Toward Development of a Label-Free Detection Technique for Microfluidic Fluorometric Peptide-Based Biosensor Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein-Markers
- Cardiac troponin I (cTpI), T9924, Sigma-Aldrich (St. Louis, MO, USA).
- Cardiac troponin T (cTpT), T0175, Sigma-Aldrich (St. Louis, MO, USA).
2.2. Construction Materials
2.3. Inorganic Luminophores
2.4. Apparatus
2.5. Peptide Aptamers
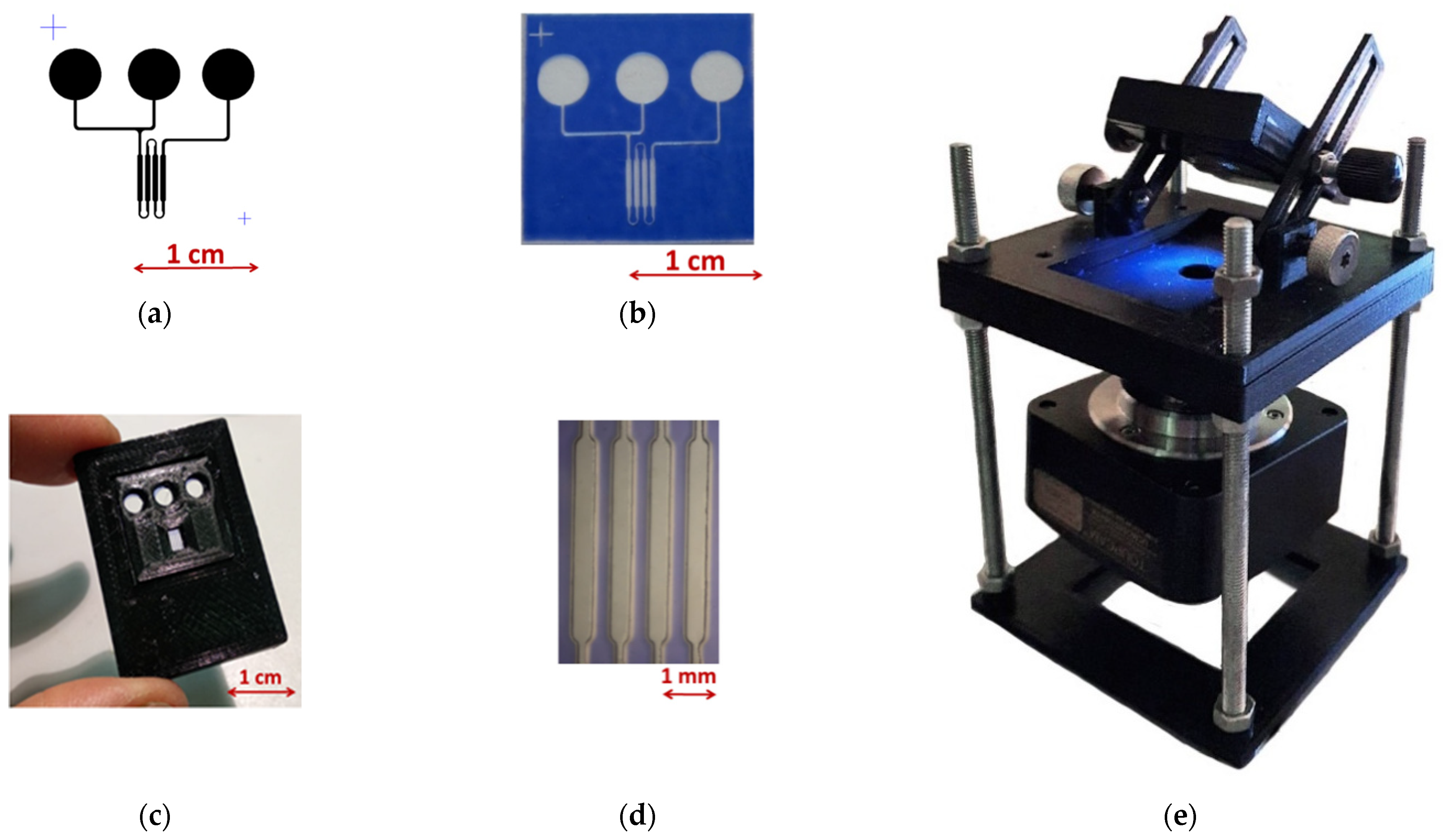
2.6. Microfluidic Chip Fabrication Process
3. Results and Discussion
3.1. Designing Peptide Aptamers for Label-Free Detection
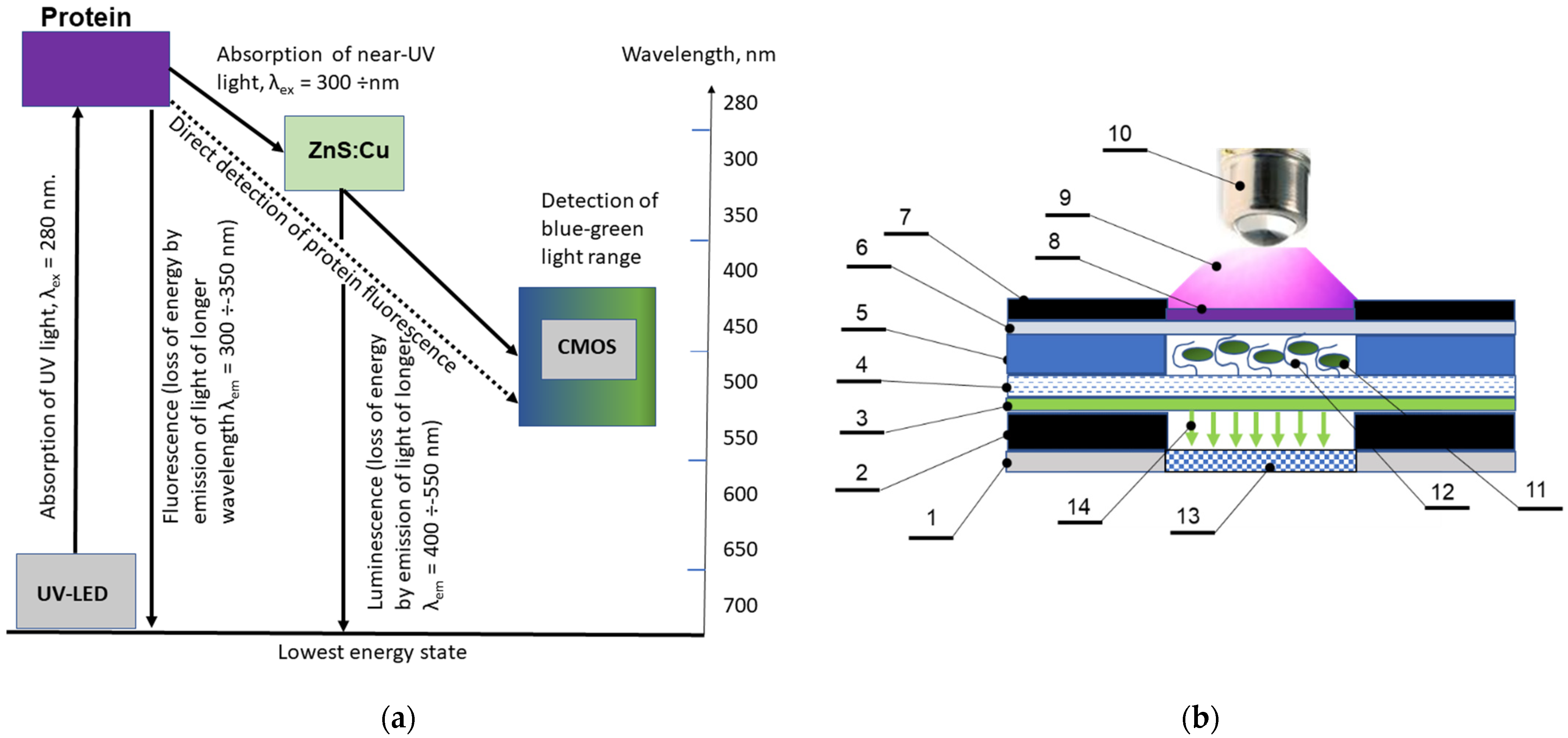
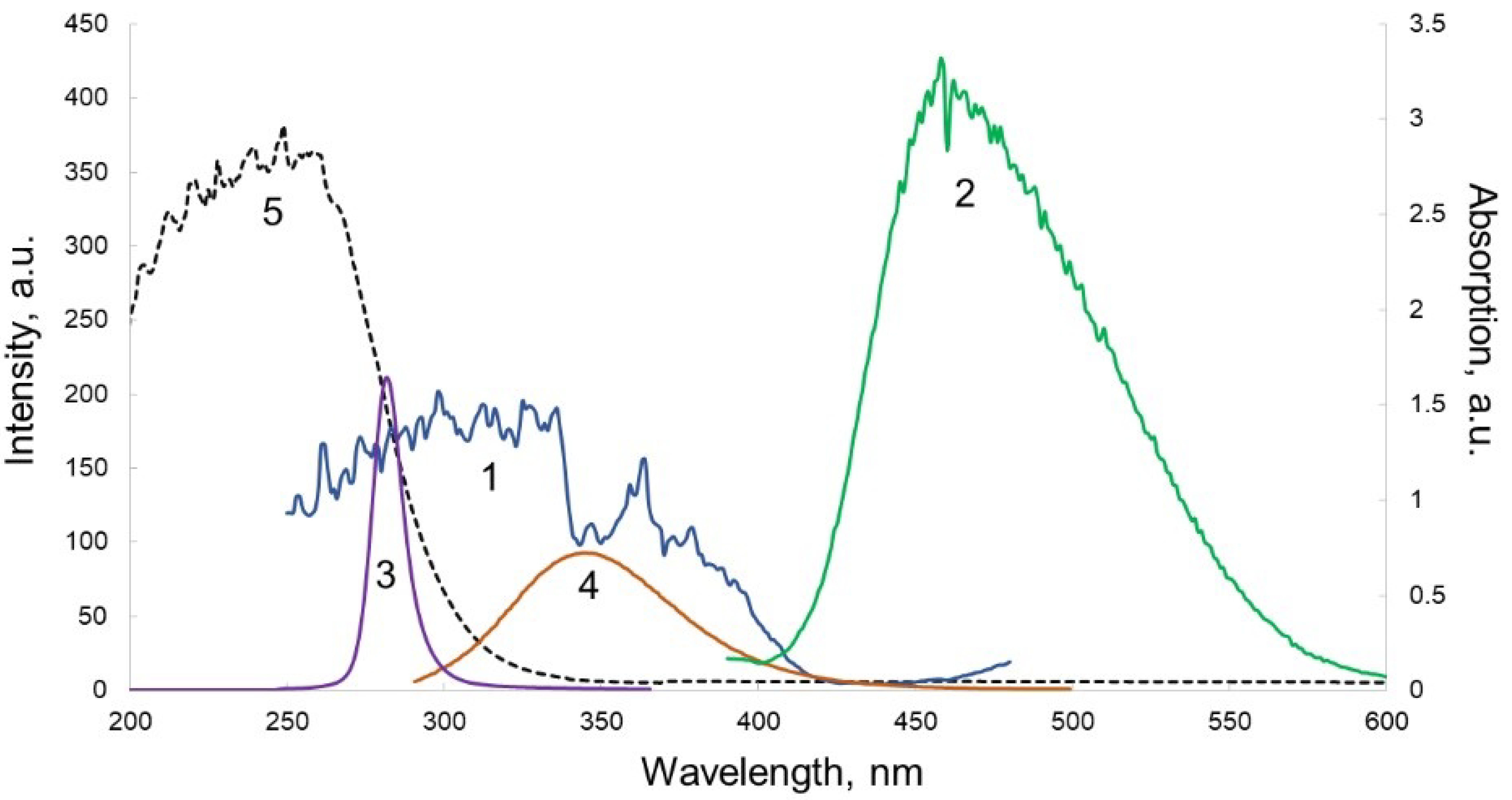
3.2. Optical Scheme of Protein Detection in Biosensor Channels
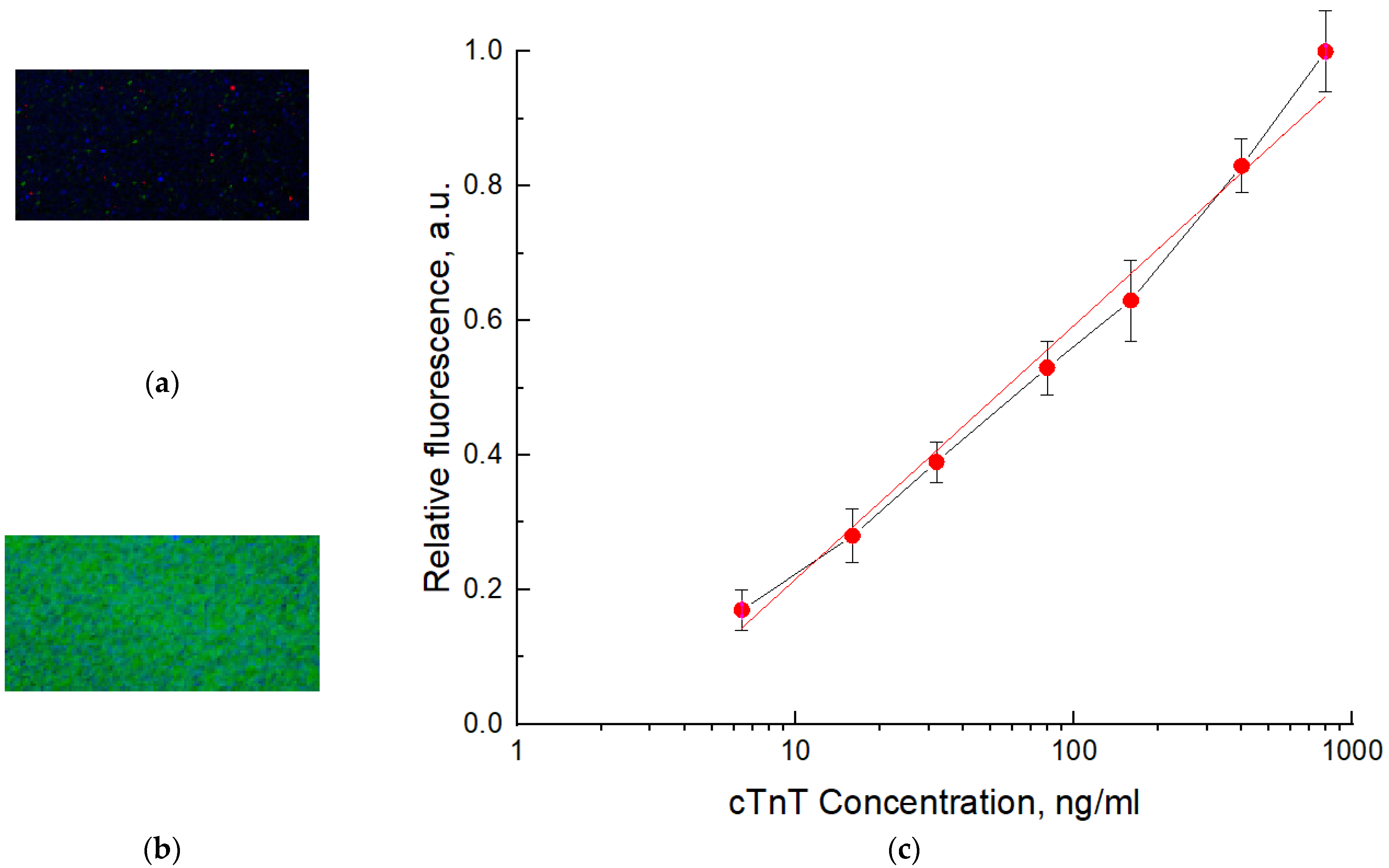
3.3. Preparation of Biosensor Model and Its Testing
4. Conclusions
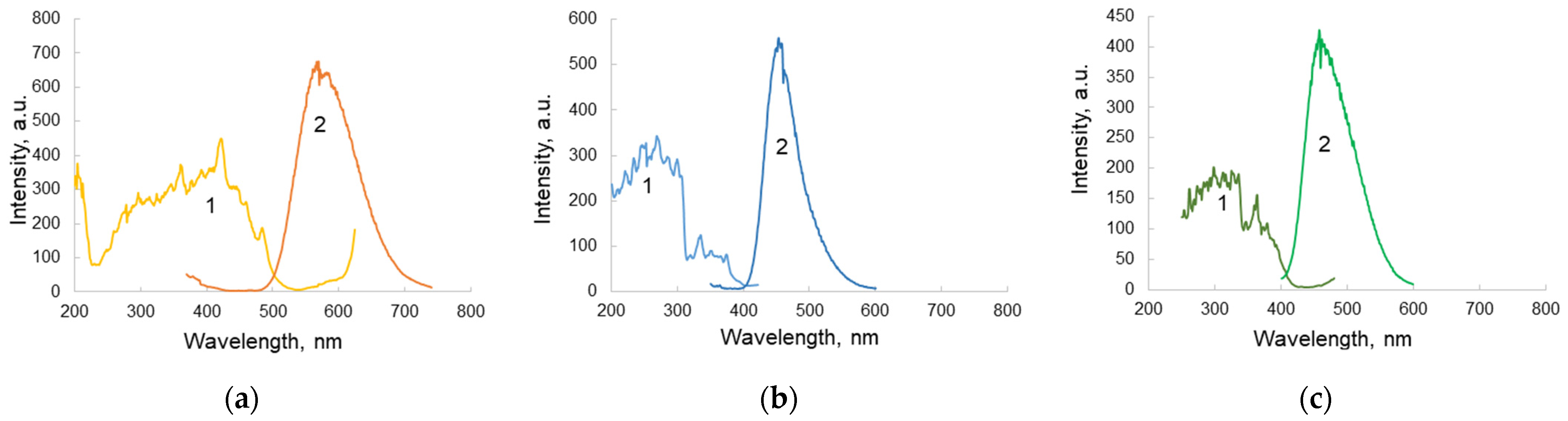
