Methods of Respiratory Virus Detection: Advances towards Point-of-Care for Early Intervention
Abstract
1. Introduction
2. Conventional Respiratory Virus Quantification Methods
3. Commercialized Respiratory Virus Diagnostic POCT Devices
3.1. Molecular Diagnostics
3.2. Immunology-Based Rapid Detection Assay
4. Advances in Respiratory Virus Testing for Rapid Diagnosis
4.1. Rapid Molecular Diagnosis
4.2. Immunoassay Paper-Based Devices/Technology
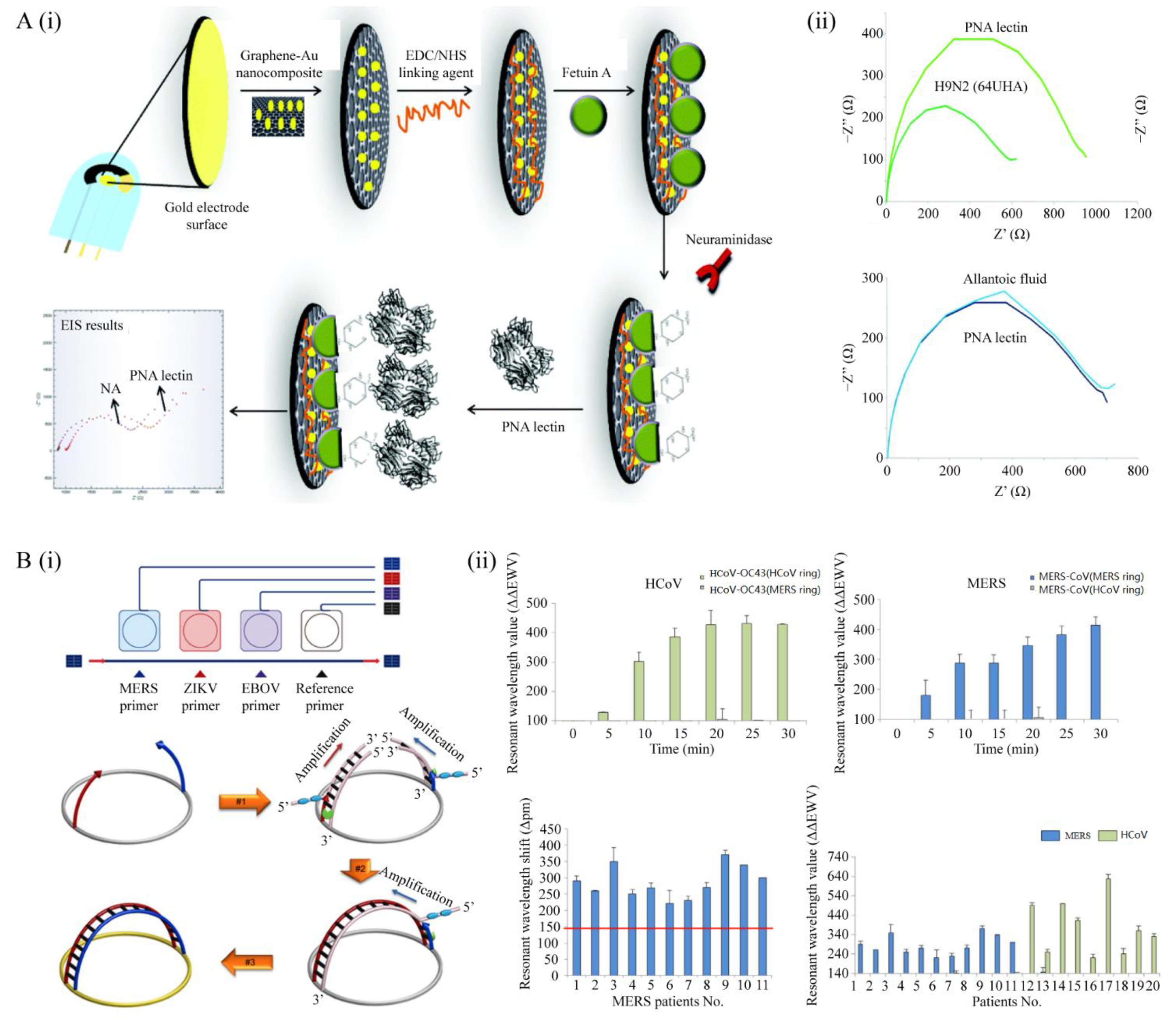
4.3. Biosensors for Respiratory Virus Detection
4.4. Microfluidic Device for Respiratory Virus Detection
4.5. Smartphone-Based Detection Technologies
5. Potential Techniques for Future POC Diagnosis
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.D.; Rappuoli, R. SARS—Beginning to understand a new virus. Nat. Rev. Microbiol. 2003, 1, 209–218. [Google Scholar] [CrossRef] [PubMed]
- The Influenza Outbreak. JAMA 2020, 323, 1621. [CrossRef]
- Zhao, H. COVID-19 drives new threat to bats in China. Science 2020, 367, 1436. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. US$675 Million Needed for New Coronavirus Preparedness and Response Global Plan; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Nguyen, T.; Duong Bang, D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef]
- Ferretti, L.; Wymant, C.; Kendall, M.; Zhao, L.; Nurtay, A.; Abeler-Dorner, L.; Parker, M.; Bonsall, D.; Fraser, C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 2020, 368. [Google Scholar] [CrossRef]
- Mahalingam, S.; Peter, J.; Xu, Z.; Bordoloi, D.; Ho, M.; Kalyanaraman, V.S.; Srinivasan, A.; Muthumani, K. Landscape of humoral immune responses against SARS-CoV-2 in patients with COVID-19 disease and the value of antibody testing. Heliyon 2021, 7, e06836. [Google Scholar] [CrossRef]
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Liao, P.; Qiu, J.F.; Lin, Y.; Cai, X.F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Gorse, G.J.; Donovan, M.M.; Patel, G.B. Antibodies to coronaviruses are higher in older compared with younger adults and binding antibodies are more sensitive than neutralizing antibodies in identifying coronavirus-associated illnesses. J. Med. Virol. 2020, 92, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Yue-ping, L.; Bo, D.; Feifei, R.; Yue, W.; Jinya, D.; Qianchuan, H. Diagnostic Indexes of a Rapid IgG/IgM Combined Antibody Test for SARS-CoV-2. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034. [Google Scholar] [CrossRef] [PubMed]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.F.; Kemball, M.E.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef]
- Leland, D.S.; Ginocchio, C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 2007, 20, 49–78. [Google Scholar] [CrossRef]
- Goldsmith, C.S.; Miller, S.E. Modern uses of electron microscopy for detection of viruses. Clin. Microbiol. Rev. 2009, 22, 552–563. [Google Scholar] [CrossRef]
