Applications of 3D Bioprinting Technology in Induced Pluripotent Stem Cells-Based Tissue Engineering
Abstract
:1. Introduction
2. Fundamental Considerations for the Use of Induced Pluripotent Stem Cells (iPSCs) in Tissue Engineering
2.1. Generation of iPSCs
2.2. Brief Discussion on the Advantages and Disadvantages of iPSCs
3. Three-Dimensional (3D) Bioprinting Techniques Integrated with iPSCs Technology
3.1. Three-Dimensional (3D) Bioprinting Techniques and Their Working Principle
3.1.1. Inkjet-Based 3D Bioprinting
3.1.2. Extrusion-Based 3D Bioprinting
3.1.3. Laser-Assisted 3D Bioprinting
3.1.4. Stereolithographic 3D Bioprinting
| Bioprinting Method | Inkjet 3D Bioprinting | Extrusion 3D Bioprinting | Laser-Assisted 3d Bioprinting | Stereolithographic 3D Bioprinting |
|---|---|---|---|---|
|
Description | Thermal, piezoelectric, or electromagnetic forces expel successive drops of bioink onto a substrate | Mechanical or pneumatic forces dispense bioink through a nozzle | Bioink and cells are suspended on the bottom of a ribbon and when vaporized by the laser pulse, are propelled to a receiving substrate | Use digital light to cure bioink in a layer by layer fashion |
|
Advantages | High speed, availability, low cost | Ability to use high viscosity bioink and print high cell density | High degree of precision and the resolution, ability to use high viscosity bioink and print high cell density | High degree of fabrication accuracy, and low printing time |
|
Disadvantages | Lack of precision in droplet placement and size, need for low viscosity bioink | Distortion of cell structure | Time consuming, high cost | Use of high-intensity UV light, lengthy postprocessing, lack of compatible materials |
| Effect on cells | >85% cell viability [79] | 40–80% viability [79] | >95% cell viability [79] | >90% cell viability [79] |
| Cost | Low | Medium | High | Medium |
| Factors | Inkjet-Based 3D Bioprinting | Extrusion-Based 3D Bioprinting | Laser-Assisted 3D Bioprinting | Stereolithographic 3D Bioprinting |
|---|---|---|---|---|
| Ink viscosity | 3.5–12 mPa/s | Up to 6 × 10 mPa/s | 1–300 mPa/s | No limitation |
| Cell density | Low, <106 cell/mL | No limitation | Medium, <108 | No limitation |
| Resolution | High | Moderate | High | High |
| Print speed | Fast | Slow | Medium | Fast |
| Cost | Low | Medium | High | Low |
| Printing Methods | Printer | Diameter of Nozzle | Bioinks | Crosslinker | Cell Source | Lineage | Function | Ref. |
|---|---|---|---|---|---|---|---|---|
| Undifferentiated iPSCs-based 3D-bioprinting | ||||||||
| Extrusion based | Felix 3.0 | 40 µm | Geltrex | None | Custom-made fibroblasts derived hiPSCs | Plurilineage | 3-germ layers | [80] |
| Extrusion based | 3D Bioploter Envision TEC | 200 µm | 5% w/v alginate, 5% w/v carboxymethyl-chitosan, 1.5% w/v agarose | CaCl2 | hiPSCs | Plurilineage | 3-germ layers (neural tissue) | [81] |
| Extrusion based | 3D Discovery regenHu | 300 µm | Nanofibrillated cellulose (NFC) alginate (60:40) NFC with HA | CaCl2 (for alginate) H2O2 (for HA) | Custom-made hiPSCs, iChons | Cartilage | Pluripotency, Chondrocytes | [82] |
| Extrusion based | Custom-built | 260 µm | 2% w/v hydroxypropyl chitin (HPCH), 0–30% Mattrigel | Temperature 37 °C | hiPSCs from human peripheral blood mononuclear cells (hPBMC) | Plurilineage | Pluripotency | [83] |
| Laser assisted | Nd:YAG 1064 laser | N/A Droplet volume 0.01–1 nL | 1 wt% HA Matrigel | - | hiPSCs | Cardiac | 3-germ layers | [84] |
4. Biofabrication Factors Pertaining to the Use of iPSCs Applied with 3D Bioprinting
4.1. Structural and Biological Biomimicry
4.2. Bioink Preparation with iPSCs
5. Applications of iPSCs-Based 3D Bioprinting
5.1. Undifferentiated iPSCs Generated by 3D Bioprinting
5.2. Differentiated iPSCs Generated by 3D Bioprinting
6. Disease Modeling on iPSCs-Based 3D Bioprinting Technology
6.1. Cardiac Disease
6.2. Alzheimer’s Disease (AD)
7. Challenges and Future Direction Associated with iPSCs-Based 3D Bioprinting Technology
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van der Sanden, B.; Dhobb, M.; Berger, F.; Wion, D. Optimizing stem cell culture. J. Cell. Biochem. 2010, 111, 801–807. [Google Scholar] [CrossRef]
- Keller, G. Embryonic stem cell differentiation: Emergence of a new era in biology and medicine. Genes Dev. 2005, 19, 1129–1155. [Google Scholar] [CrossRef] [Green Version]
- Murphy, M.B.; Moncivais, K.; Caplan, A. Mesenchymal stem cells: Environmentally responsive therapeutics for regenerative medicine. Exp. Mol. Med. 2013, 45, e54. [Google Scholar] [CrossRef] [Green Version]
