected Long-Term Co-Culture of Human Intestine, Liver, Skin and Kidney Equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 2018, 22, 310–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreß, S.; Schaller-Ammann, R.; Feiel, J.; Priedl, J.; Kasper, C.; Egger, D. 3D Printing of Cell Culture Devices: Assessment and Prevention of the Cytotoxicity of Photopolymers for Stereolithography. Materials 2020, 13, 3011. [Google Scholar] [CrossRef] [PubMed]
- Toepke, M.W.; Beebe, D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Amoyav, B.; Goldstein, Y.; Steinberg, E.; Benny, O. 3D Printed Microfluidic Devices for Drug Release Assays. Pharmceutics 2020, 13, 13. [Google Scholar] [CrossRef]
- Gong, H.; Beauchamp, M.; Perry, S.T.; Woolley, A.T.; Nordin, G.P. Optical approach to resin formulation for 3D printed microfluidics. RSC Adv. 2015, 5, 106621–106632. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goldstein, Y.; Spitz, S.; Turjeman, K.; Selinger, F.; Barenholz, Y.; Ertl, P.; Benny, O.; Bavli, D. Breaking the Third Wall: Implementing 3D-Printing Techniques to Expand the Complexity and Abilities of Multi-Organ-on-a-Chip Devices. Micromachines 2021, 12, 627. https://doi.org/10.3390/mi12060627
Goldstein Y, Spitz S, Turjeman K, Selinger F, Barenholz Y, Ertl P, Benny O, Bavli D. Breaking the Third Wall: Implementing 3D-Printing Techniques to Expand the Complexity and Abilities of Multi-Organ-on-a-Chip Devices. Micromachines. 2021; 12(6):627. https://doi.org/10.3390/mi12060627
Chicago/Turabian StyleGoldstein, Yoel, Sarah Spitz, Keren Turjeman, Florian Selinger, Yechezkel Barenholz, Peter Ertl, Ofra Benny, and Danny Bavli. 2021. "Breaking the Third Wall: Implementing 3D-Printing Techniques to Expand the Complexity and Abilities of Multi-Organ-on-a-Chip Devices" Micromachines 12, no. 6: 627. https://doi.org/10.3390/mi12060627
APA StyleGoldstein, Y., Spitz, S., Turjeman, K., Selinger, F., Barenholz, Y., Ertl, P., Benny, O., & Bavli, D. (2021). Breaking the Third Wall: Implementing 3D-Printing Techniques to Expand the Complexity and Abilities of Multi-Organ-on-a-Chip Devices. Micromachines, 12(6), 627. https://doi.org/10.3390/mi12060627