- Connor, E.M.; Loeb, M.R. A hemadsorption method for detection of colonies of Haemophilus influenzae type b expressing fimbriae. J. Infect. Dis. 1983, 148, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Gleaves, C.A.; Smith, T.F.; Shuster, E.A.; Pearson, G.R. Rapid detection of cytomegalovirus in mrc-5-cells inoculated with urine specimens by using low-speed centrifugation and monoclonal-antibody to an early antigen. J. Clin. Microbiol. 1984, 19, 917–919. [Google Scholar] [CrossRef] [PubMed]
- Espy, M.J.; Smith, T.F.; Harmon, M.W.; Kendal, A.P. Rapid detection of influenza virus by shell vial assay with monoclonal antibodies. J. Clin. Microbiol. 1986, 24, 677–679. [Google Scholar] [CrossRef]
- Navarro-Mari, J.M.; Sanbonmatsu-Gamez, S.; Perez-Ruiz, M.; De la Rosa-Fraile, M. Rapid detection of respiratory viruses by shell vial assay using simultaneous culture of HEp-2, LLC-MK2, and MDCK cells in a single vial. J. Clin. Microbiol. 1999, 37, 2346–2347. [Google Scholar] [CrossRef]
- Pesaresi, M.; Pirani, F.; Tagliabracci, A.; Valsecchi, M.; Procopio, A.D.; Busardo, F.P.; Graciotti, L. SARS-CoV-2 identification in lungs, heart and kidney specimens by transmission and scanning electron microscopy. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5186–5188. [Google Scholar] [CrossRef]
- Hopfer, H.; Herzig, M.C.; Gosert, R.; Menter, T.; Hench, J.; Tzankov, A.; Hirsch, H.H.; Miller, S.E. Hunting coronavirus by transmission electron microscopy—A guide to SARS-CoV-2-associated ultrastructural pathology in COVID-19 tissues. Histopathology 2021, 78, 358–370. [Google Scholar] [CrossRef]
- Nagayama, K.; Danev, R. Phase contrast electron microscopy: Development of thin-film phase plates and biological applications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 2153–2162. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Johnson, K.M.; Lange, J.V.; Webb, P.A.; Murphy, F.A. Isolation and partial characterisation of a new virus causing acute haemorrhagic fever in Zaire. Lancet 1977, 1, 569–571. [Google Scholar] [CrossRef]
- Drosten, C.; Gunther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.; et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with 2019 Novel Coronavirus Disease, United States. Emerg. Infect. Dis. 2020, 26. [Google Scholar] [CrossRef] [PubMed]
- Khanna, M.; Kumar, P.; Chugh, L.; Prasad, A.K.; Chhabra, S.K. Evaluation of influenza virus detection by direct enzyme immunoassay (EIA) and conventional methods in asthmatic patients. J. Commun. Dis. 2001, 33, 163–169. [Google Scholar] [PubMed]
- Burke, D.S.; Nisalak, A.; Ussery, M.A. Antibody capture immunoassay detection of japanese encephalitis virus immunoglobulin m and g antibodies in cerebrospinal fluid. J. Clin. Microbiol. 1982, 16, 1034–1042. [Google Scholar] [CrossRef]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef]
- Zhang, M.; Ye, J.; He, J.-S.; Zhang, F.; Ping, J.; Qian, C.; Wu, J. Visual detection for nucleic acid-based techniques as potential on-site detection methods. A review. Anal. Chim. Acta 2020, 1099, 1–15. [Google Scholar] [CrossRef]
- van Elden, L.J.R.; Nijhuis, M.; Schipper, P.; Schuurman, R.; van Loon, A.M. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 2001, 39, 196–200. [Google Scholar] [CrossRef]