- Mathur, A.; Loskill, P.; Shao, K.; Huebsch, N.; Hong, S.; Marcus, S.G.; Marks, N.; Mandegar, M.; Conklin, B.R.; Lee, L.P.; et al. Human iPSC-based Cardiac Microphysiological System For Drug Screening Applications. Sci. Rep. 2015, 5, srep08883. [Google Scholar] [CrossRef] [Green Version]
- DeQuach, J.A.; Yuan, S.H.; Goldstein, L.S.; Christman, K.L. Decellularized Porcine Brain Matrix for Cell Culture and Tissue Engineering Scaffolds. Tissue Eng. Part A 2011, 17, 2583–2592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinoza, J.L.; Elbadry, M.I.; Chonabayashi, K.; Yoshida, Y.; Katagiri, T.; Harada, K.; Nakagawa, N.; Zaimoku, Y.; Imi, T.; Takamatsu, H.; et al. Hematopoiesis by iPSC-derived hematopoietic stem cells of aplastic anemia that escape cytotoxic T-cell attack. Blood Adv. 2018, 2, 390–400. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.-T.; Ye, L.; Xie, F.; Beyer, A.I.; Muench, M.; Wang, J.; Chen, Z.; Liu, H.; Chen, S.-J.; Kan, Y.W. Respecifying human iPSC-derived blood cells into highly engraftable hematopoietic stem and progenitor cells with a single factor. Proc. Natl. Acad. Sci. USA 2018, 115, 2180–2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atchison, L.; Abutaleb, N.O.; Snyder-Mounts, E.; Gete, Y.; Ladha, A.; Ribar, T.; Cao, K.; Truskey, G.A. iPSC-Derived Endothelial Cells Affect Vascular Function in a Tissue-Engineered Blood Vessel Model of Hutchinson-Gilford Progeria Syndrome. Stem Cell Rep. 2020, 14, 325–337. [Google Scholar] [CrossRef] [Green Version]
- de Peppo, G.M.; Marcos-Campos, I.; Kahler, D.J.; Alsalman, D.; Shang, L.; Vunjak-Novakovic, G.; Marolt, D. Engineering bone tissue substitutes from human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 8680–8685. [Google Scholar] [CrossRef] [Green Version]
- Ware, B.R.; Berger, D.R.; Khetani, S.R.; Wei, M.; Fujioka, M.; Yamano, S.; Shimomura, E.; Ishii, N.; Kakehashi, A.; Takeshita, M.; et al. Prediction of Drug-Induced Liver Injury in Micropatterned Co-cultures Containing iPSC-Derived Human Hepatocytes. Toxicol. Sci. 2015, 145, 252–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worthington, K.S.; Wiley, L.A.; Kaalberg, E.E.; Collins, M.M.; Mullins, R.F.; Stone, E.M.; Tucker, B.A. Two-photon polymerization for production of human iPSC-derived retinal cell grafts. Acta Biomater. 2017, 55, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hu, J.; He, T.-C. iPSC-based treatment of age-related macular degeneration (AMD): The path to success requires more than blind faith. Genes Dis. 2017, 4, 41–42. [Google Scholar] [CrossRef] [PubMed]
- Chichagova, V.; Hilgen, G.; Ghareeb, A.; Georgiou, M.; Carter, M.; Sernagor, E.; Lako, M.; Armstrong, L. Human iPSC differentiation to retinal organoids in response to IGF1 and BMP4 activation is line- and method-dependent. Stem Cells 2020, 38, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.-C.; Chuang, J.-H.; Buddhakosai, W.; Wu, W.-J.; Lee, C.-J.; Chen, W.-S.; Yang, Y.-P.; Li, M.-C.; Peng, C.-H.; Chen, S.-J. Elongation of Axon Extension for Human iPSC-Derived Retinal Ganglion Cells by a Nano-Imprinted Scaffold. Int. J. Mol. Sci. 2017, 18, 2013. [Google Scholar] [CrossRef] [Green Version]
- Calejo, M.T.; Saari, J.; Vuorenpää, H.; Vuorimaa-Laukkanen, E.; Kallio, P.; Aalto-Setälä, K.; Miettinen, S.; Skottman, H.; Kellomäki, M.; Juuti-Uusitalo, K. Co-culture of human induced pluripotent stem cell-derived retinal pigment epithelial cells and endothelial cells on double collagen-coated honeycomb films. Acta Biomater. 2019, 101, 327–343. [Google Scholar] [CrossRef]
- Sharma, A.; Sances, S.; Workman, M.; Svendsen, C.N. Multi-lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell 2020, 26, 309–329. [Google Scholar] [CrossRef]
- Murphy, S.V.; De Coppi, P.; Atala, A. Opportunities and challenges of translational 3D bioprinting. Nat. Biomed. Eng. 2020, 4, 370–380. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Ahadian, S.; Xu, C.; Montazerian, H.; Ko, H.; Nasiri, R.; Barros, N.; Khademhosseini, A. Bioinks and bioprinting technologies to make heterogeneous and biomimetic tissue constructs. Mater. Today Bio 2019, 1, 100008. [Google Scholar] [CrossRef]
- Tavafoghi, M.; Darabi, M.A.; Mahmoodi, M.; Tutar, R.; Xu, C.; Mirjafari, A.; Billi, F.; Swieszkowski, W.; Nasrollahi, F.; Ahadian, S.; et al. Multimaterial bioprinting and combination of processing techniques towards the fabrication of biomimetic tissues and organs. Biofabrication 2021, 13, 042002. [Google Scholar] [CrossRef] [PubMed]
- Demirci, U.; Montesano, G. Single cell epitaxy by acoustic picolitre droplets. Lab Chip 2007, 7, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gao, L.; Ma, L.; Luo, Y.; Yang, H.; Cui, Z. 3D Bioprinting: A Novel Avenue for Manufacturing Tissues and Organs. Engineering 2019, 5, 777–794. [Google Scholar] [CrossRef]