- The fluorescence of a number of polymer materials was studied under excitation in the range of 260–280 nm, in order to select materials with minimal background fluorescence under radiation. The results showed that the selected materials meet the necessary requirements.
- A material for the preparation of the inlet window of the biosensor, transparent for UV radiation at 275 nm, was selected. The most fitting proved to be a PP film, 100 µm thick.
- A peptide aptamer constructed using the «Protein 3D» software complementary to troponin T was designed. On its basis, its non-fluorescent digital twin was designed, which retained its three-dimensional complementarity to target protein, which has been shown with capillary electrophoresis-on-a-chip experiments.
- For the outlet window transmitting fluorescence of troponin T for the channel depth of 50 µm in the range of 300–350 nm and absorbing incidental UV radiation at 275 nm, the most fitting material was glass. The cover glass plates were used for the preparation of this element.
- Comparison of solid-state inorganic luminophores in order to select the optimal material for use in a planar optical element with an excitation range of 300–350 nm (Troponin T emission range) and an emission range of 450–550 nm (optimal for a receiving device) showed that a luminophore with a ZnS:Cu composition was the most consistent with the tasks.
- A laboratory sample of the biosensor was designed and manufactured. Technology for applying a luminophore to glass was developed.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Daar, A.; Singer, P.; Leah Persad, D.; Pramming, S.K.; Matthews, D.R.; Beaglehole, R.; Bernstein, A.; Borysiewicz, L.K.; Colagiuri, S.; Ganguly, N.; et al. Grand challenges in chronic non-communicable diseases. Nature 2007, 450, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Hlapčić, I.; Belamarić, D.; Bosnar, M.; Kifer, D.; Dugac, A.V.; Rumora, L. Combination of systemic inflammatory biomarkers in assessment of chronic obstructive pulmonary disease: Diagnostic performance and identification of networks and clusters. Diagnostics 2020, 10, 1029. [Google Scholar] [CrossRef]
- Reverdatto, S.; Burz, D.S.; Shekhtman, A. Peptide aptamers: Development and applications. Curr. Med. Chem. 2015, 15, 1082–1101. [Google Scholar] [CrossRef] [PubMed]
- Madjid, M.; Safavi-Naeini, P.; Solomon, S.D.; Vardeny, O. Potential effects of coronaviruses on the cardiovascular system: A Review. JAMA Cardiol. 2020, 5, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Bax, J.J.; Baumgartner, H.; Ceconi, C.; Dean, V.; Fagard, R.; Funck-Brentano, C.; Kolh, P. Third universal definition of myocardial infarction. J. Am. Coll. Cardiol. 2012, 60, 1581–1598. [Google Scholar]
- Johns Hopkins Medicine. Diagnosis and Screening for Cardiovascular Conditions. Available online: https://www.hopkinsmedicine.org/health/treatment-tests-and-therapies/diagnosis-and-screening-for-cardiovascular-conditions (accessed on 21 December 2020).