- Fiercebiotech. FDA Grants Roche Coronavirus Test Emergency Green Light within 24 Hours. 2020. Available online: https://www.fiercebiotech.com/medtech/fda-grants-roche-coronavirus-test-emergency-green-light-within-24-hours (accessed on 11 June 2021).
- Baron, E.J.; Persing, D.H. Invited Product Profile - GeneXpert Xpress System for Respiratory Testing State-of-the-Art Molecular Testing for Any Health Care Setting. Point Care 2019, 18, 66–71. [Google Scholar] [CrossRef]
- Navidad, J.F.; Griswold, D.J.; Gradus, M.S.; Bhattacharyya, S. Evaluation of Luminex xTAG Gastrointestinal Pathogen Analyte-Specific Reagents for High-Throughput, Simultaneous Detection of Bacteria, Viruses, and Parasites of Clinical and Public Health Importance. J. Clin. Microbiol. 2013, 51, 3018–3024. [Google Scholar] [CrossRef]
- Hazelton, B.; Gray, T.; Ho, J.; Ratnamohan, V.M.; Dwyer, D.E.; Kok, J. Detection of influenza A and B with the Alere i Influenza A & B: A novel isothermal nucleic acid amplification assay. Influenza Other Respir Viruses 2015, 9, 151–154. [Google Scholar] [CrossRef] [PubMed]
- de la Tabla, V.O.; Antequera, P.; Masia, M.; Ros, P.; Martin, C.; Gazquez, G.; Bunuel, F.; Sanchez, V.; Robledano, C.; Gutierrez, F. Clinical evaluation of rapid point-of-care testing for detection of novel influenza A (H1N1) virus in a population-based study in Spain. Clin. Microbiol. Infect. 2010, 16, 1358–1361. [Google Scholar] [CrossRef]
- Ling, L.; Kaplan, S.E.; Lopez, J.C.; Stiles, J.; Lu, X.; Tang, Y.W. Parallel Validation of Three Molecular Devices for Simultaneous Detection and Identification of Influenza A and B and Respiratory Syncytial Viruses. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- Loeffelholz, M.J.; Alland, D.; Butler-Wu, S.M.; Pandey, U.; Perno, C.F.; Nava, A.; Carroll, K.C.; Mostafa, H.; Davies, E.; McEwan, A.; et al. Multicenter Evaluation of the Cepheid Xpert Xpress SARS-CoV-2 Test. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Zhen, W.; Smith, E.; Manji, R.; Schron, D.; Berry, G.J. Clinical Evaluation of Three Sample-to-Answer Platforms for Detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Dunn, J.; Obuekwe, J.; Baun, T.; Rogers, J.; Patel, T.; Snow, L. Prompt detection of influenza A and B viruses using the BD Veritor (TM) System Flu A plus B, Quidele (R) Sofia (R) Influenza A plus B FIA, and Alere BinaxNOW (R) Influenza A&B compared to real-time reverse transcription-polymerase chain reaction (RT-PCR). Diagn. Microbiol. Infect. Disease 2014, 79, 10–13. [Google Scholar] [CrossRef]
- Fuenzalida, L.; Blanco, S.; Prat, C.; Vivancos, M.; Dominguez, M.J.; Mòdol, J.M.; Rodrigo, C.; Ausina, V. Utility of the rapid antigen detection BinaxNOW Influenza A&B test for detection of novel influenza A (H1N1) virus. Clin. Microbiol. Infect. 2010, 16, 1574–1576. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Diseases 2020. [Google Scholar] [CrossRef]
- BioMedomics. COVID-19 IgM/IgG Rapid Test. 2020. Available online: https://www.biomedomics.com/products/infectious-disease/covid-19-rt/ (accessed on 11 June 2021).