- Soman, S.S.; VijayaVenkataRaman, S. Applications of 3D Bioprinted-Induced Pluripotent Stem Cells in Healthcare. Int. J. Bioprinting 2020, 6, 280. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Yang, S. Advances in Reprogramming Somatic Cells to Induced Pluripotent Stem Cells. Stem Cell Rev. Rep. 2010, 6, 367–380. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, R.; Plath, K. The roles of the reprogramming factors Oct4, Sox2 and Klf4 in resetting the somatic cell epigenome during induced pluripotent stem cell generation. Genome Biol. 2012, 13, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Ng, J.-H.; Heng, J.-C.D.; Ng, H.H. Molecules that Promote or Enhance Reprogramming of Somatic Cells to Induced Pluripotent Stem Cells. Cell Stem Cell 2009, 4, 301–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mali, P.; Ye, Z.; Hommond, H.H.; Yu, X.; Lin, J.; Chen, G.; Zou, J.; Cheng, L. Improved Efficiency and Pace of Generating Induced Pluripotent Stem Cells from Human Adult and Fetal Fibroblasts. Stem Cells 2008, 26, 1998–2005. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Desponts, C.; Do, J.T.; Hahm, H.S.; Schöler, H.R.; Ding, S. Induction of Pluripotent Stem Cells from Mouse Embryonic Fibroblasts by Oct4 and Klf4 with Small-Molecule Compounds. Cell Stem Cell 2008, 3, 568–574. [Google Scholar] [CrossRef] [Green Version]
- Huangfu, D.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Snitow, M.; Chen, A.E.; Melton, D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008, 26, 795–797. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Aasen, T.; Raya, A.; Barrero, M.J.; Garreta, E.; Consiglio, A.; Gonzalez, F.; Vassena, R.; Bilić, J.; Pekarik, V.; Tiscornia, G.; et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008, 26, 1276–1284. [Google Scholar] [CrossRef]
- HHaase, A.; Olmer, R.; Schwanke, K.; Wunderlich, S.; Merkert, S.; Hess, C.; Zweigerdt, R.; Gruh, I.; Meyer, J.; Wagner, S.; et al. Generation of Induced Pluripotent Stem Cells from Human Cord Blood. Cell Stem Cell 2009, 5, 434–441. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.; Zhan, H.; Mali, P.; Dowey, S.; Williams, D.M.; Jang, Y.-Y.; Dang, C.V.; Spivak, J.L.; Moliterno, A.R.; Cheng, L. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood 2009, 114, 5473–5480. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.B.; Greber, B.; Araúzo-Bravo, M.J.; Meyer, J.; Park, K.I.; Zaehres, H.; Schöler, H.R. Direct reprogramming of human neural stem cells by OCT4. Nature 2009, 461, 649–653. [Google Scholar] [CrossRef]
- Utikal, J.; Maherali, N.; Kulalert, W.; Hochedlinger, K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 2009, 122, 3502–3510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Zhou, J.; Shi, G.; Ma, Y.; Yang, Y.; Gu, J.; Yu, H.; Jin, S.; Wei, Z.; Chen, F.; et al. Pluripotency can be rapidly and efficiently induced in human amniotic fluid-derived cells. Hum. Mol. Genet. 2009, 18, 4340–4349. [Google Scholar] [CrossRef] [Green Version]
- Sugii, S.; Kida, Y.; Kawamura, T.; Suzuki, J.; Vassena, R.; Yin, Y.-Q.; Lutz, M.K.; Berggren, W.T.; Belmonte, J.C.I.; Evans, R.M. Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 3558–3563. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Zhang, S.; Chen, X.; Li, G.; Wang, Y. Hepatic Differentiation of Mouse Embryonic Stem Cells in Three-Dimensional Polymer Scaffolds. Tissue Eng. Part A 2010, 16, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Seki, T.; Yuasa, S.; Oda, M.; Egashira, T.; Yae, K.; Kusumoto, D.; Nakata, H.; Tohyama, S.; Hashimoto, H.; Kodaira, M.; et al. Generation of Induced Pluripotent Stem Cells from Human Terminally Differentiated Circulating T Cells. Cell Stem Cell 2010, 7, 11–14. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, S.; Brennand, K.; Panopoulos, A.D.; Herrerías, A.; Gage, F.H.; Izpisua-Belmonte, J.C. High-Efficient Generation of Induced Pluripotent Stem Cells from Human Astrocytes. PLoS ONE 2010, 5, e15526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunisato, A.; Wakatsuki, M.; Shinba, H.; Ota, T.; Ishida, I.; Nagao, K. Direct Generation of Induced Pluripotent Stem Cells from Human Nonmobilized Blood. Stem Cells Dev. 2011, 20, 159–168. [Google Scholar] [CrossRef]
- Song, B.; Niclis, J.C.; Alikhan, M.A.; Sakkal, S.; Sylvain, A.; Kerr, P.G.; Laslett, A.; Bernard, C.C.A.; Ricardo, S.D. Generation of Induced Pluripotent Stem Cells from Human Kidney Mesangial Cells. J. Am. Soc. Nephrol. 2011, 22, 1213–1220. [Google Scholar] [CrossRef]