- Deng, A.; Matloff, D.; Lin, C.E.; Probst, D.; Broniak, T.; Alsuwailem, M.; Belle, J.T.L. Development toward a triple-marker biosensor for diagnosing cardiovascular disease. Crit. Rev. Biomed. Eng. 2019, 47, 169–178. [Google Scholar] [CrossRef]
- Aldous, S.J. Cardiac biomarkers in acute myocardial infarction. Int. J. Cardiol. 2013, 164, 282–294. [Google Scholar] [CrossRef]
- Mythili, S.; Mythili, S. Diagnostic markers of acute myocardial infarction. Biomed. Rep. 2015, 3, 743–748. [Google Scholar] [CrossRef]
- Nagel, B.; Dellweg, H.; Gierasch, L.M. Glossary for chemists of terms used in biotechnology (IUPAC Recommendations 1992). Pure Appl. Chem. 2013, 64, 143–168. [Google Scholar] [CrossRef]
- Szunerits, S.; Mishyn, V.; Grabowska, I.; Boukherroub, R. Electrochemical cardiovascular platforms: Current state of the art and beyond. Biosens. Bioelectron. 2019, 131, 287–298. [Google Scholar] [CrossRef]
- Dhara, K.; Mahapatra, D.R. Review on electrochemical sensing strategies for C-reactive protein and cardiac troponin I detection. Microchem. J. 2020, 156, 104857. [Google Scholar] [CrossRef]
- Sharma, A.; Bhardwaj, J.; Jang, J. Label-free, highly sensitive electrochemical aptasensors using polymer-modified reduced graphene oxide for cardiac biomarker detection. ACS Omega 2020, 5, 3924–3931. [Google Scholar] [CrossRef]
- Vasudevan, M.; Tai, M.J.; Perumal, V.; Gopinath, S.C.; Murthe, S.S.; Ovinis, M.; Mohamed, N.M.; Joshi, N. Highly sensitive and selective acute myocardial infarction detection using aptamer-tethered MoS2 nanoflower and screen-printed electrodes. Biotechnol. Appl. Biochem. 2020, 1–10. [Google Scholar] [CrossRef]
- Mi, X.; Li, H.; Tan, R.; Tu, Y. Dual-modular aptasensor for detection of cardiac troponin I based on mesoporous silica films by electrochemiluminescence/electrochemical impedance spectroscopy. Anal. Chem. 2020, 92, 14640–14647. [Google Scholar] [CrossRef]
- Negahdary, M.; Heli, H. An electrochemical troponin I peptisensor using a triangular icicle-like gold nanostructure. Biochem. Eng. J. 2019, 151, 107326. [Google Scholar] [CrossRef]
- Negahdary, M.; Behjati-Ardakani, M.; Heli, H. An electrochemical troponin T aptasensor based on the use of a macroporous gold nanostructure. Microchim. Acta 2019, 186, 377. [Google Scholar] [CrossRef]
- Shanmugam, N.R.; Muthukumar, S.; Tanak, A.S.; Prasad, S. Multiplexed electrochemical detection of three cardiac biomarkers cTnI, cTnT and BNP using nanostructured ZnO-sensing platform. Future Cardiol. 2018, 14, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Kirste, R.; Rohrbaugh, N.; Bryan, I.; Bryan, Z.; Collazo, R.; Ivanisevic, A. Electronic biosensors based on III-nitride semiconductors. Ann. Rev. Anal. Chem. 2015, 8, 149–169. [Google Scholar] [CrossRef] [PubMed]
- Sarangadharan, I.; Regmi, A.; Chen, Y.W.; Hsu, C.P.; Chen, P.C.; Chang, W.H.; Lee, G.Y.; Chyi, J.I.; Shiesh, S.C.; Lee, G.B.; et al. High sensitivity cardiac troponin I detection in physiological environment using AlGaN/GaN high electron mobility transistor (HEMT) biosensors. Biosens. Bioelectron. 2018, 100, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Tai, T.Y.; Li, K.H.; Gopinathan, P.; Chung, Y.D.; Sarangadharan, I.; Ma, H.P.; Huang, P.C.; Shiesh, S.C.; Wang, Y.L.; et al. An integrated microfluidic system with field-effect-transistor sensor arrays for detecting multiple cardiovascular biomarkers from clinical samples. Biosens. Bioelectron. 2019, 129, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Jang, J. Flexible electrical aptasensor using dielectrophoretic assembly of graphene oxide and its subsequent reduction for cardiac biomarker detection. Sci. Rep. 2019, 9, 1–10. [Google Scholar]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar]