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef]
- FDA. Coronavirus (COVID-19) Update: FDA Authorizes First COVID-19 Test for Self-Testing at Home. 2020. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-covid-19-test-self-testing-home (accessed on 11 June 2021).
- Ahn, S.J.; Baek, Y.H.; Lloren, K.K.S.; Choi, W.S.; Jeong, J.H.; Antigua, K.J.C.; Kwon, H.I.; Park, S.J.; Kim, E.H.; Kim, Y.I.; et al. Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. BMC Infect. Dis. 2019, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Wang, H.; Cao, Z.; Jin, H.; Chi, H.; Zhao, J.; Yu, B.; Yan, F.; Hu, X.; Wu, F.; et al. A Rapid and Specific Assay for the Detection of MERS-CoV. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Choi, N.; Moon, J.-I.; Choo, J. Determination of total iron-binding capacity of transferrin using metal organic framework-based surface-enhanced Raman scattering spectroscopy. J. Raman Spectrosc. 2021, 52, 506–515. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Wang, X.; Wang, K.; Zhu, Y.; Rong, Z.; Wang, W.; Xiao, R.; Wang, S. Magnetic SERS Strip for Sensitive and Simultaneous Detection of Respiratory Viruses. ACS Appl. Mater. Interfaces 2019, 11, 19495–19505. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Multiplex Paper-Based Colorimetric DNA Sensor Using Pyrrolidinyl Peptide Nucleic Acid-Induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. [Google Scholar] [CrossRef]
- Xiao, M.; Huang, L.; Dong, X.; Xie, K.; Shen, H.; Huang, C.; Xiao, W.; Jin, M.; Tang, Y. Integration of a 3D-printed read-out platform with a quantum dot-based immunoassay for detection of the avian influenza A (H7N9) virus. Analyst 2019, 144, 2594–2603. [Google Scholar] [CrossRef]
- Anik, U.; Tepeli, Y.; Sayhi, M.; Nsiri, J.; Diouani, M.F. Towards the electrochemical diagnostic of influenza virus: Development of a graphene-Au hybrid nanocomposite modified influenza virus biosensor based on neuraminidase activity. Analyst 2018, 143, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Layqah, L.A.; Eissa, S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta 2019, 186. [Google Scholar] [CrossRef] [PubMed]
- Dunajova, A.A.; Gal, M.; Tomcikova, K.; Sokolova, R.; Kolivoska, V.; Vaneckova, E.; Kielar, F.; Kostolansky, F.; Vareckova, E.; Naumowicz, M. Ultrasensitive impedimetric imunosensor for influenza A detection. J. Electroanal. Chem. 2020, 858, 5. [Google Scholar] [CrossRef]
- Koo, B.; Hong, K.H.; Jin, C.E.; Kim, J.Y.; Kim, S.-H.; Shin, Y. Arch-shaped multiple-target sensing for rapid diagnosis and identification of emerging infectious pathogens. Biosens. Bioelectron. 2018, 119, 79–85. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, R.; Li, Y.; Kong, W.; Guo, X.; Yang, Y.; Wu, F.; Liu, W.; Song, H.; Hao, R. Rapid detection of multiple respiratory viruses based on microfluidic isothermal amplification and a real-time colorimetric method. Lab Chip 2018, 18, 3507–3515. [Google Scholar] [CrossRef]