- Zhou, T.; Benda, C.; Dunzinger, S.; Huang, Y.; Ho, J.C.; Yang, J.; Wang, Y.; Zhang, Y.; Zhuang, Q.; Li, Y.; et al. Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 2012, 7, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Benda, C.; Duzinger, S.; Huang, Y.; Li, X.; Li, Y.; Guo, X.; Cao, G.; Chen, S.; Hao, L.; et al. Generation of induced pluripotent stem cells from urine. J. Am. Soc. Nephrol. 2011, 22, 1221–1228. [Google Scholar] [CrossRef]
- Li, W.; Wei, W.; Zhu, S.; Zhu, J.; Shi, Y.; Lin, T.; Hao, E.; Hayek, A.; Deng, H.; Ding, S. Generation of Rat and Human Induced Pluripotent Stem Cells by Combining Genetic Reprogramming and Chemical Inhibitors. Cell Stem Cell 2009, 4, 16–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maherali, N.; Hochedlinger, K. Guidelines and Techniques for the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell 2008, 3, 595–605. [Google Scholar] [CrossRef] [Green Version]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B 2009, 85, 348–362. [Google Scholar] [CrossRef] [Green Version]
- Stadtfeld, M.; Nagaya, M.; Utikal, J.; Weir, G.; Hochedlinger, K. Induced Pluripotent Stem Cells Generated Without Viral Integration. Science 2008, 322, 945–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okita, K.; Nakagawa, M.; Hyenjong, H.; Ichisaka, T.; Yamanaka, S. Generation of Mouse Induced Pluripotent Stem Cells without Viral Vectors. Science 2008, 322, 949–953. [Google Scholar] [CrossRef]
- Somers, A.; Jean, J.-C.; Sommer, C.A.; Omari, A.; Ford, C.C.; Mills, J.A.; Ying, L.; Sommer, A.G.; Jean, J.M.; Smith, B.W.; et al. Generation of Transgene-Free Lung Disease-Specific Human Induced Pluripotent Stem Cells Using a Single Excisable Lentiviral Stem Cell Cassette. Stem Cells 2010, 28, 1728–1740. [Google Scholar] [CrossRef] [Green Version]
- Woltjen, K.; Michael, I.P.; Mohseni, P.; Desai, R.; Mileikovsky, M.; Hämäläinen, R.; Cowling, R.; Wang, W.; Liu, P.; Gertsenstein, M.; et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 2009, 458, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-K.; Davaa, E.; Myung, C.-S.; Park, J.-S. Enhanced siRNA delivery using cationic liposomes with new polyarginine-conjugated PEG-lipid. Int. J. Pharm. 2010, 392, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.-H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.; Smith, Z.D.; Meissner, A.; et al. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [Green Version]
- Kim, C. iPSC technology-Powerful hand for disease modeling and therapeutic screen. BMB Rep. 2015, 48, 256–265. [Google Scholar] [CrossRef] [Green Version]
- Guha, P.; Morgan, J.W.; Mostoslavsky, G.; Rodrigues, N.P.; Boyd, A.S. Lack of Immune Response to Differentiated Cells Derived from Syngeneic Induced Pluripotent Stem Cells. Cell Stem Cell 2013, 12, 407–412. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, W.; Laurent, T.; Ding, S. Small molecules, big roles—the chemical manipulation of stem cell fate and somatic cell reprogramming. J. Cell Sci. 2012, 125 Pt 23, 5609–5620. [Google Scholar] [CrossRef] [Green Version]
- Fong, H.; Wang, C.; Knoferle, J.; Walker, D.; Balestra, M.E.; Tong, L.M.; Leung, L.; Ring, K.L.; Seeley, W.W.; Karydas, A.; et al. Genetic Correction of Tauopathy Phenotypes in Neurons Derived from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2013, 1, 226–234. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Pak, C.; Han, Y.; Ahlenius, H.; Zhang, Z.; Chanda, S.; Marro, S.; Patzke, C.; Acuna, C.; Covy, J.; et al. Rapid Single-Step Induction of Functional Neurons from Human Pluripotent Stem Cells. Neuron 2013, 78, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Hamanaka, S.; Yamaguchi, T.; Kobayashi, T.; Kato-Itoh, M.; Yamazaki, S.; Sato, H.; Umino, A.; Wakiyama, Y.; Arai, M.; Sanbo, M.; et al. Generation of Germline-Competent Rat Induced Pluripotent Stem Cells. PLoS ONE 2011, 6, e22008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Thornhill, S.I.; Howe, S.J.; Ulaganathan, M.; Schambach, A.; Sinclair, J.; Kinnon, C.; Gaspar, H.B.; Antoniou, M.; Thrasher, A.J. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood J. Am. Soc. Hematol. 2007, 110, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Ghaleb, A.M.; O Nandan, M.; Chanchevalap, S.; Dalton, W.B.; Hisamuddin, I.M.; Yang, V.W. Krüppel-like factors 4 and 5: The yin and yang regulators of cellular proliferation. Cell Res. 2005, 15, 92–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochedlinger, K.; Yamada, Y.; Beard, C.; Jaenisch, R. Ectopic Expression of Oct-4 Blocks Progenitor-Cell Differentiation and Causes Dysplasia in Epithelial Tissues. Cell 2005, 121, 465–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Liu, M.; Li, Y.; Liu, Y.; Li, S.; Ge, R. Dry and wet deposition of polycyclic aromatic hydrocarbons and comparison with typical media in urban system of Shanghai, China. Atmos. Environ. 2016, 144, 175–181. [Google Scholar] [CrossRef]