- Chen, J.; Ran, F.; Chen, Q.; Luo, D.; Ma, W.; Han, T.; Wang, C.; Wang, C. A fluorescent biosensor for cardiac biomarker myoglobin detection based on carbon dots and deoxyribonuclease I-aided target recycling signal amplification. RSC Adv. 2019, 9, 4463–4468. [Google Scholar] [CrossRef]
- Khan, S.; Hasan, A.; Attar, F.; Sharifi, M.; Siddique, R.; Mraiche, F.; Falahati, M. Gold nanoparticle-based platforms for diagnosis and treatment of myocardial infarction. ACS Biomater. Sci. Eng. 2020, 6, 6460–6477. [Google Scholar] [CrossRef] [PubMed]
- El-Said, W.A.; Fouad, D.M.; El-Safty, S.A. Ultrasensitive label-free detection of cardiac biomarker myoglobin based on surface-enhanced Raman spectroscopy. Sens. Actuators B Chem. 2016, 228, 401–409. [Google Scholar] [CrossRef]
- Diware, M.S.; Cho, H.M.; Chegal, W.; Cho, Y.J.; Kim, D.S.; Kim, K.S.; Paek, S.H. Ultrasensitive, label-free detection of cardiac biomarkers with optical SIS sensor. Biosens. Bioelectron. 2017, 87, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Li, J.; Mou, L.; Liu, Y.; Deng, J.; Qian, W.; Sun, J.; Cha, R.; Jiang, X. An automated and portable microfluidic chemiluminescence immunoassay for quantitative detection of biomarkers. Lab Chip 2017, 17, 2225–2234. [Google Scholar] [CrossRef]
- Bhalla, N.; Chung, D.W.Y.; Chang, Y.J.; Uy, K.J.S.; Ye, Y.Y.; Chin, T.Y.; Pijanowska, D.G. Microfluidic platform for enzyme-linked and magnetic particle-based immunoassay. Micromachines 2013, 4, 257–271. [Google Scholar] [CrossRef]
- Sanjay, S.T.; Fu, G.; Dou, M.; Xu, F.; Liu, R.; Qi, H.; Li, X. Biomarker detection for disease diagnosis using cost-effective microfluidic platforms. Analyst 2015, 140, 7062–7081. [Google Scholar] [CrossRef] [PubMed]
- Soum, V.; Park, S.; Brilian, A.I.; Kwon, O.S.; Shin, K. Programmable paper-based microfluidic devices for biomarker detections. Micromachines 2019, 10, 516. [Google Scholar] [CrossRef]
- Oliveira, R.A.D.; Materon, E.M.; Melendez, M.E.; Carvalho, A.L.; Faria, R.C. Disposable microfluidic immunoarray device for sensitive breast cancer biomarker detection. ACS Appl. Mater. Interfaces 2017, 9, 27433–27440. [Google Scholar] [CrossRef] [PubMed]
- Honikel, M.M.; Lin, C.E.; Probst, D.; Belle, J.T. Facilitating earlier diagnosis of cardiovascular disease through point-of-care biosensors: A review. Crit. Rev. Biomed. Eng. 2018, 46, 53–82. [Google Scholar] [CrossRef]
- Aziz, S.B.; Brza, M.A.; Nofal, M.M.; Abdulwahid, R.T.; Hussen, S.A.; Hussein, A.M.; Karim, W.O. A comprehensive review on optical properties of polymer electrolytes and composites. Materials 2020, 13, 3675. [Google Scholar] [CrossRef]
- Maheshwari, N.; Chatterjee, G.; Rao, V.R. A technology overview and applications of Bio-MEMS. J. ISSS 2014, 3, 39–59. [Google Scholar]