- Xiong, H.; Ye, X.; Li, Y.; Wang, L.; Zhang, J.; Fang, X.; Kong, J. Rapid Differential Diagnosis of Seven Human Respiratory Coronaviruses Based on Centrifugal Microfluidic Nucleic Acid Assay. Anal. Chem. 2020, 92, 14297–14302. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.; Liu, Y.; Wang, H.; Li, S.; Lin, Y.; Chen, L.; Zhao, Y.; Chao, S.; Huang, X.; Ge, S.; et al. A High-Throughput, Multi-Index Isothermal Amplification Platform for Rapid Detection of 19 Types of Common Respiratory Viruses Including SARS-CoV-2. Engineering 2020, 6, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chen, Y.; Tang, Y.; Cheng, G.; Yu, X.; He, H.; Cao, G.; Lu, H.; Liu, Z.; Zheng, S.-Y. Smartphone-Based Point-of-Care Microfluidic Platform Fabricated with a ZnO Nanorod Template for Colorimetric Virus Detection. ACS Sens. 2019, 4, 3298–3307. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Ganguli, A.; Nguyen, J.; Brisbin, R.; Shanmugam, K.; Hirschberg, D.L.; Wheeler, M.B.; Bashir, R.; Nash, D.M.; Cunningham, B.T. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip 2020, 20, 1621–1627. [Google Scholar] [CrossRef]
- Chen, L.; Yadav, V.; Zhang, C.; Huo, X.; Wang, C.; Senapati, S.; Chang, H.C. Elliptical Pipette Generated Large Microdroplets for POC Visual ddPCR Quantification of Low Viral Load. Anal. Chem. 2021, 93, 6456–6462. [Google Scholar] [CrossRef]
- Chen, H.; Park, S.G.; Choi, N.; Moon, J.I.; Dang, H.; Das, A.; Lee, S.; Kim, D.G.; Chen, L.; Choo, J. SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens. Bioelectron. 2020, 167, 112496. [Google Scholar] [CrossRef]
- Li, Z.; Leustean, L.; Inci, F.; Zheng, M.; Demirci, U.; Wang, S. Plasmonic-based platforms for diagnosis of infectious diseases at the point-of-care. Biotechnol. Adv. 2019, 37, 107440. [Google Scholar] [CrossRef]
- Soler, M.; Estevez, M.C.; Cardenosa-Rubio, M.; Astua, A.; Lechuga, L.M. How Nanophotonic Label-Free Biosensors Can Contribute to Rapid and Massive Diagnostics of Respiratory Virus Infections: COVID-19 Case. ACS Sens. 2020, 5, 2663–2678. [Google Scholar] [CrossRef]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020. [Google Scholar] [CrossRef] [PubMed]




| Method | Principle | Time Required |
|---|---|---|
| Virus plaque test | infectivity detection | several days |
| Immunofluorescent plaque test | infectivity detection | several days |
| qPCR | viral nucleic acid detection | several hours |
| ELISA | viral protein detection | several hours |
| Hemagglutination assay | viral protein detection | several hours |
| Viral flow cytometry | viral particle detection | several hours |
| Transmission electron microscopy | viral particle detection | several weeks |
| Company | Kit | Virus | Principle | Sample Type | Sensitivity | Specificity | Time |
|---|---|---|---|---|---|---|---|
| Abbott | SD BIOLINE Influenza Antigen | Influenza virus type A&B and H1N1 | antigen test | human nasal swab, throat swab, nasopharyngeal swab or nasal/nasopharyngeal aspirate | 91.8% | 98.9% | 5 min |