- Park, I.-H.; Arora, N.; Huo, H.; Maherali, N.; Ahfeldt, T.; Shimamura, A.; Lensch, M.W.; Cowan, C.; Hochedlinger, K.; Daley, G.Q. Disease-Specific Induced Pluripotent Stem Cells. Cell 2008, 134, 877–886. [Google Scholar] [CrossRef] [Green Version]
- Wobus, A.M.; Löser, P. Present state and future perspectives of using pluripotent stem cells in toxicology research. Arch. Toxicol. 2011, 85, 79–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pappas, J.J.; Yang, P.C. Human ESC vs. iPSC—Pros and Cons. J. Cardiovasc. Transl. Res. 2008, 1, 96–99. [Google Scholar] [CrossRef]
- Choi, S.M.; Kim, Y.; Shim, J.S.; Park, J.T.; Wang, R.-H.; Leach, S.D.; Liu, J.O.; Deng, C.; Ye, Z.; Jang, Y.-Y. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology 2013, 57, 2458–2468. [Google Scholar] [CrossRef] [Green Version]
- Chun, Y.S.; Byun, K.; Lee, B. Induced pluripotent stem cells and personalized medicine: Current progress and future perspectives. Anat. Cell Biol. 2011, 44, 245–255. [Google Scholar] [CrossRef] [Green Version]
- Gu, B.K.; Choi, D.J.; Park, S.J.; Kim, Y.J.; Kim, C.H. 3D bioprinting technologies for tissue engineering applications. Adv. Exp. Med. Biol. 2018, 1078, 15–28. [Google Scholar]
- Kim, S.-J.; Park, J.; Byun, H.; Park, Y.-W.; Major, L.G.; Lee, D.Y.; Choi, Y.S.; Shin, H. Hydrogels with an embossed surface: An all-in-one platform for mass production and culture of human adipose-derived stem cell spheroids. Biomaterials 2019, 188, 198–212. [Google Scholar] [CrossRef]
- Bejleri, D.; Streeter, B.W.; Nachlas, A.L.Y.; Brown, M.E.; Gaetani, R.; Christman, K.L.; Davis, M.E. A Bioprinted Cardiac Patch Composed of Cardiac-Specific Extracellular Matrix and Progenitor Cells for Heart Repair. Adv. Healthc. Mater. 2018, 7, e1800672. [Google Scholar] [CrossRef]
- Dhawan, A.; Kennedy, P.M.; Rizk, E.B.; Ozbolat, I.T. Three-dimensional bioprinting for bone and cartilage restoration in orthopaedic surgery. J. Am. Acad. Orthop. Surg. 2019, 27, e215–e226. [Google Scholar] [CrossRef]
- Wang, X.; Dai, X.; Zhang, X.; Ma, C.; Li, X.; Xu, T.; Lan, Q. 3D bioprinted glioma cell-laden scaffolds enriching glioma stem cells via epithelial-mesenchymal transition. J. Biomed. Mater. Res. Part A 2019, 107, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.G.; Kim, H.; Kwon, J.; Choi, Y.J.; Jang, J.; Cho, D.W. Application of 3D bioprinting in the prevention and the therapy for human diseases. Signal Transduct. Target. Ther. 2021, 6, 177. [Google Scholar] [CrossRef]
- Ong, C.S.; Fukunishi, T.; Nashed, A.; Blazeski, A.; Zhang, H.; Hardy, S.; DiSilvestre, D.; Vricella, L.; Conte, J.; Tung, L.; et al. Creation of cardiac tissue exhibiting mechanical integration of spheroids using 3D bioprinting. J. Vis. Exp. 2017, 125, e55438. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, A.A.; Vig, K.; Baganizi, D.R.; Sahu, R.; Dixit, S.; Dennis, V.; Singh, S.R.; Pillai, S.R. Future Prospects for Scaffolding Methods and Biomaterials in Skin Tissue Engineering: A Review. Int. J. Mol. Sci. 2016, 17, 1974. [Google Scholar] [CrossRef]
- Bishop, E.S.; Mostafa, S.; Pakvasa, M.; Luu, H.H.; Lee, M.J.; Wolf, J.M.; Ameer, G.A.; He, T.-C.; Reid, R.R. 3-D bioprinting technologies in tissue engineering and regenerative medicine: Current and future trends. Genes Dis. 2017, 4, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Xiong, J.; Wang, D.; Zhang, J.; Yang, L.; Sun, S.; Liang, Y. 3D Bioprinting of Hydrogels for Cartilage Tissue Engineering. Gels 2021, 7, 144. [Google Scholar] [CrossRef]
- Murphy, S.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Reid, J.A.; Mollica, P.A.; Johnson, G.D.; Ogle, R.C.; Bruno, R.D.; Sachs, P.C. Accessible bioprinting: Adaptation of a low-cost 3D-printer for precise cell placement and stem cell differentiation. Biofabrication 2016, 8, 025017. [Google Scholar] [CrossRef] [Green Version]
- Gu, Q.; Tomaskovic-Crook, E.; Wallace, G.G.; Crook, J.M. 3D Bioprinting Human Induced Pluripotent Stem Cell Constructs for In Situ Cell Proliferation and Successive Multilineage Differentiation. Adv. Healthc. Mater. 2017, 6, 1700175. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.; Hägg, D.A.; Forsman, A.; Ekholm, J.; Nimkingratana, P.; Brantsing, C.; Kalogeropoulos, T.; Zaunz, S.; Concaro, S.; Brittberg, M.; et al. Cartilage Tissue Engineering by the 3D Bioprinting of iPS Cells in a Nanocellulose/Alginate Bioink. Sci. Rep. 2017, 7, 658. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, X.; Li, L.; Chen, Z.-N.; Gao, G.; Yao, R.; Sun, W. 3d printing human induced pluripotent stem cells with novel hydroxypropyl chitin bioink: Scalable expansion and uniform aggregation. Biofabrication 2018, 10, 044101. [Google Scholar] [CrossRef]