- Chilukoti, G.R.; Periyasamy, A.P. Ultra High Molecular Weight Polyethylene for Medical Applications. 2012. Available online: https://www.researchgate.net/profile/Govardhan-Rao-Chilukoti/publication/298626009_Ultra_high_molecular_weight_polyethylene_for_medical_applications/links/589038ee92851c9794c4ecd3/Ultra-high-molecular-weight-polyethylene-for-medical-applications.pdf (accessed on 21 December 2020).
- Umar, A.; Khairil, J.B.A.K.; Nor, A.B. A review of the properties and applications of poly (methyl methacrylate) (PMMA). Polym. Rev. 2015, 55, 678–705. [Google Scholar]
- Chanturia, V.A.; Dvoichenkova, G.P.; Morozov, V.V.; Koval’chuk, O.E.; Podkamenny, Y.A.; Yakovlev, V.N. Experimental justification of luminophore composition for indication of diamonds in X-ray luminescence separation of kimberlite ore. J. Min. Sci. 2018, 54, 458–465. [Google Scholar] [CrossRef]
- Cary Eclipse Fluorescence Spectrophotometer. Available online: https://www.agilent.com/en/product/molecular-spectroscopy/fluorescence-spectroscopy/fluorescence-systems/cary-eclipse-fluorescence-spectrophotometer (accessed on 22 December 2020).
- Protein Data Bank. Available online: https://www.rcsb.org/ (accessed on 22 December 2020).
- Visualizer of Supramolecular Biostructures «Protein 3D». Available online: http://protein-3d.ru/index_e.html (accessed on 21 December 2020).
- Karasev, V. Data on the application of the molecular vector machine model: A database of protein pentafragments and computer software for predicting and designing secondary protein structures. Data Brief 2020, 28, 104815. [Google Scholar] [CrossRef]
- Sitkov, N.O.; Zimina, T.M.; Karasev, V.A.; Lemozerskii, V.E.; Kolobov, A.A. Design of Peptide Ligands (Aptamers) for Determination of Myeloperoxidase Level in Blood Using Biochips. In Proceedings of the 2020 IEEE Conference of Russian Young Researchers in Electrical and Electronic Engineering, EIConRus, St. Petersburg and Moscow, Russia, 27–30 January 2020; Volume 2020, pp. 1599–1603. [Google Scholar]
- Li, J.; Tan, S.; Chen, X.; Zhang, C.Y.; Zhang, Y. Peptide aptamers with biological and therapeutic applications. Curr. Med. Chem. 2011, 18, 4215–4222. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 2009, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Groner, B.; Borghouts, C.; Kunz, C. Peptide aptamer libraries. Comb. Chem. High Throughput Screen. 2008, 11, 135–145. [Google Scholar] [CrossRef]
- Antes, I. DynaDock: A new molecular dynamics-based algorithm for protein-peptide docking including receptor flexibility. Proteins 2010, 78, 1084–1104. [Google Scholar] [CrossRef]
- Obarska-Kosinska, A.; Iacoangeli, A.; Lepore, R.; Tramontano, A. PepComposer: Computational design of peptides binding to a given protein surface. Nucleic Acids Res. 2016, 44, 522–528. [Google Scholar] [CrossRef]
- Lee, J.F.; Hesselberth, J.R.; Meyers, L.A.; Ellington, A.D. Aptamer database. Nucleic Acids Res. 2004, 32, D95–D100. [Google Scholar] [CrossRef]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on aptamer research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef] [PubMed]
- Agnew, H.D.; Coppock, M.B.; Idso, M.N.; Lai, B.T.; Liang, J.X.; McCarthy-Torrens, A.M.; Warren, C.M.; Heath, J.R. Protein-catalyzed capture agents. Chem. Rev. 2019, 119, 9950–9970. [Google Scholar] [CrossRef] [PubMed]
- Gompetrs, R.; Renner, E.; Mehta, M. Enabling technologies for innovative new materials. Am. Lab. 2005, 37, 12–14. [Google Scholar]