| Abbott | SD BIOLINE Influenza Ultra | Influenza virus type A&B | antigen test | nasopharyngeal swab, nasopharygeal aspirate | nasopharyngeal swab Flu A: 88.5% Flu B: 91.5% nasopharyngeal aspirate Flu A: 93.9% Flu B: 91.7% | nasopharyngeal swab Flu A: 97.4% Flu B: 97.4% nasopharyngeal aspirate Flu A: 97.7% Flu B: 97.7% | 5–8 min |
| Abbott | CLEARVIEW® EXACT INFLUENZA A&B | Influenza virus type A&B | antigen test | nasopharyngeal swab | Flu A: 81.7% Flu B: 88.6% | Flu A: 98.5% Flu B: 97.4% | 15 min |
| Abbott | BINAXNOW® INFLUENZA A&B | Influenza virus type A&B | chromatographic immunoassay | nasopharyngeal swab, nasopharygeal aspirate | Flu A: 70–89% Flu B: 50–69% | Flu A: 90–99% Flu B: 94–100% | 15 min |
| BIOFIRE | The BioFire® FilmArray® Respiratory (RP&RP2) Panels | VIRUSES: Adenovirus; Coronavirus HKU1; Coronavirus NL63; Coronavirus 229E; Coronavirus OC43; Human Metapneumovirus; Human Rhinovirus/Enterovirus; Influenza A; Influenza A/H1; Influenza A/H3; Influenza A/H1-2009; Influenza B; Parainfluenza Virus 1/2/3/4; Respiratory Syncytial Virus BACTERIA:Bordetella parapertussis; Bordetella pertussis; Chlamydia pneumoniae; Mycoplasma pneumoniae * Available only on the BioFire Respiratory Panel 2 | Real-time RT-PCR molecular test | nasopharyngeal swab in transport media | 97.1% | 99.3% | 45 min |
| BIOFIRE | The BioFire® FilmArray® Respiratory EZ (RP EZ) Panel | VIRUSES:Adenovirus; Coronavirus; Human Metapneumovirus; Human Rhinovirus/Enterovirus; Influenza A; Influenza A/H1; Influenza A/H3; Influenza A/H1-2009; Influenza B; Parainfluenza Virus; Respiratory BACTERIA: Bordetella pertussis; Chlamydophila pneumoniae; Mycoplasma pneumoniae | Real-time RT-PCR molecular test | nasopharyngeal swab | BAL: 96.2% Sputum: 96.3% | BAL: 98.3% Sputum:97.2% | 45 min |
| Cepheid | Xpert® Xpress SARS-CoV-2 | SARS-CoV-2 | Real-time RT-PCR molecular test results | nasal swab, nasopharyngeal swab, aspirate specimens | 100% | 100% | 45 min |
| Cepheid | Xpert®Xpress Flu/RSV | Detects viral RNA, enabling better detection of seasonal mutations of the flu virus (A,b,RSV) | Real-time RT-PCR molecular test results | nasal swab, nasopharyngeal swab specimens | Flu A: 98.1% Flu B: 100% RSV: 98.4% | Flu A: 98.1% Flu B: 99.1% RSV: 99.3% | 20 min |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, S.; Lin, S.; Zhang, H.; Liang, L.; Shen, S. Methods of Respiratory Virus Detection: Advances towards Point-of-Care for Early Intervention. Micromachines 2021, 12, 697. https://doi.org/10.3390/mi12060697
Lu S, Lin S, Zhang H, Liang L, Shen S. Methods of Respiratory Virus Detection: Advances towards Point-of-Care for Early Intervention. Micromachines. 2021; 12(6):697. https://doi.org/10.3390/mi12060697
Chicago/Turabian StyleLu, Siming, Sha Lin, Hongrui Zhang, Liguo Liang, and Shien Shen. 2021. "Methods of Respiratory Virus Detection: Advances towards Point-of-Care for Early Intervention" Micromachines 12, no. 6: 697. https://doi.org/10.3390/mi12060697
APA StyleLu, S., Lin, S., Zhang, H., Liang, L., & Shen, S. (2021). Methods of Respiratory Virus Detection: Advances towards Point-of-Care for Early Intervention. Micromachines, 12(6), 697. https://doi.org/10.3390/mi12060697