- Beg, S.; Almalki, W.H.; Malik, A.; Farhan, M.; Aatif, M.; Rahman, Z.; Alruwaili, N.K.; Alrobaian, M.; Tarique, M.; Rahman, M. 3D printing for drug delivery and biomedical applications. Drug Discov. Today 2020, 25, 1668–1681. [Google Scholar] [CrossRef]
- Mironov, V.; Visconti, R.P.; Kasyanov, V.; Forgacs, G.; Drake, C.J.; Markwald, R.R. Organ printing: Tissue spheroids as building blocks. Biomaterials 2009, 30, 2164–2174. [Google Scholar] [CrossRef] [Green Version]
- Ingber, D.E.; Mow, V.C.; Butler, D.; Niklason, L.; Huard, J.; Mao, J.; Yannas, I.; Kaplan, D.; Vunjak-Novakovic, G. Tissue Engineering and Developmental Biology: Going Biomimetic. Tissue Eng. 2006, 12, 3265–3283. [Google Scholar] [CrossRef] [PubMed]
- Derby, B. Printing and Prototyping of Tissues and Scaffolds. Science 2012, 338, 921–926. [Google Scholar] [CrossRef] [Green Version]
- Boland, T.; Xu, T.; Damon, B.; Cui, X. Application of inkjet printing to tissue engineering. Biotechnol. J. 2006, 1, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Giraud, M.N.; Guex, A.G.; Tevaearai, H.T. Cell therapies for heart function recovery: Focus on myocardial tissue engineering and nanotechnologies. Cardiol. Res. Pract. 2012, 2012, 971614. [Google Scholar] [CrossRef]
- Benwood, C.; Chrenek, J.; Kirsch, R.L.; Masri, N.Z.; Richards, H.; Teetzen, K.; Willerth, S.M. Natural Biomaterials and Their Use as Bioinks for Printing Tissues. Bioengineering 2021, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Starly, B.; Daly, A.C.; Burdick, J.A.; Groll, J.; Skeldon, G.; Shu, W.; Sakai, Y.; Shinohara, M.; Nishikawa, M.; et al. The bioprinting roadmap. Biofabrication 2020, 12, 022002. [Google Scholar] [CrossRef] [PubMed]
- Warren, D.; Tomaskovic-Crook, E.; Wallace, G.G.; Crook, J.M. Engineering in vitro human neural tissue analogs by 3D bioprinting and electrostimulation. APL Bioeng. 2021, 5, 020901. [Google Scholar] [CrossRef] [PubMed]
- Romanazzo, S.; Nemec, S.; Roohani, I. iPSC Bioprinting: Where are We at? Materials 2019, 12, 2453. [Google Scholar] [CrossRef] [Green Version]
- Koch, L.; Deiwick, A.; Franke, A.; Schwanke, K.; Haverich, A.; Zweigerdt, R.; Chichkov, B. Laser bioprinting of human induced pluripotent stem cells—the effect of printing and biomaterials on cell survival, pluripotency, and differentiation. Biofabrication 2018, 10, 035005. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Smits, I.P.M.; De La Vega, L.; Lee, C.; Willerth, S.M. 3D Bioprinting Pluripotent Stem Cell Derived Neural Tissues Using a Novel Fibrin Bioink Containing Drug Releasing Microspheres. Front. Bioeng. Biotechnol. 2020, 8, 57. [Google Scholar] [CrossRef]
- Salaris, F.; Rosa, A. Construction of 3D in vitro models by bioprinting human pluripotent stem cells: Challenges and opportunities. Brain Res. 2019, 1723, 146393. [Google Scholar] [CrossRef]
- Liu, J.; Chen, W.; Zhao, Z.; Xu, H.H. Reprogramming of mesenchymal stem cells derived from iPSCs seeded on biofunctionalized calcium phosphate scaffold for bone engineering. Biomaterials 2013, 34, 7862–7872. [Google Scholar] [CrossRef] [Green Version]
- Kupfer, M.E.; Lin, W.-H.; Ravikumar, V.; Qiu, K.; Wang, L.; Gao, L.; Bhuiyan, D.B.; Lenz, M.; Ai, J.; Mahutga, R.R.; et al. In Situ Expansion, Differentiation, and Electromechanical Coupling of Human Cardiac Muscle in a 3D Bioprinted, Chambered Organoid. Circ. Res. 2020, 127, 207–224. [Google Scholar] [CrossRef]
- Maiullari, F.; Costantini, M.; Milan, M.; Pace, V.; Chirivì, M.; Maiullari, S.; Rainer, A.; Baci, D.; Marei, H.E.-S.; Seliktar, D. A multi-cellular 3d bioprinting approach for vascularized heart tissue engineering based on huvecs and ipsc-derived cardiomyocytes. Sci. Rep. 2018, 8, 13532. [Google Scholar] [CrossRef]
- Gao, G.; Lee, J.H.; Jang, J.; Lee, D.H.; Kong, J.-S.; Kim, B.S.; Choi, Y.-J.; Jang, W.B.; Hong, Y.J.; Kwon, S.-M.; et al. Tissue Engineered Bio-Blood-Vessels Constructed Using a Tissue-Specific Bioink and 3D Coaxial Cell Printing Technique: A Novel Therapy for Ischemic Disease. Adv. Funct. Mater. 2017, 27, 1700798. [Google Scholar] [CrossRef]
- Noor, N.; Shapira, A.; Edri, R.; Gal, I.; Wertheim, L.; Dvir, T. 3d printing of personalized thick and perfusable cardiac patches and hearts. Adv. Sci. 2019, 6, 1900344. [Google Scholar] [CrossRef] [Green Version]
- Arai, K.; Kitsuka, T.; Nakayama, K. Scaffold-based and scaffold-free cardiac constructs for drug testing. Biofabrication 2021, 13, 042001. [Google Scholar] [CrossRef] [PubMed]