- Sim, A.Y.L.; Minary, P.; Levitt, M. Modelling nucleic acids. Curr. Opin. Struct. Biol. 2012, 22, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; McCammon, J.A. Molecular dynamics symulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.H.; Lim, J.; Jee, J.E.; Aw, J.H.; Lee, S.S. Peptide-peptide co-assembly: A design strategy for functional detection of C-peptide, a biomarker of diabetic neuropathy. Int. J. Mol. Sci. 2020, 21, 9671. [Google Scholar] [CrossRef] [PubMed]
- Sitkov, N.O.; Karasev, V.A.; Luchinin, V.V.; Zimina, T.M. Development of biosensors for express-detection of protein markers of diseases in blood using peptide biorecognition elements. AIP Conf. Proc. 2019, 2140, 020072. [Google Scholar]
- Takeda, S.; Yamashita, A.; Maeda, K.; Maeda, Y. Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature 2003, 424, 35–41. [Google Scholar] [CrossRef]
- Ghisaidoobe, A.B.; Chung, S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: A focus on Förster resonance energy transfer techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef]
- Jang, D.; Chae, G.; Shin, S. Analysis of surface plasmon resonance curves with a novel sigmoid-asymmetric fitting algorithm. Sensors 2015, 15, 25385–25398. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.; Souza, E.; Ferreira, D.; Zanforlin, D.; Bezerra, W.; Borba, M.A.; Arruda, M.; Lopes, K.; Nascimento, G.; Martins, D.; et al. A sensitive and selective label-free electrochemical DNA biosensor for the detection of specific dengue virus serotype 3 sequences. Sensors 2015, 15, 15562–15577. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Anderson, M.; Bernards, M.T.; Hunt, H.K. PEG functionalization of whispering gallery mode optical microresonator biosensors to minimize non-specific adsorption during targeted, label-free sensing. Sensors 2015, 15, 18040–18060. [Google Scholar] [CrossRef] [PubMed]
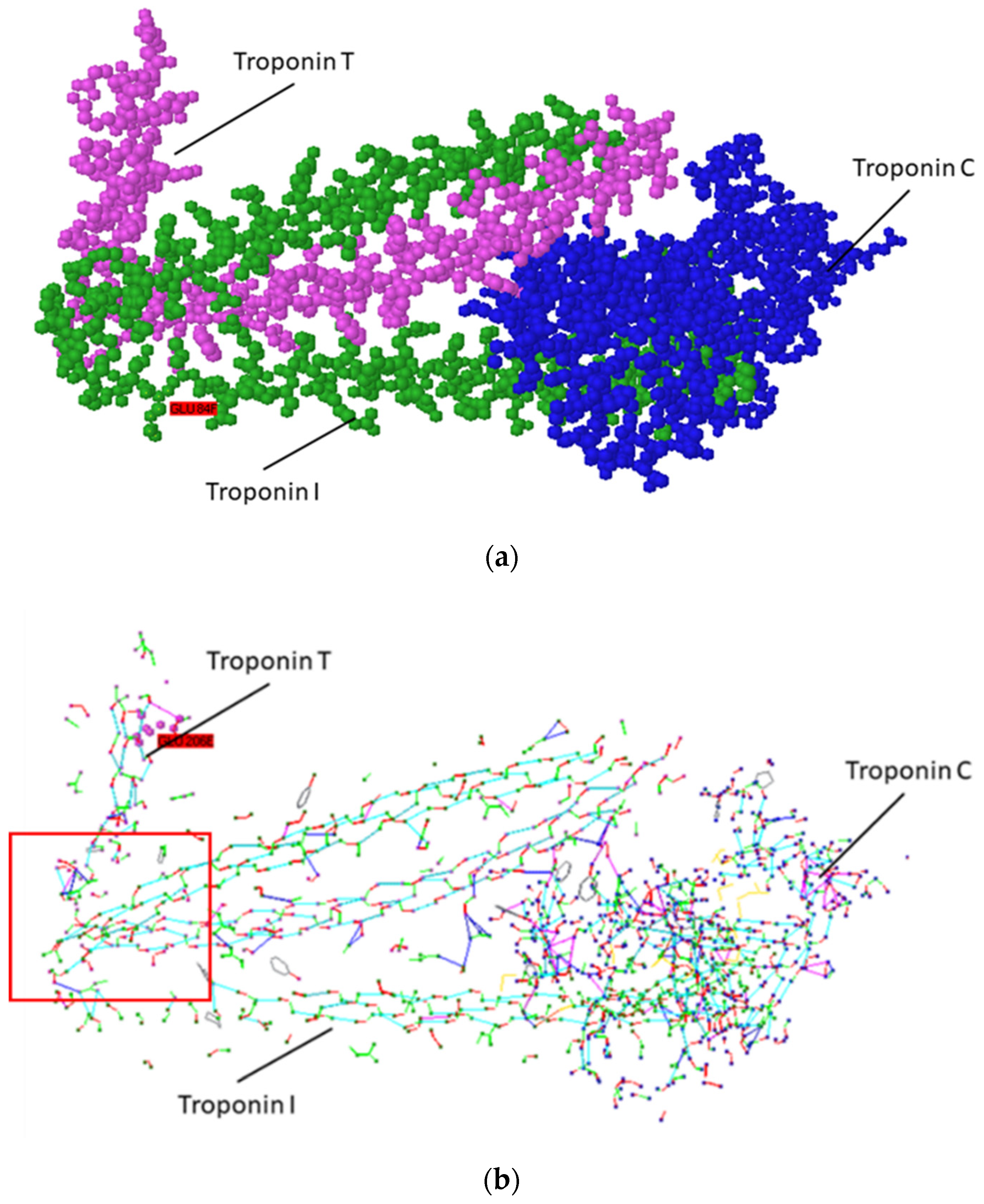
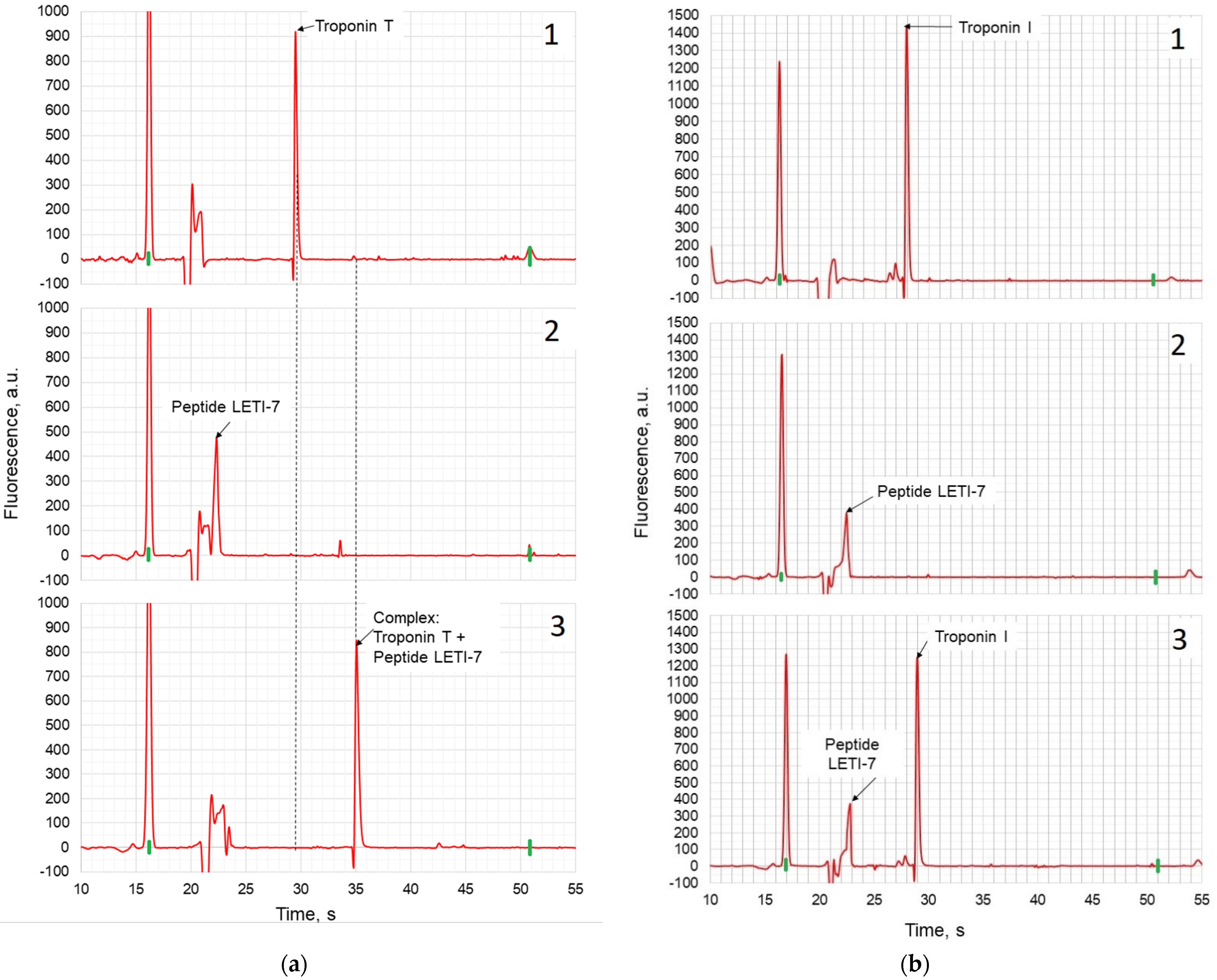
| Name of Peptide | Structure | Number of Amino Acid Residues |
|---|---|---|
| LETI-2 | HLNEDQLREKAKELWQTIYNLEAEKFDLQEKFKQQKE | 38 |
| LETI-7 | TLNEDQLREKAKELAQTIANLEAEKIDLQEKAKQQKYE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sitkov, N.; Zimina, T.; Kolobov, A.; Karasev, V.; Romanov, A.; Luchinin, V.; Kaplun, D. Toward Development of a Label-Free Detection Technique for Microfluidic Fluorometric Peptide-Based Biosensor Systems. Micromachines 2021, 12, 691. https://doi.org/10.3390/mi12060691
Sitkov N, Zimina T, Kolobov A, Karasev V, Romanov A, Luchinin V, Kaplun D. Toward Development of a Label-Free Detection Technique for Microfluidic Fluorometric Peptide-Based Biosensor Systems. Micromachines. 2021; 12(6):691. https://doi.org/10.3390/mi12060691
Chicago/Turabian StyleSitkov, Nikita, Tatiana Zimina, Alexander Kolobov, Vladimir Karasev, Alexander Romanov, Viktor Luchinin, and Dmitry Kaplun. 2021. "Toward Development of a Label-Free Detection Technique for Microfluidic Fluorometric Peptide-Based Biosensor Systems" Micromachines 12, no. 6: 691. https://doi.org/10.3390/mi12060691
APA StyleSitkov, N., Zimina, T., Kolobov, A., Karasev, V., Romanov, A., Luchinin, V., & Kaplun, D. (2021). Toward Development of a Label-Free Detection Technique for Microfluidic Fluorometric Peptide-Based Biosensor Systems. Micromachines, 12(6), 691. https://doi.org/10.3390/mi12060691