- Bakirci, E.; Toprakhisar, B.; Zeybek, M.C.; Ince, G.O.; Koc, B. Cell sheet based bioink for 3D bioprinting applications. Biofabrication 2017, 9, 024105. [Google Scholar] [CrossRef]
- Ma, X.; Qu, X.; Zhu, W.; Li, Y.; Yuan, S.; Zhang, H.; Liu, J.; Wang, P.; Lai, C.S.E.; Zanella, F.; et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. USA 2016, 113, 2206–2211. [Google Scholar] [CrossRef] [Green Version]
- Joung, D.; Truong, V.; Neitzke, C.C.; Guo, S.-Z.; Walsh, P.J.; Monat, J.R.; Meng, F.; Park, S.H.; Dutton, J.R.; Parr, A.M.; et al. 3D Printed Stem-Cell Derived Neural Progenitors Generate Spinal Cord Scaffolds. Adv. Funct. Mater. 2018, 28, 1801850. [Google Scholar] [CrossRef]
- Abaci, H.E.; Guo, Z.; Coffman, A.; Gillette, B.; Lee, W.-H.; Sia, S.K.; Christiano, A.M. Human Skin Constructs with Spatially Controlled Vasculature Using Primary and iPSC-Derived Endothelial Cells. Adv. Healthc. Mater. 2016, 5, 1800–1807. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; David, B.T.; Trawczynski, M.; Fessler, R.G. Advances in Pluripotent Stem Cells: History, Mechanisms, Technologies, and Applications. Stem Cell Rev. Rep. 2020, 16, 3–32. [Google Scholar] [CrossRef] [Green Version]
- Alonzo, M.; AnilKumar, S.; Roman, B.; Tasnim, N.; Joddar, B. 3D Bioprinting of cardiac tissue and cardiac stem cell therapy. Transl. Res. 2019, 211, 64–83. [Google Scholar] [CrossRef]
- Zimmermann, W.-H.; Cesnjevar, R. Cardiac Tissue Engineering: Implications for Pediatric Heart Surgery. Pediatr. Cardiol. 2009, 30, 716–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Low, L.A.; Tagle, D.A. Tissue chips—innovative tools for drug development and disease modeling. Lab Chip 2017, 17, 3026–3036. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-Organized Formation of Polarized Cortical Tissues from ESCs and Its Active Manipulation by Extrinsic Signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarawneh, R.; Holtzman, D.M. The Clinical Problem of Symptomatic Alzheimer Disease and Mild Cognitive Impairment. Cold Spring Harb. Perspect. Med. 2012, 2, a006148. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Association. 2020 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2020, 16, 391–460. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Virumbrales, M.; Moreno, C.; Kruglikov, I.; Marazuela, P.; Sproul, A.; Jacob, S.; Zimmer, M.; Paull, D.; Zhang, B.; Schadt, E.E.; et al. CRISPR/Cas9-Correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer’s PSEN2 N141I neurons. Acta Neuropathol. Commun. 2017, 5, 77. [Google Scholar] [CrossRef]
- Gao, G.; Cui, X. Three-dimensional bioprinting in tissue engineering and regenerative medicine. Biotechnol. Lett. 2016, 38, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.S.; Yesantharao, P.; Huang, C.Y.; Mattson, G.; Boktor, J.; Fukunishi, T.; Zhang, H.; Hibino, N. 3D bioprinting using stem cells. Pediatr. Res. 2018, 83, 223–231. [Google Scholar] [CrossRef] [Green Version]
- Fetah, K.; Tebon, P.; Goudie, M.J.; Eichenbaum, J.; Ren, L.; Barros, N.; Nasiri, R.; Ahadian, S.; Ashammakhi, N.; Dokmeci, M.R.; et al. The emergence of 3D bioprinting in organ-on-chip systems. Prog. Biomed. Eng. 2019, 1, 012001. [Google Scholar] [CrossRef]
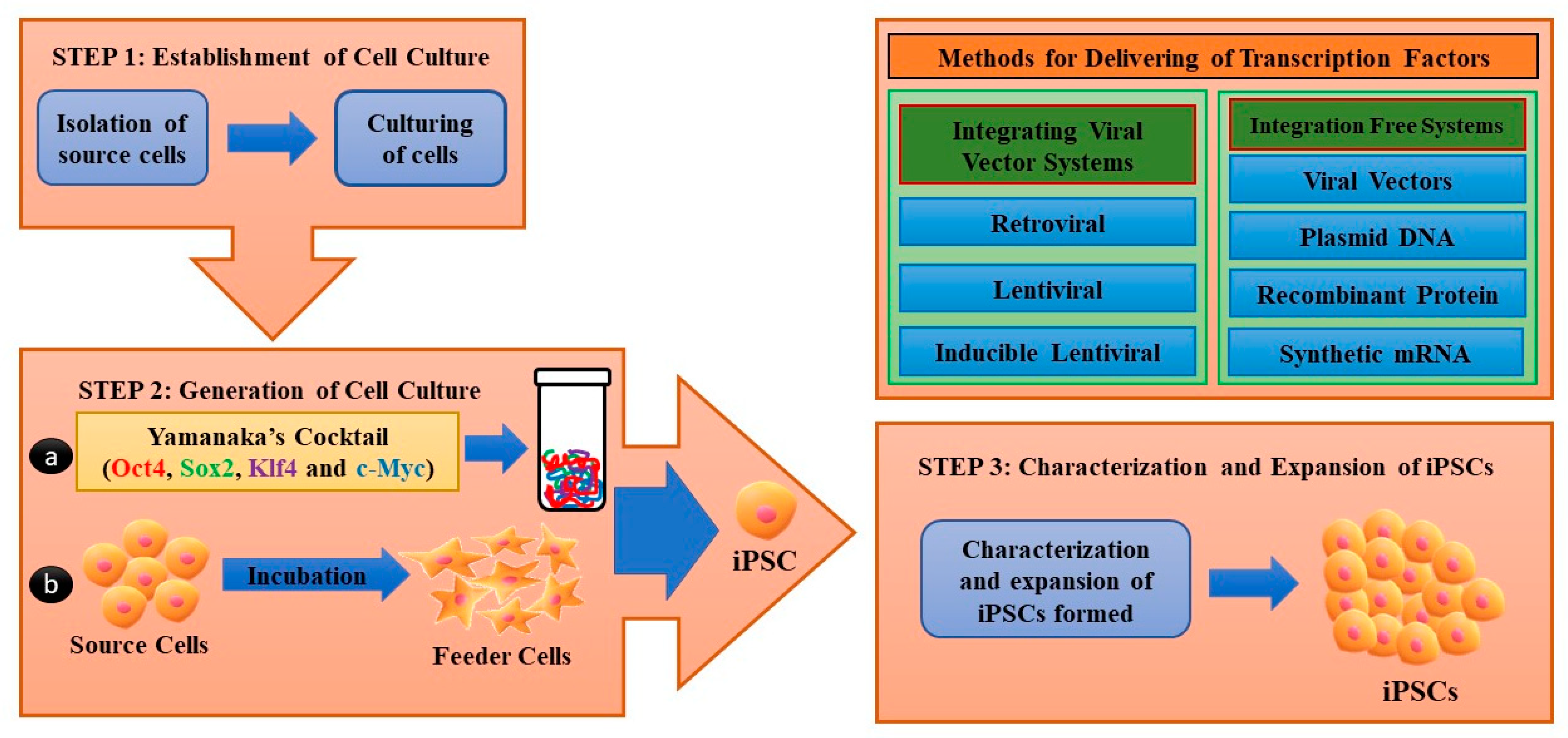

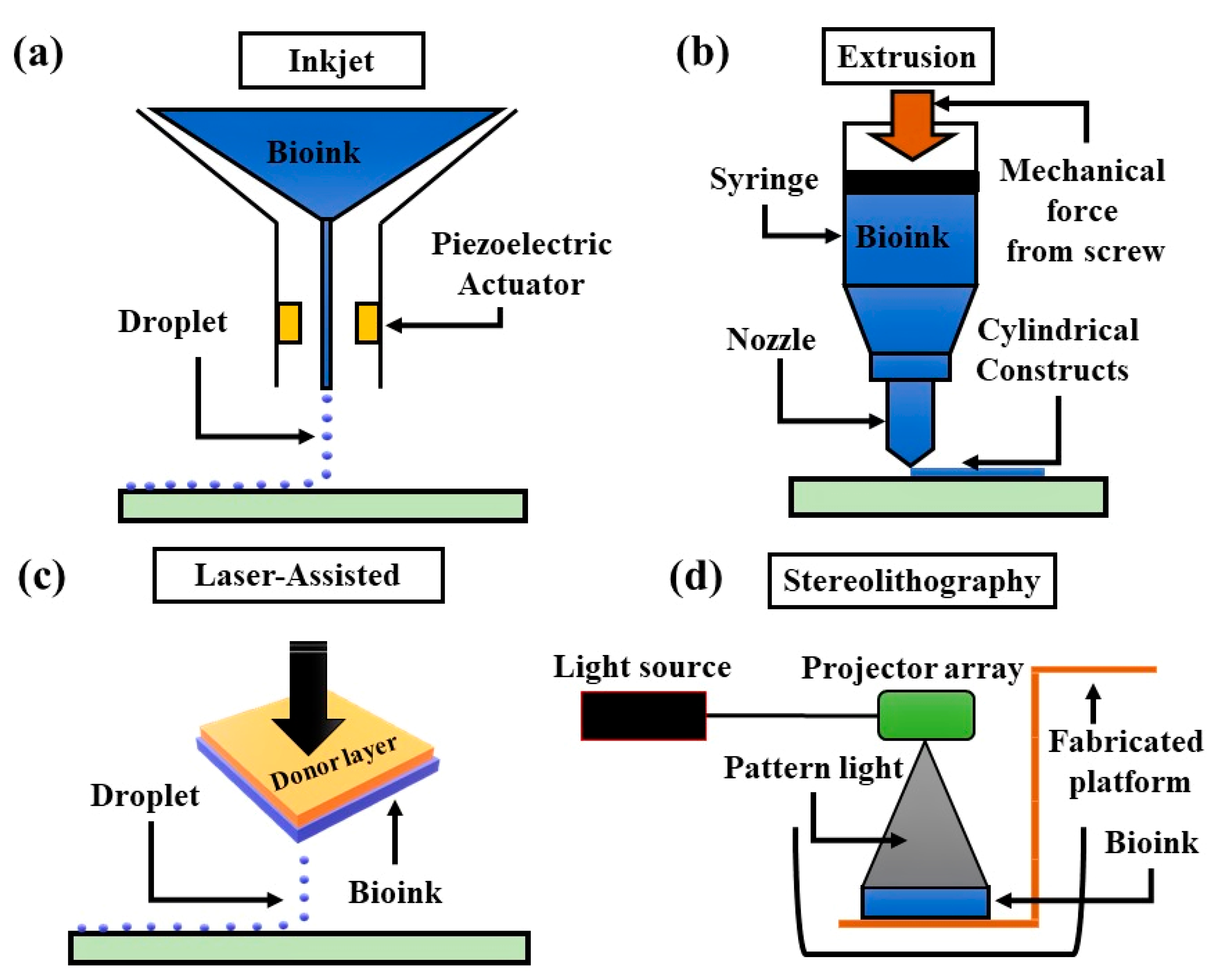
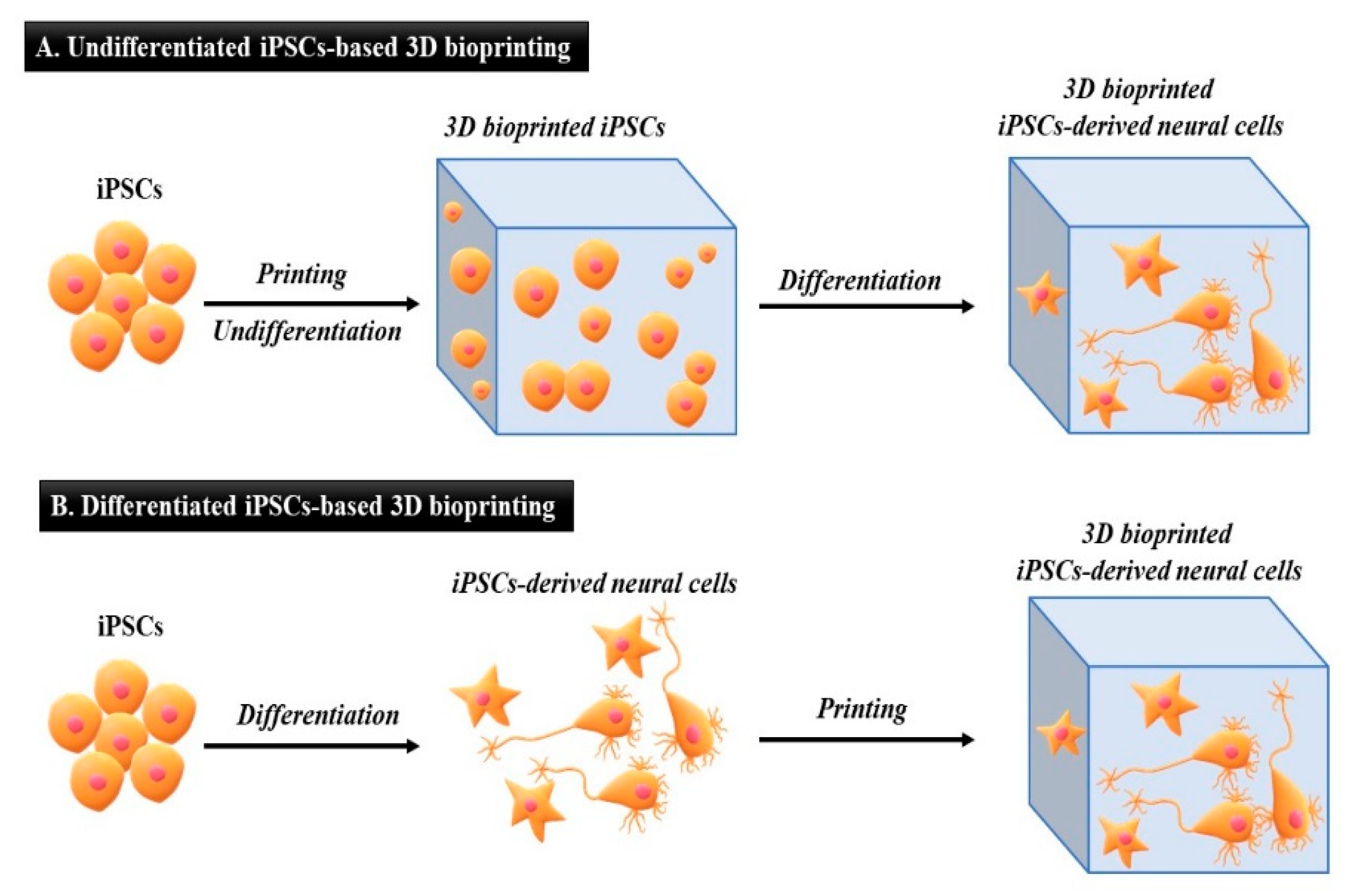
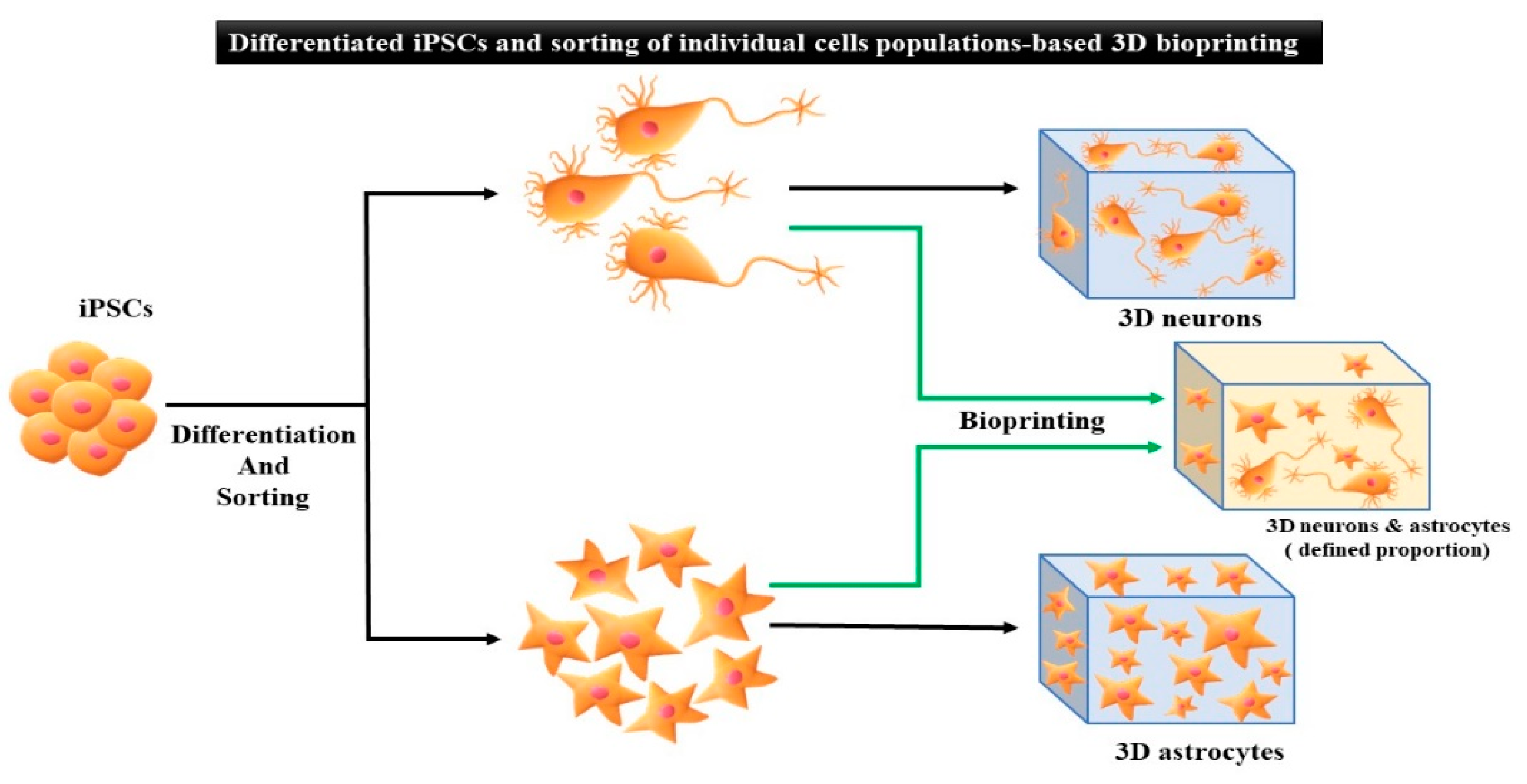



| Factors/Chemical (Small Molecules) | Function | Replacing Transcriptions Factors | References |
|---|---|---|---|
| Nanog | Embryonic stem cells (ESCs)-specific transcription factor | Together with Lin28, able of replacing Klf4 and c-Myc | [25] |
| Lin28 | Embryonic stem cells (ESCs)-specific RNA-binding protein | Together with Nanog, able of replacing Klf4 and c-Myc | [25] |
| Esrrb | Orphan nuclear receptor | Klf4 | [26] |
| SV40 LT (T) | SV40 large T antigen used for cell transformation | Klf4; N-Myc and Lin28, Nanog | [27] |
| BIX-01294 | Inhibitor of G9a histone methyltransferase | Sox2, Oct4 | [28] |
| VPA | Inhibitor of histone deacetylase | Klf4 and c-Myc | [29] |
| Type of Cells | Reprogramming Factors | References |
|---|---|---|
| Fibroblast | Oct4, Sox2, Klf4, c-Myc | [30] |
| Oct4, Sox2, Lin28, Nanog | [25] | |
| Keratinocytes | Oct4, Sox2, Klf4, c-Myc | [31] |
| Cord blood endothelial cells | Oct4, Sox2, Lin28, Nanog | [32] |
| Cord blood stem cells | Oct4, Sox2, Klf4, c-Myc | [33] |
| Neural stem cells | Oct4 | [34] |
| Melanocytes | Oct4, Sox2, Klf4, c-Myc | [35] |
| Amniotic cells | [36] | |
| Adipose derived stem cells | [37] | |
| Hepatocytes | [38] | |
| Circulating T cells | [39] | |
| Astrocytes | [40] | |
| Peripheral blood | [41] | |
| Kidney mesangial cells | [42] | |
| Urine cells | Oct4, Sox2 | [43,44] |
| Methods | Reprograming | Factors | Type of Cell | References |
|---|---|---|---|---|
|
Integrating | Retroviral transduction Lentiviral | Oct4, Sox2, Klf4, c-Myc | Mouse fibroblast | [30] |
| (Oct4, Sox2, Klf4, c-Myc) + (VPA) | Neonatal | [29] | ||
| Oct4, Sox2, Klf4, c-Myc | Human fibroblast | [25] | ||
| Inducible lentiviral | (Oct4, Klf4) + parnate + CHIR99021 | Neonatal | [45] | |
| Oct4, Sox2, Klf4, c-Myc | Human fibroblast | [46] | ||
|
Non-integrating | Sendai virus | Oct4, Sox2, Klf4, c-Myc | Human fibroblast | [47] |
| Adeno viral transduction | Mouse fibroblast | [48] | ||
| Plasmid DNA transfer | Fibroblast | [49] | ||
| lox p lentivirus | Fibroblast | [50] | ||
| PiggyBAC | Fibroblast | [51] | ||
| Polyarginine tagged polypeptide | Neonatal fibroblast | [52] | ||
| RNA modified synthetic mRNA | Human fibroblast | [53] |
| Advantages | Disadvantages | |
|---|---|---|
| Due to characteristics of iPSCs | Eliminates ethical issues | Premature aging |
| Reduced chances of immunorejection [55] | High rate of apoptosis | |
| Differentiation to any cell type | Low rate of reprogramming | |
| Reduced risks of clinical trials | Low-level DNA damage repair [56] | |
| Consistent phenotypes for disease modeling [57] | Sensitive to ionizing radiation [58] | |
|
Due to technology of development | Possible preservation | Tumourogenesis [59] |
| Continuous cell supply | Insertional mutagenesis [49,60] | |
| Possible preservation | Tumourogenesis [49] | |
| Availability and accessibility of source cells | Chances of development of diseases due to factors used [61,62,63,64] | |
| Personalization of treatment [65] | Suboptimal standardization [66] | |
|
Applications | High-throughput screening of drugs and toxicity prediction [67,68] | Complex assessment |
| Reduced cost | Complex diseases become difficult to be modeled | |
| Gene correction therapies add to the benefits from iPSCs [65] | Immature cells cause problems during cell line development |
| Tissue | Cell | Bioinks | Cross-Linker | Printer | Ref. |
|---|---|---|---|---|---|
| Cartilage | hiPSC-derived chondrocytes | NFC/A * NFC/HA * | CaCl2 | 3D Discovery (regenHu, Switzerland) | [96] |
| iPSC source: chondrocytes | |||||
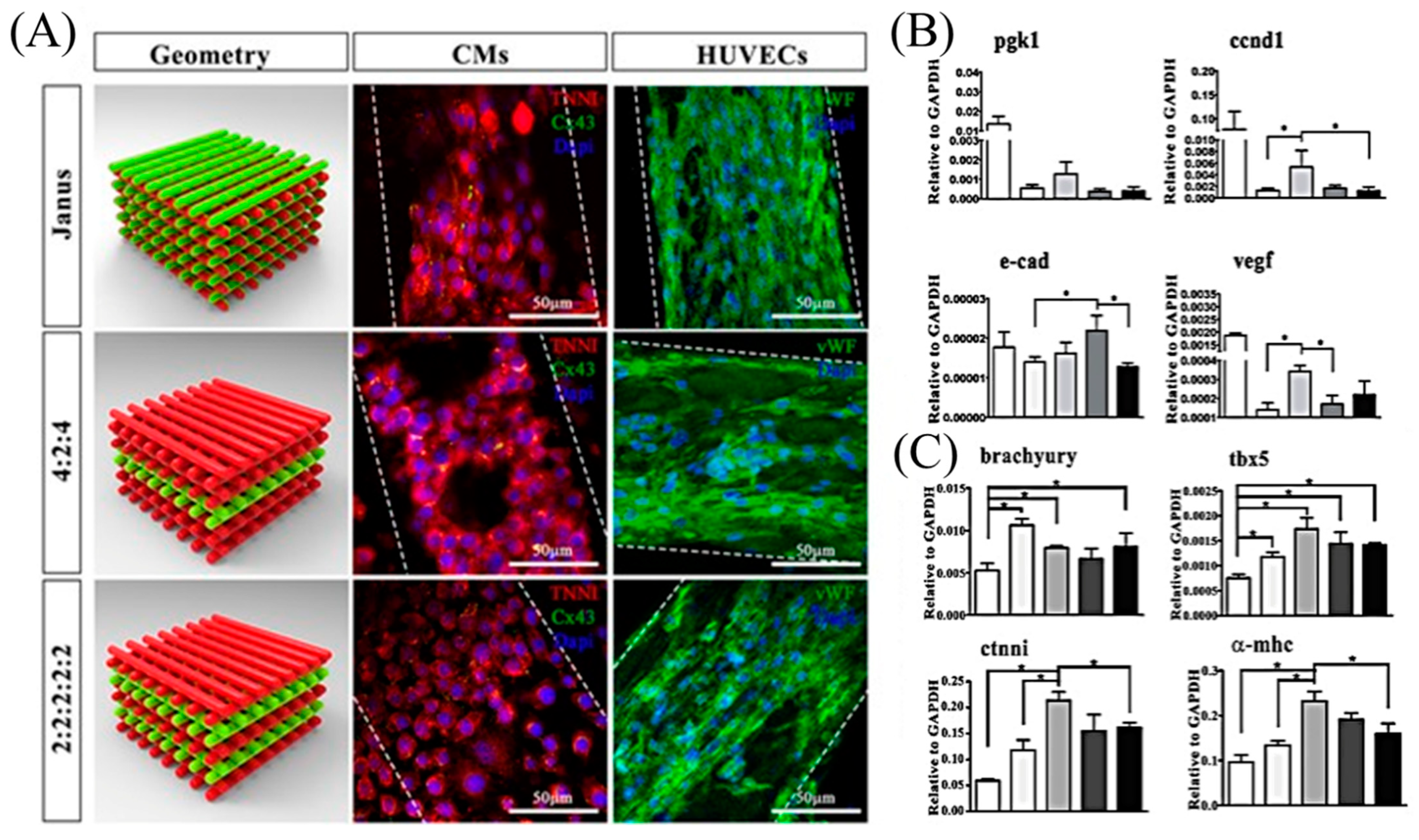
| Heart | hiPSC-derived CM, SMC, EC | GelMA | † Multiphoton excitation | Custom-built multiphoton laser-scanning 3D printer | [100,101] |
| iPSC source: cardiac fibroblasts | |||||
| HUVEC and iPSC-CM | Alginate and PEG-fibrinogen hydrogel | CaCl2 and UV | Custom designed MPH for the simultaneous extrusion of multiple bioinks | [99] | |
| iPSC source: mouse embryonic fibroblasts | |||||
| CM and EC derived from the same iPSC | Decellularized omental tissue printed in supporting medium | 37 °C for 45 min | 3D Discovery (regenHu) | [102] | |
| iPSC source: omental stromal cells | |||||
| iPSC-CM, HUVEC and NHDF | Scaffold free | - | Regenova (Cyfuse Biomedical K.K.) | [103] | |
| Human skin fibroblasts | Scaffold free | - | Novogen MMX (Organova) | [104] | |
| Hepatic tissue | iPSC-HPC | GMHA *, GelMA | UV polymerization | Custom extraction based 3D printer | [105] |
| iPSC source: human perinatal and foreskin fibroblast | |||||
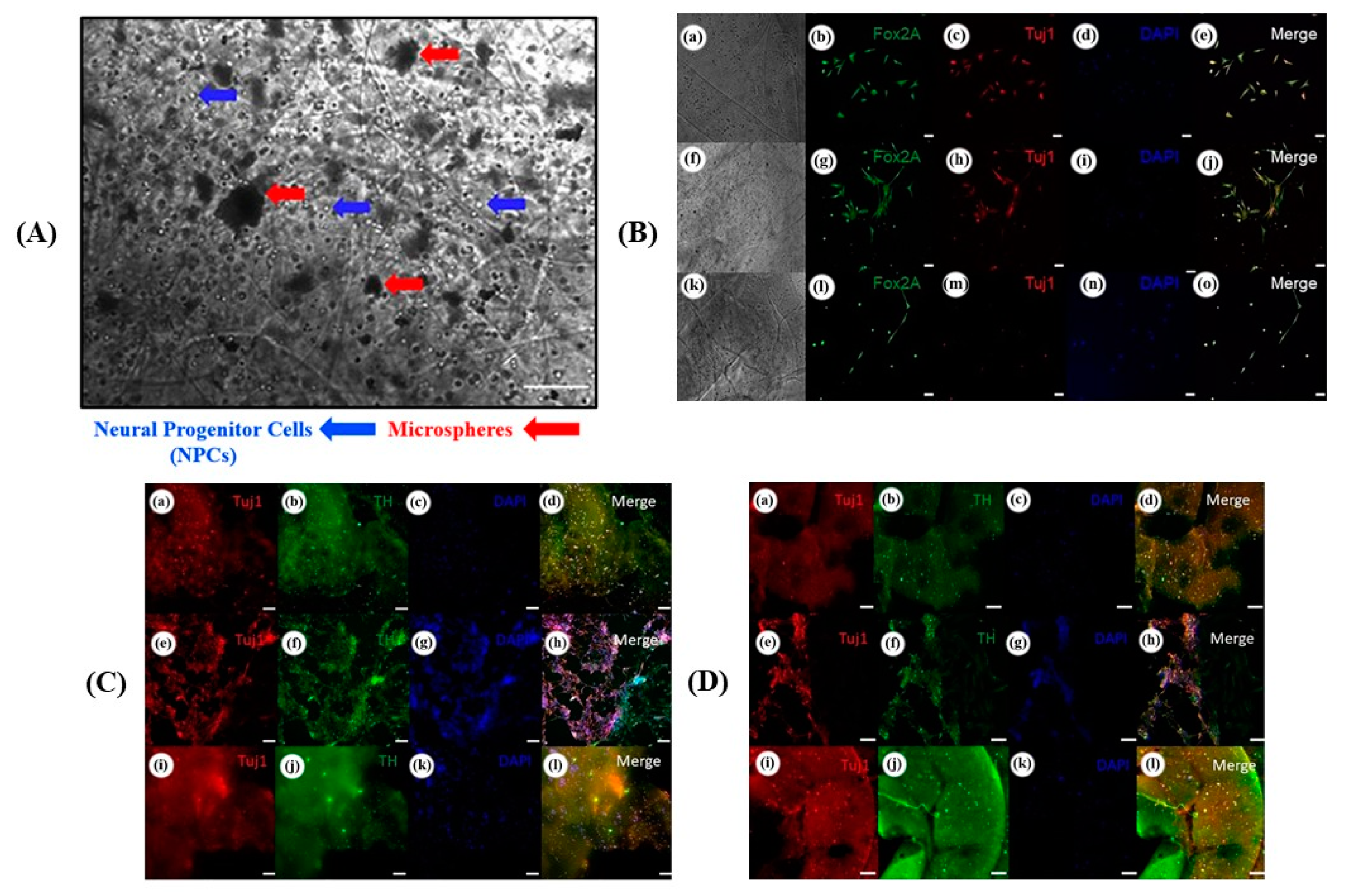
| Neural tissue | SNPC and OPC | Matrigel as cell laden bioink AG/MC * as supporting ink | Temperature, CaCl2 or BaCl2 | Custom microextrusion-based 3D printer | [106] |
| iPSC source: † UMN-X7 and UMN-3F10 | |||||
| Skin | iPSC-derived endothelial cells | Alginate molds | CaCl2 | † Object24 3D-Printer (Stratasys) | [107] |
| iPSC source: human fibroblast from foreskin |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shukla, A.K.; Gao, G.; Kim, B.S. Applications of 3D Bioprinting Technology in Induced Pluripotent Stem Cells-Based Tissue Engineering. Micromachines 2022, 13, 155. https://doi.org/10.3390/mi13020155
Shukla AK, Gao G, Kim BS. Applications of 3D Bioprinting Technology in Induced Pluripotent Stem Cells-Based Tissue Engineering. Micromachines. 2022; 13(2):155. https://doi.org/10.3390/mi13020155
Chicago/Turabian StyleShukla, Arvind Kumar, Ge Gao, and Byoung Soo Kim. 2022. "Applications of 3D Bioprinting Technology in Induced Pluripotent Stem Cells-Based Tissue Engineering" Micromachines 13, no. 2: 155. https://doi.org/10.3390/mi13020155
APA StyleShukla, A. K., Gao, G., & Kim, B. S. (2022). Applications of 3D Bioprinting Technology in Induced Pluripotent Stem Cells-Based Tissue Engineering. Micromachines, 13(2), 155. https://doi.org/10.3390/mi13020155








